Cathepsin D—Managing the Delicate Balance
Abstract
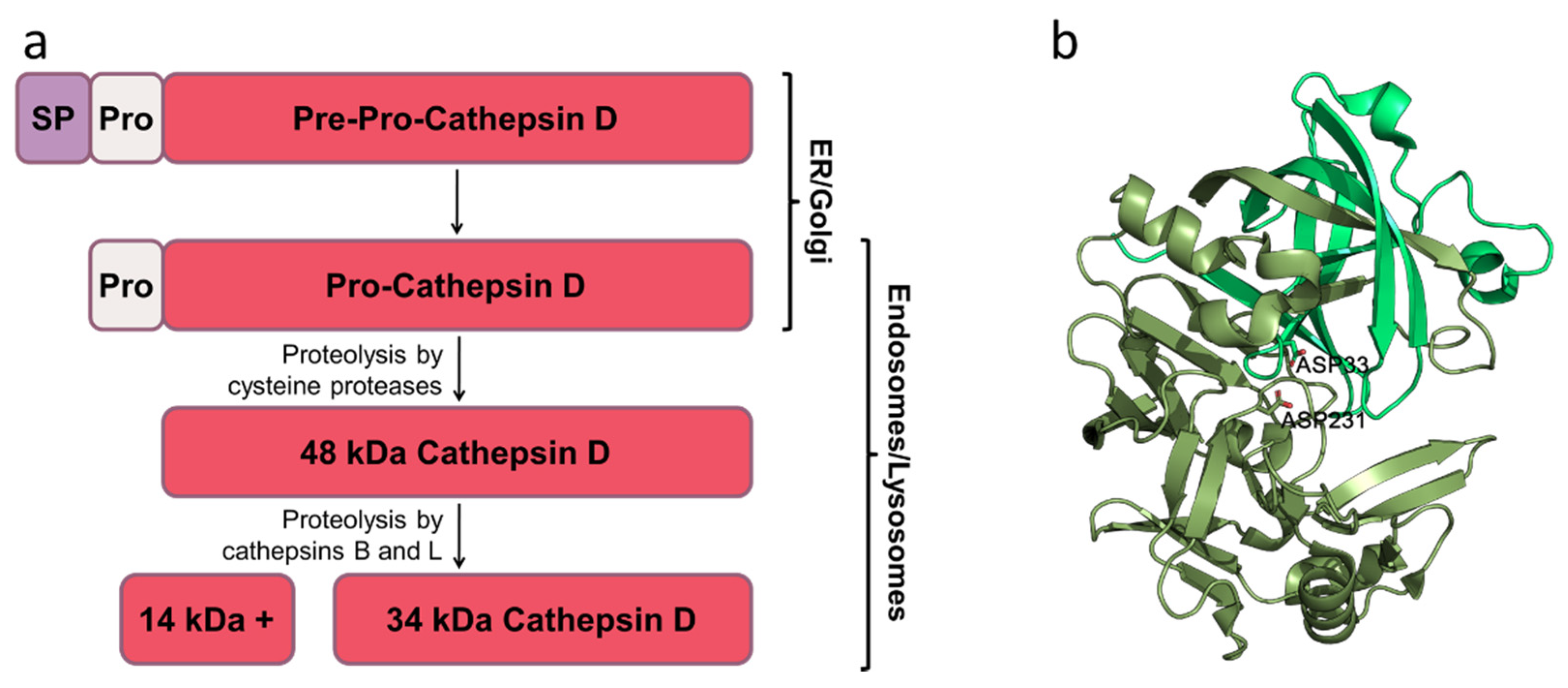
:1. Introduction
2. Vital Functions of Cathepsin D
3. Cathepsin D Down-Regulation in Neurodegenerative Diseases
4. Treatment to Restore Cathepsin D
5. Excessive Levels of Cathepsin D in Neurodegenerative Disorders
6. Excessive Levels of Cathepsin D in Disorders Associated with Diabetes
7. Excessive Levels of Cathepsin D in Malignant Tumors
8. Cathepsin D Inhibitors
9. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
References
- Johansson, A.C.; Appelqvist, H.; Nilsson, C.; Kågedal, K.; Roberg, K.; Öllinger, K. Regulation of apoptosis-associated lysosomal membrane permeabilization. Apoptosis 2010, 15, 527–540. [Google Scholar] [CrossRef] [Green Version]
- Trivedi, P.C.; Bartlett, J.J.; Pulinilkunnil, T. Lysosomal Biology and Function: Modern View of Cellular Debris Bin. Cells 2020, 9, 1131. [Google Scholar] [CrossRef]
- Ishida, Y.; Nayak, S.; Mindell, J.A.; Grabe, M. A model of lysosomal pH regulation. J. Gen. Physiol. 2013, 141, 705–720. [Google Scholar] [CrossRef] [Green Version]
- Patel, S.; Homaei, A.; El-Seedi, H.R.; Akhtar, N. Cathepsins: Proteases that are vital for survival but can also be fatal. Biomed. Pharmacother. 2018, 105, 526–532. [Google Scholar] [CrossRef] [PubMed]
- Novinec, M.; Lenarčič, B.; Turk, B. Cysteine cathepsin activity regulation by glycosaminoglycans. Biomed Res. Int. 2014, 2014, 309718. [Google Scholar] [CrossRef] [PubMed]
- Soond, S.M.; Kozhevnikova, M.V.; Zamyatnin, A.A. “Patchiness” and basic cancer research: Unravelling the proteases. Cell Cycle 2019, 18, 1687–1701. [Google Scholar] [CrossRef] [PubMed]
- Soond, S.M.; Kozhevnikova, M.V.; Frolova, A.S.; Savvateeva, L.V.; Plotnikov, E.Y.; Townsend, P.A.; Han, Y.-P.; Zamyatnin, A.A. Lost or Forgotten: The nuclear cathepsin protein isoforms in cancer. Cancer Lett. 2019, 462, 43–50. [Google Scholar] [CrossRef]
- Petushkova, A.I.; Zamyatnin, A.A. Redox-Mediated Post-Translational Modifications of Proteolytic Enzymes and Their Role in Protease Functioning. Biomolecules 2020, 10, 650. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pišlar, A.; Kos, J. Cysteine cathepsins in neurological disorders. Mol. Neurobiol. 2014, 49, 1017–1030. [Google Scholar] [CrossRef] [PubMed]
- Petushkova, A.I.; Savvateeva, L.V.; Korolev, D.O.; Zamyatnin, A.A. Cysteine Cathepsins: Potential Applications in Diagnostics and Therapy of Malignant Tumors. Biochemistry 2019, 84, 746–761. [Google Scholar] [CrossRef]
- Rudzińska, M.; Parodi, A.; Soond, S.M.; Vinarov, A.Z.; Korolev, D.O.; Morozov, A.O.; Daglioglu, C.; Tutar, Y.; Zamyatnin, A.A. The Role of Cysteine Cathepsins in Cancer Progression and Drug Resistance. Int. J. Mol. Sci. 2019, 20, 3602. [Google Scholar] [CrossRef]
- Soond, S.M.; Savvateeva, L.V.; Makarov, V.A.; Gorokhovets, N.V.; Townsend, P.A.; Zamyatnin, A.A. Making Connections: p53 and the Cathepsin Proteases as Co-Regulators of Cancer and Apoptosis. Cancers 2020, 12, 3476. [Google Scholar] [CrossRef]
- Soond, S.M.; Kozhevnikova, M.V.; Townsend, P.A.; Zamyatnin, A.A. Integrative p53, micro-RNA and Cathepsin Protease Co-Regulatory Expression Networks in Cancer. Cancers 2020, 12, 3454. [Google Scholar] [CrossRef]
- Oberle, C.; Huai, J.; Reinheckel, T.; Tacke, M.; Rassner, M.; Ekert, P.G.; Buellesbach, J.; Borner, C. Lysosomal membrane permeabilization and cathepsin release is a Bax/Bak-dependent, amplifying event of apoptosis in fibroblasts and monocytes. Cell Death Differ. 2010, 17, 1167–1178. [Google Scholar] [CrossRef] [Green Version]
- Lehesjoki, A.-E.; Gardiner, M. Progressive Myoclonus Epilepsy. In Jasper’s Basic Mechanisms of the Epilepsies; Oxford University Press: Oxford, UK, 2012; pp. 878–886. [Google Scholar]
- Cavailles, V.; Augereau, P.; Rochefort, H. Cathepsin D gene is controlled by a mixed promoter, and estrogens stimulate only TATA-dependent transcription in breast cancer cells. Proc. Natl. Acad. Sci. USA 1993, 90, 203–207. [Google Scholar] [CrossRef] [Green Version]
- Markmann, S.; Thelen, M.; Cornils, K.; Schweizer, M.; Brocke-Ahmadinejad, N.; Willnow, T.; Heeren, J.; Gieselmann, V.; Braulke, T.; Kollmann, K. Lrp1/LDL Receptor Play Critical Roles in Mannose 6-Phosphate-Independent Lysosomal Enzyme Targeting. Traffic 2015, 16, 743–759. [Google Scholar] [CrossRef]
- Zaidi, N.; Maurer, A.; Nieke, S.; Kalbacher, H. Cathepsin D: A cellular roadmap. Biochem. Biophys. Res. Commun. 2008, 376, 5–9. [Google Scholar] [CrossRef] [PubMed]
- Laurent-Matha, V.; Derocq, D.; Prébois, C.; Katunuma, N.; Liaudet-Coopman, E. Processing of human cathepsin D is independent of its catalytic function and auto-activation: Involvement of cathepsins L and B. J. Biochem. 2006, 139, 363–371. [Google Scholar] [CrossRef] [PubMed]
- Butler, V.J.; Cortopassi, W.A.; Argouarch, A.R.; Ivry, S.L.; Craik, C.S.; Jacobson, M.P.; Kao, A.W. Progranulin Stimulates the In Vitro Maturation of Pro-Cathepsin D at Acidic pH. J. Mol. Biol. 2019, 431, 1038–1047. [Google Scholar] [CrossRef] [PubMed]
- Butler, V.J.; Cortopassi, W.A.; Gururaj, S.; Wang, A.L.; Pierce, O.M.; Jacobson, M.P.; Kao, A.W. Multi-Granulin Domain Peptides Bind to Pro-Cathepsin D and Stimulate Its Enzymatic Activity More Effectively Than Progranulin in Vitro. Biochemistry 2019, 58, 2670–2674. [Google Scholar] [CrossRef]
- Metcalf, P.; Fusek, M. Two crystal structures for cathepsin D: The lysosomal targeting signal and active site. EMBO J. 1993, 12, 1293–1302. [Google Scholar] [CrossRef]
- Martínez-Alarcón, D.; Saborowski, R.; Rojo-Arreola, L.; García-Carreño, F. Is digestive cathepsin D the rule in decapod crustaceans? Comp. Biochem. Physiol. B Biochem. Mol. Biol. 2018, 215, 31–38. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pranjol, Z.I.; Whatmore, J.L. Cathepsin D in the Tumor Microenvironment of Breast and Ovarian Cancers. Adv. Exp. Med. Biol. 2020, 1259, 1–16. [Google Scholar]
- Wang, Y.; Wu, Q.; Anand, B.G.; Karthivashan, G.; Phukan, G.; Yang, J.; Thinakaran, G.; Westaway, D.; Kar, S. Significance of cytosolic cathepsin D in Alzheimer’s disease pathology: Protective cellular effects of PLGA nanoparticles against β-amyloid-toxicity. Neuropathol. Appl. Neurobiol. 2020, 46, 686–706. [Google Scholar] [CrossRef] [PubMed]
- Dean, R.T. Lysosomes and protein degradation. Ciba Found. Symp. 1979, 139–149. [Google Scholar]
- Turk, B.; Dolenc, I.; Lenarcic, B.; Krizaj, I.; Turk, V.; Bieth, J.G.; Björk, I. Acidic pH as a physiological regulator of human cathepsin L activity. Eur. J. Biochem. 1999, 259, 926–932. [Google Scholar] [CrossRef]
- Benes, P.; Vetvicka, V.; Fusek, M. Cathepsin D--many functions of one aspartic protease. Crit. Rev. Oncol. Hematol. 2008, 68, 12–28. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cocchiaro, P.; Fox, C.; Tregidgo, N.W.; Howarth, R.; Wood, K.M.; Situmorang, G.R.; Pavone, L.M.; Sheerin, N.S.; Moles, A. Lysosomal protease cathepsin D; A new driver of apoptosis during acute kidney injury. Sci. Rep. 2016, 6, 27112. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Qin, Z.H.; Wang, Y.; Kegel, K.B.; Kazantsev, A.; Apostol, B.L.; Thompson, L.M.; Yoder, J.; Aronin, N.; DiFiglia, M. Autophagy regulates the processing of amino terminal huntingtin fragments. Hum. Mol. Genet. 2003, 12, 3231–3244. [Google Scholar] [CrossRef] [Green Version]
- Aufschnaiter, A.; Kohler, V.; Büttner, S. Taking out the garbage: Cathepsin D and calcineurin in neurodegeneration. Neural Regen. Res. 2017, 12, 1776–1779. [Google Scholar]
- Qiao, L.; Hamamichi, S.; Caldwell, K.A.; Caldwell, G.A.; Yacoubian, T.A.; Wilson, S.; Xie, Z.L.; Speake, L.D.; Parks, R.; Crabtree, D.; et al. Lysosomal enzyme cathepsin D protects against alpha-synuclein aggregation and toxicity. Mol. Brain 2008, 1, 17. [Google Scholar] [CrossRef] [Green Version]
- Aghdassi, A.A.; John, D.S.; Sendler, M.; Ulrich Weiss, F.; Reinheckel, T.; Mayerle, J.; Lerch, M.M. Cathepsin d regulates cathepsin b activation and disease severity predominantly in inflammatory cells during experimental pancreatitis. J. Biol. Chem. 2018, 293, 1018–1029. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yamac, A.H.; Sevgili, E.; Kucukbuzcu, S.; Nasifov, M.; Ismailoglu, Z.; Kilic, E.; Ercan, C.; Jafarov, P.; Uyarel, H.; Bacaksiz, A. Rolle der Cathepsin-D-Aktivität bei schweren unerwünschten kardiovaskulären Ereignissen und neu auftretender Herzinsuffizienz nach ST-Hebungs-Infarkt. Herz 2015, 40, 912–920. [Google Scholar] [CrossRef] [PubMed]
- Gonçalves, I.; Hultman, K.; Dunér, P.; Edsfeldt, A.; Hedblad, B.; Fredrikson, G.N.; Björkbacka, H.; Nilsson, J.; Bengtsson, E. High levels of cathepsin D and cystatin B are associated with increased risk of coronary events. Open Heart 2016, 3, e000353. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Houštecká, R.; Hadzima, M.; Fanfrlík, J.; Brynda, J.; Pallová, L.; Hánová, I.; Mertlíková-Kaiserová, H.; Lepšík, M.; Horn, M.; Smrčina, M.; et al. Biomimetic Macrocyclic Inhibitors of Human Cathepsin D: Structure-Activity Relationship and Binding Mode Analysis. J. Med. Chem. 2020, 63, 1576–1596. [Google Scholar] [CrossRef] [PubMed]
- Whitaker, J.N.; Terry, L.C.; Whetsell, W.O. Immunocytochemical localization of cathepsin D in rat neural tissue. Brain Res. 1981, 216, 109–124. [Google Scholar] [CrossRef]
- Castino, R.; Davies, J.; Beaucourt, S.; Isidoro, C.; Murphy, D. Autophagy is a prosurvival mechanism in cells expressing an autosomal dominant familial neurohypophyseal diabetes insipidus mutant vasopressin transgene. FASEB J. 2005, 19, 1021–1023. [Google Scholar] [CrossRef] [Green Version]
- Wang, J.; Davis, S.; Zhu, M.; Miller, E.A.; Ferro-Novick, S. Autophagosome formation: Where the secretory and autophagy pathways meet. Autophagy 2017, 13, 973–974. [Google Scholar] [CrossRef] [Green Version]
- Marques, A.R.A.; Di Spiezio, A.; Thießen, N.; Schmidt, L.; Grötzinger, J.; Lüllmann-Rauch, R.; Damme, M.; Storck, S.E.; Pietrzik, C.U.; Fogh, J.; et al. Enzyme replacement therapy with recombinant pro-CTSD (cathepsin D) corrects defective proteolysis and autophagy in neuronal ceroid lipofuscinosis. Autophagy 2020, 16, 811–825. [Google Scholar] [CrossRef]
- Nakashima, A.; Cheng, S.B.; Ikawa, M.; Yoshimori, T.; Huber, W.J.; Menon, R.; Huang, Z.; Fierce, J.; Padbury, J.F.; Sadovsky, Y.; et al. Evidence for lysosomal biogenesis proteome defect and impaired autophagy in preeclampsia. Autophagy 2020, 16, 1771–1785. [Google Scholar] [CrossRef]
- Zhang, X.; Wei, M.; Fan, J.; Yan, W.; Zha, X.; Song, H.; Wan, R.; Yin, Y.; Wang, W. Ischemia-induced upregulation of autophagy preludes dysfunctional lysosomal storage and associated synaptic impairments in neurons. Autophagy 2020, 1–24. [Google Scholar] [CrossRef] [PubMed]
- Hossain, M.I.; Marcus, J.M.; Lee, J.H.; Garcia, P.L.; Singh, V.; Shacka, J.J.; Zhang, J.; Gropen, T.I.; Falany, C.N.; Andrabi, S.A. Restoration of CTSD (cathepsin D) and lysosomal function in stroke is neuroprotective. Autophagy 2020, 1–19. [Google Scholar] [CrossRef]
- Iwama, H.; Mehanna, S.; Imasaka, M.; Hashidume, S.; Nishiura, H.; Yamamura, K.-I.; Suzuki, C.; Uchiyama, Y.; Hatano, E.; Ohmuraya, M. Cathepsin B and D deficiency in the mouse pancreas induces impaired autophagy and chronic pancreatitis. Sci. Rep. 2021, 11, 6596. [Google Scholar] [CrossRef] [PubMed]
- Dai, J.; Zhang, Q.; Wan, C.; Liu, J.; Zhang, Q.; Yu, Y.; Wang, J. Significances of viable synergistic autophagy-associated cathepsin B and cathepsin D (CTSB/CTSD) as potential biomarkers for sudden cardiac death. BMC Cardiovasc. Disord. 2021, 21, 233. [Google Scholar] [CrossRef] [PubMed]
- Shibata, M.; Kanamori, S.; Isahara, K.; Ohsawa, Y.; Konishi, A.; Kametaka, S.; Watanabe, T.; Ebisu, S.; Ishido, K.; Kominami, E.; et al. Participation of cathepsins B and D in apoptosis of PC12 cells following serum deprivation. Biochem. Biophys. Res. Commun. 1998, 251, 199–203. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Huang, Y.; Xie, Q.; Zhang, J.; Zhan, Z. A novel bis-aryl urea compound inhibits tumor proliferation via cathepsin D-associated apoptosis. Anticancer. Drugs 2020, 31, 500–506. [Google Scholar] [CrossRef] [PubMed]
- Ivankovic, D.; Chau, K.Y.; Schapira, A.H.V.; Gegg, M.E. Mitochondrial and lysosomal biogenesis are activated following PINK1/parkin-mediated mitophagy. J. Neurochem. 2016, 136, 388–402. [Google Scholar] [CrossRef] [PubMed]
- Groß, R.; Bauer, R.; Krüger, F.; Rücker-Braun, E.; Olari, L.R.; Ständker, L.; Preising, N.; Rodríguez, A.A.; Conzelmann, C.; Gerbl, F.; et al. A Placenta Derived C-Terminal Fragment of β-Hemoglobin with Combined Antibacterial and Antiviral Activity. Front. Microbiol. 2020, 11, 508. [Google Scholar] [CrossRef]
- Xu, X.; Zheng, Y.; Huang, Y.; Chen, J.; Gong, Z.; Li, Y.; Lu, C.; Lai, W.; Xu, Q. Cathepsin D contributes to the accumulation of advanced glycation end products during photoaging. J. Dermatol. Sci. 2018, 90, 263–275. [Google Scholar] [CrossRef]
- Oliveira, C.S.F.; Pereira, H.; Alves, S.; Castro, L.; Baltazar, F.; Chaves, S.R.; Preto, A.; Côrte-Real, M. Cathepsin D protects colorectal cancer cells from acetate-induced apoptosis through autophagyindependent degradation of damaged mitochondria. Cell Death Dis. 2015, 6, e1788. [Google Scholar] [CrossRef] [Green Version]
- Moles, A.; Tarrats, N.; Fernández-Checa, J.C.; Marí, M. Cathepsins B and D drive hepatic stellate cell proliferation and promote their fibrogenic potential. Hepatology 2009, 49, 1297–1307. [Google Scholar] [CrossRef] [Green Version]
- Guo, D.-Z.; Xiao, L.; Liu, Y.-J.; Shen, C.; Lou, H.-F.; Lv, Y.; Pan, S.-Y. Cathepsin D deficiency delays central nervous system myelination by inhibiting proteolipid protein trafficking from late endosome/lysosome to plasma membrane. Exp. Mol. Med. 2018, 50, e457. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mutka, A.L.; Haapanen, A.; Käkelä, R.; Lindfors, M.; Wright, A.K.; Inkinen, T.; Hermansson, M.; Rokka, A.; Corthals, G.; Jauhiainen, M.; et al. Murine cathepsin D deficiency is associated with dysmyelination/myelin disruption and accumulation of cholesteryl esters in the brain. J. Neurochem. 2010, 112, 193–203. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, Y.-J.; Zhang, T.; Chen, S.; Cheng, D.; Wu, C.; Wang, X.; Duan, D.; Zhu, L.; Lou, H.; Gong, Z.; et al. The noncanonical role of the protease cathepsin D as a cofilin phosphatase. Cell Res. 2021, 1–13. [Google Scholar] [CrossRef]
- Siintola, E.; Partanen, S.; Strömme, P.; Haapanen, A.; Haltia, M.; Maehlen, J.; Lehesjoki, A.E.; Tyynelä, J. Cathepsin D deficiency underlies congenital human neuronal ceroid-lipofuscinosis. Brain 2006, 129, 1438–1445. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tian, L.; Zhang, K.; Tian, Z.-Y.; Wang, T.; Shang, D.-S.; Li, B.; Liu, D.-X.; Fang, W.-G.; Wang, Z.-Y.; Chen, Y.-H. Decreased expression of cathepsin D in monocytes is related to the defective degradation of amyloid-β in Alzheimer’s disease. J. Alzheimers. Dis. 2014, 42, 511–520. [Google Scholar] [CrossRef]
- Saftig, P.; Hetman, M.; Schmahl, W.; Weber, K.; Heine, L.; Mossmann, H.; Köster, A.; Hess, B.; Evers, M.; von Figura, K. Mice deficient for the lysosomal proteinase cathepsin D exhibit progressive atrophy of the intestinal mucosa and profound destruction of lymphoid cells. EMBO J. 1995, 14, 3599–3608. [Google Scholar] [CrossRef]
- Follo, C.; Ozzano, M.; Mugoni, V.; Castino, R.; Santoro, M.; Isidoro, C. Knock-down of cathepsin D affects the retinal pigment epithelium, impairs swim-bladder ontogenesis and causes premature death in zebrafish. PLoS ONE 2011, 6, e21908. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vidoni, C.; Follo, C.; Savino, M.; Melone, M.A.B.; Isidoro, C. The Role of Cathepsin D in the Pathogenesis of Human Neurodegenerative Disorders. Med. Res. Rev. 2016, 36, 845–870. [Google Scholar] [CrossRef]
- Matilla-Dueñas, A.; Corral-Juan, M.; Rodríguez-Palmero Seuma, A.; Vilas, D.; Ispierto, L.; Morais, S.; Sequeiros, J.; Alonso, I.; Volpini, V.; Serrano-Munuera, C.; et al. Rare Neurodegenerative Diseases: Clinical and Genetic Update. Adv. Exp. Med. Biol. 2017, 1031, 443–496. [Google Scholar]
- Bunk, J.; Prieto Huarcaya, S.; Drobny, A.; Dobert, J.P.; Walther, L.; Rose-John, S.; Arnold, P.; Zunke, F. Cathepsin D Variants Associated with Neurodegenerative Diseases Show Dysregulated Functionality and Modified α-Synuclein Degradation Properties. Front. Cell Dev. Biol. 2021, 9, 581805. [Google Scholar] [CrossRef]
- Steinfeld, R.; Reinhardt, K.; Schreiber, K.; Hillebrand, M.; Kraetzner, R.; Brück, W.; Saftig, P.; Gärtner, J. Cathepsin D deficiency is associated with a human neurodegenerative disorder. Am. J. Hum. Genet. 2006, 78, 988–998. [Google Scholar] [CrossRef] [Green Version]
- Qureshi, Y.H.; Patel, V.M.; Berman, D.E.; Kothiya, M.J.; Neufeld, J.L.; Vardarajan, B.; Tang, M.; Reyes-Dumeyer, D.; Lantigua, R.; Medrano, M.; et al. An Alzheimer’s Disease-Linked Loss-of-Function CLN5 Variant Impairs Cathepsin D Maturation, Consistent with a Retromer Trafficking Defect. Mol. Cell. Biol. 2018, 38, e00011-18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thottath, J.; Vellarikkal, S.K.; Jayarajan, R.; Verma, A.; Manamel, M.; Singh, A.; Rajendran, V.R.; Sivasubbu, S.; Scaria, V. A novel cathepsin D mutation in 2 siblings with late infantile neuronal ceroid lipofuscinosis. Neurol. Genet. 2019, 5, e302. [Google Scholar] [CrossRef] [Green Version]
- Hersheson, J.; Burke, D.; Clayton, R.; Anderson, G.; Jacques, T.S.; Mills, P.; Wood, N.W.; Gissen, P.; Clayton, P.; Fearnley, J.; et al. Cathepsin D deficiency causes juvenile-onset ataxia and distinctive muscle pathology. Neurology 2014, 83, 1873–1875. [Google Scholar] [CrossRef] [Green Version]
- Fritchie, K.; Siintola, E.; Armao, D.; Lehesjoki, A.-E.; Marino, T.; Powell, C.; Tennison, M.; Booker, J.M.; Koch, S.; Partanen, S.; et al. Novel mutation and the first prenatal screening of cathepsin D deficiency (CLN10). Acta Neuropathol. 2009, 117, 201–208. [Google Scholar] [CrossRef]
- Amritraj, A.; Wang, Y.; Revett, T.J.; Vergote, D.; Westaway, D.; Kar, S. Role of Cathepsin d in u18666a-induced neuronal cell death potential implication in Niemann-Pick type c disease pathogenesis. J. Biol. Chem. 2013, 288, 3136–3152. [Google Scholar] [CrossRef] [Green Version]
- Witt, M.; Thiemer, R.; Meyer, A.; Schmitt, O.; Wree, A. Main Olfactory and Vomeronasal Epithelium are Differently Affected in Niemann-Pick Disease Type C1. Int. J. Mol. Sci. 2018, 19, 3563. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kettwig, M.; Ohlenbusch, A.; Jung, K.; Steinfeld, R.; Gärtner, J. Cathepsin D Polymorphism C224T in Childhood-Onset Neurodegenerative Disorders: No Impact for Childhood Dementia. J. Pediatr. Genet. 2018, 7, 14–18. [Google Scholar]
- Kim, H.N.; Seo, B.-R.; Kim, H.; Koh, J.-Y. Cilostazol restores autophagy flux in bafilomycin A1-treated, cultured cortical astrocytes through lysosomal reacidification: Roles of PKA, zinc and metallothionein 3. Sci. Rep. 2020, 10, 9175. [Google Scholar] [CrossRef] [PubMed]
- Pomilio, C.; Gorojod, R.M.; Riudavets, M.; Vinuesa, A.; Presa, J.; Gregosa, A.; Bentivegna, M.; Alaimo, A.; Alcon, S.P.; Sevlever, G.; et al. Microglial autophagy is impaired by prolonged exposure to β-amyloid peptides: Evidence from experimental models and Alzheimer’s disease patients. GeroScience 2020, 42, 613–632. [Google Scholar] [CrossRef]
- Suire, C.N.; Leissring, M.A. Cathepsin D: A Candidate Link between Amyloid β-protein and Tauopathy in Alzheimer Disease. J. Exp. Neurol. 2021, 2, 10–15. [Google Scholar]
- Kim, J.-W.; Jung, S.-Y.; Kim, Y.; Heo, H.; Hong, C.-H.; Seo, S.-W.; Choi, S.-H.; Son, S.-J.; Lee, S.; Chang, J. Identification of Cathepsin D as a Plasma Biomarker for Alzheimer’s Disease. Cells 2021, 10, 138. [Google Scholar] [CrossRef]
- Lehri-Boufala, S.; Ouidja, M.O.; Barbier-Chassefière, V.; Hénault, E.; Raisman-Vozari, R.; Garrigue-Antar, L.; Papy-Garcia, D.; Morin, C. New roles of glycosaminoglycans in α-synuclein aggregation in a cellular model of Parkinson disease. PLoS ONE 2015, 10, e0116641. [Google Scholar] [CrossRef]
- Sevlever, D.; Jiang, P.; Yen, S.H.C. Cathepsin D is the main lysosomal enzyme involved in the degradation of α-synuclein and generation of its carboxy-terminally truncated species. Biochemistry 2008, 47, 9678–9687. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McGlinchey, R.P.; Lee, J.C. Cysteine cathepsins are essential in lysosomal degradation of α-synuclein. Proc. Natl. Acad. Sci. USA 2015, 112, 9322–9327. [Google Scholar] [CrossRef] [Green Version]
- Aufschnaiter, A.; Habernig, L.; Kohler, V.; Diessl, J.; Carmona-Gutierrez, D.; Eisenberg, T.; Keller, W.; Büttner, S. The coordinated action of calcineurin and cathepsin D protects against α-synuclein toxicity. Front. Mol. Neurosci. 2017, 10, 207. [Google Scholar] [CrossRef] [Green Version]
- Aflaki, E.; Stubblefield, B.K.; McGlinchey, R.P.; McMahon, B.; Ory, D.S.; Sidransky, E. A characterization of Gaucher iPS-derived astrocytes: Potential implications for Parkinson’s disease. Neurobiol. Dis. 2020, 134, 104647. [Google Scholar] [CrossRef] [PubMed]
- Moors, T.E.; Paciotti, S.; Ingrassia, A.; Quadri, M.; Breedveld, G.; Tasegian, A.; Chiasserini, D.; Eusebi, P.; Duran-Pacheco, G.; Kremer, T.; et al. Characterization of Brain Lysosomal Activities in GBA-Related and Sporadic Parkinson’s Disease and Dementia with Lewy Bodies. Mol. Neurobiol. 2019, 56, 1344–1355. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McNeill, A.; Magalhaes, J.; Shen, C.; Chau, K.Y.; Hughes, D.; Mehta, A.; Foltynie, T.; Cooper, J.M.; Abramov, A.Y.; Gegg, M.; et al. Ambroxol improves lysosomal biochemistry in glucocerebrosidase mutation-linked Parkinson disease cells. Brain 2014, 137, 1481–1495. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fernandes, H.J.R.; Hartfield, E.M.; Christian, H.C.; Emmanoulidou, E.; Zheng, Y.; Booth, H.; Bogetofte, H.; Lang, C.; Ryan, B.J.; Sardi, S.P.; et al. ER Stress and Autophagic Perturbations Lead to Elevated Extracellular α-Synuclein in GBA-N370S Parkinson’s iPSC-Derived Dopamine Neurons. Stem Cell Rep. 2016, 6, 342–356. [Google Scholar] [CrossRef] [Green Version]
- Puska, G.; Lutz, M.I.; Molnar, K.; Regelsberger, G.; Ricken, G.; Pirker, W.; Laszlo, L.; Kovacs, G.G. Lysosomal response in relation to α-synuclein pathology differs between Parkinson’s disease and multiple system atrophy. Neurobiol. Dis. 2018, 114, 140–152. [Google Scholar] [CrossRef]
- Matrone, C.; Dzamko, N.; Madsen, P.; Nyegaard, M.; Pohlmann, R.; Søndergaard, R.V.; Lassen, L.B.; Andresen, T.L.; Halliday, G.M.; Jensen, P.H.; et al. Mannose 6-Phosphate Receptor Is Reduced in -Synuclein Overexpressing Models of Parkinsons Disease. PLoS ONE 2016, 11, e0160501. [Google Scholar] [CrossRef] [Green Version]
- Nyuzuki, H.; Ito, S.; Nagasaki, K.; Nitta, Y.; Matsui, N.; Saitoh, A.; Matsui, H. Degeneration of dopaminergic neurons and impaired intracellular trafficking in Atp13a2 deficient zebrafish. IBRO Rep. 2020, 9, 1–8. [Google Scholar] [CrossRef]
- Kang, J.; Kim, J.W.; Heo, H.; Lee, J.; Park, K.Y.; Yoon, J.H.; Chang, J. Identification of BAG2 and Cathepsin D as Plasma Biomarkers for Parkinson’s Disease. Clin. Transl. Sci. 2021, 14, 606–616. [Google Scholar] [CrossRef]
- Parnetti, L.; Balducci, C.; Pierguidi, L.; de Carlo, C.; Peducci, M.; D’Amore, C.; Padiglioni, C.; Mastrocola, S.; Persichetti, E.; Paciotti, S.; et al. Cerebrospinal fluid β-glucocerebrosidase activity is reduced in Dementia with Lewy Bodies. Neurobiol. Dis. 2009, 34, 484–486. [Google Scholar] [CrossRef]
- Xicoy, H.; Peñuelas, N.; Vila, M.; Laguna, A. Autophagic- and Lysosomal-Related Biomarkers for Parkinson’s Disease: Lights and Shadows. Cells 2019, 8, 1317. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nazmi, A.; Field, R.H.; Griffin, E.W.; Haugh, O.; Hennessy, E.; Cox, D.; Reis, R.; Tortorelli, L.; Murray, C.L.; Lopez-Rodriguez, A.B.; et al. Chronic neurodegeneration induces type I interferon synthesis via STING, shaping microglial phenotype and accelerating disease progression. Glia 2019, 67, 1254–1276. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, F.; Xu, H.-D.; Guan, J.-J.; Hou, Y.-S.; Gu, J.-H.; Zhen, X.-C.; Qin, Z.-H. Rotenone impairs autophagic flux and lysosomal functions in Parkinson’s disease. Neuroscience 2015, 284, 900–911. [Google Scholar] [CrossRef]
- Arrant, A.E.; Onyilo, V.C.; Unger, D.E.; Roberson, E.D. Progranulin gene therapy improves lysosomal dysfunction and microglial pathology associated with frontotemporal dementia and neuronal ceroid lipofuscinosis. J. Neurosci. 2018, 38, 2341–2358. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lu, S.; Sung, T.; Lin, N.; Abraham, R.T.; Jessen, B.A. Lysosomal adaptation: How cells respond to lysosomotropic compounds. PLoS ONE 2017, 12, e0173771. [Google Scholar] [CrossRef] [Green Version]
- Głombik, K.; Stachowicz, A.; Trojan, E.; Olszanecki, R.; Ślusarczyk, J.; Suski, M.; Chamera, K.; Budziszewska, B.; Lasoń, W.; Basta-Kaim, A. Evaluation of the effectiveness of chronic antidepressant drug treatments in the hippocampal mitochondria—A proteomic study in an animal model of depression. Prog. Neuro-Psychopharmacol. Biol. Psychiatry 2017, 78, 51–60. [Google Scholar] [CrossRef]
- Choi, S.R.; Britigan, B.E.; Moran, D.M.; Narayanasamy, P. Gallium nanoparticles facilitate phagosome maturation and inhibit growth of virulent Mycobacterium tuberculosis in macrophages. PLoS ONE 2017, 12, e0177987. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gao, L.; Li, X.; Meng, S.; Ma, T.; Wan, L.; Xu, S. Chlorogenic acid alleviates Aβ25-35-induced autophagy and cognitive impairment via the mTOR/TFEB signaling pathway. Drug Des. Devel. Ther. 2020, 14, 1705–1716. [Google Scholar] [CrossRef]
- Yang, S.Y.; Gegg, M.; Chau, D.; Schapira, A. Glucocerebrosidase activity, cathepsin D and monomeric α-synuclein interactions in a stem cell derived neuronal model of a PD associated GBA1 mutation. Neurobiol. Dis. 2020, 134, 104620. [Google Scholar] [CrossRef] [PubMed]
- Goetzl, E.J.; Boxer, A.; Schwartz, J.B.; Abner, E.L.; Petersen, R.C.; Miller, B.L.; Kapogiannis, D. Altered lysosomal proteins in neural-derived plasma exosomes in preclinical Alzheimer disease. Neurology 2015, 85, 40–47. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Morena, F.; Argentati, C.; Trotta, R.; Crispoltoni, L.; Stabile, A.; Pistilli, A.; di Baldassarre, A.; Calafiore, R.; Montanucci, P.; Basta, G.; et al. A Comparison of Lysosomal Enzymes Expression Levels in Peripheral Blood of Mild- and Severe-Alzheimer’s Disease and MCI Patients: Implications for Regenerative Medicine Approaches. Int. J. Mol. Sci. 2017, 18, 1806. [Google Scholar] [CrossRef]
- Guerreiro, G.; Diaz Jaques, C.E.; Wajner, M.; Vargas, C.R. Elevated levels of BDNF and cathepsin-d as possible peripheral markers of neurodegeneration in plasma of patients with glutaric acidemia type I. Int. J. Dev. Neurosci. 2020, 80, 42–49. [Google Scholar] [CrossRef]
- Scaini, G.; Tonon, T.; de Souza, C.F.M.; Schuk, P.F.; Ferreira, G.C.; Neto, J.S.; Amorin, T.; Schwartz, I.V.D.; Streck, E.L. Serum Markers of Neurodegeneration in Maple Syrup Urine Disease. Mol. Neurobiol. 2017, 54, 5709–5719. [Google Scholar] [CrossRef]
- Chai, Y.L.; Chong, J.R.; Weng, J.; Howlett, D.; Halsey, A.; Lee, J.H.; Attems, J.; Aarsland, D.; Francis, P.T.; Chen, C.P.; et al. Lysosomal cathepsin D is upregulated in Alzheimer’s disease neocortex and may be a marker for neurofibrillary degeneration. Brain Pathol. 2019, 29, 63–74. [Google Scholar] [CrossRef]
- Kobayashi, S.; Zhao, F.; Kobayashi, T.; Hagiwara, M.; Kaminaris, A.; Li, C.; Cai, F.; Huang, Y.; Liang, Q. Hyperglycemia-induced cardiomyocyte death is mediated by lysosomal membrane injury and aberrant expression of cathepsin D. Biochem. Biophys. Res. Commun. 2020, 523, 239–245. [Google Scholar] [CrossRef] [PubMed]
- Hoes, M.F.; Tromp, J.; Ouwerkerk, W.; Bomer, N.; Oberdorf-Maass, S.U.; Samani, N.J.; Ng, L.L.; Lang, C.C.; van der Harst, P.; Hillege, H.; et al. The role of cathepsin D in the pathophysiology of heart failure and its potentially beneficial properties: A translational approach. Eur. J. Heart Fail. 2020, 22, 2102–2111. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Chen, B.; Zhang, X.; Tan, L.; Wang, D.W. Increased Cathepsin D Correlates with Clinical Parameters in Newly Diagnosed Type 2 Diabetes. Dis. Markers 2017, 2017, 5286408. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vĕtvicka, V.; Vĕktvicková, J.; Fusek, M. Effect of human procathepsin D on proliferation of human cell lines. Cancer Lett. 1994, 79, 131–135. [Google Scholar] [CrossRef]
- Pranjol, M.Z.I.; Gutowski, N.; Hannemann, M.; Whatmore, J. The potential role of the proteases cathepsin D and cathepsin L in the progression and metastasis of epithelial ovarian cancer. Biomolecules 2015, 5, 3260–3279. [Google Scholar] [CrossRef] [Green Version]
- Hu, L.; Roth, J.M.; Brooks, P.; Luty, J.; Karpatkin, S. Thrombin up-regulates cathepsin D which enhances angiogenesis, growth, and metastasis. Cancer Res. 2008, 68, 4666–4673. [Google Scholar] [CrossRef] [Green Version]
- Zhu, L.; Wada, M.; Usagawa, Y.; Yasukochi, Y.; Yokoyama, A.; Wada, N.; Sakamoto, M.; Maekawa, T.; Miyazaki, R.; Yonenaga, E.; et al. Overexpression of cathepsin D in malignant melanoma. Fukuoka Igaku Zasshi 2013, 104, 370–375. [Google Scholar] [PubMed]
- Vetvicka, V.; Fusek, M. Cathepsin D: Autoantibody profiling as a diagnostic marker for cancers. World J. Clin. Oncol. 2013, 4, 1–3. [Google Scholar] [CrossRef]
- Su, S.; Zhu, X.; Lin, L.; Chen, X.; Wang, Y.; Zi, J.; Dong, Y.; Xie, Y.; Zhu, Y.; Zhang, J.; et al. Lowering endogenous cathepsin D abundance results in reactive oxygen species accumulation and cell senescence. Mol. Cell. Proteom. 2017, 16, 1217–1232. [Google Scholar] [CrossRef] [Green Version]
- Glondu, M.; Coopman, P.; Laurent-Matha, V.; Garcia, M.; Rochefort, H.; Liaudet-Coopman, E. A mutated cathepsin-D devoid of its catalytic activity stimulates the growth of cancer cells. Oncogene 2001, 20, 6920–6929. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hopp, T.A.; Weiss, H.L.; Hilsenbeck, S.G.; Cui, Y.; Allred, D.C.; Horwitz, K.B.; Fuqua, S.A.W. Breast cancer patients with progesterone receptor PR-A-rich tumors have poorer disease-free survival rates. Clin. Cancer Res. 2004, 10, 2751–2760. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pateetin, P.; Pisitkun, T.; McGowan, E.; Boonyaratanakornkit, V. Differential quantitative proteomics reveals key proteins related to phenotypic changes of breast cancer cells expressing progesterone receptor A. J. Steroid Biochem. Mol. Biol. 2020, 198, 105560. [Google Scholar] [CrossRef]
- Westley, B.; Rochefort, H. A secreted glycoprotein induced by estrogen in human breast cancer cell lines. Cell 1980, 20, 353–362. [Google Scholar] [CrossRef]
- Vignon, F.; Capony, F.; Ghambon, M.; Freiss, G.; Garcia, M.; Rochefort, H. Autocrine growth stimulation of the mcf 7 breast cancer cells by the estrogen-regulated 52 K protein. Endocrinology 1986, 118, 1537–1545. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bannoud, N.; Carvelli, F.L.; Troncoso, M.; Sartor, T.; Vargas-Roig, L.M.; Sosa, M. Cation-dependent mannose-6-phosphate receptor expression and distribution are influenced by estradiol in MCF-7 breast cancer cells. PLoS ONE 2018, 13, e0201844. [Google Scholar] [CrossRef]
- Lyu, L.; Jin, X.; Li, Z.; Liu, S.; Li, Y.; Su, R.; Su, H. TBBPA regulates calcium-mediated lysosomal exocytosis and thereby promotes invasion and migration in hepatocellular carcinoma. Ecotoxicol. Environ. Saf. 2020, 192, 110255. [Google Scholar] [CrossRef]
- Ha, Y.; Fang, Y.; Romecin Duran, P.A.; Tolosa, E.J.; Moser, C.D.; Fernandez-Zapico, M.E.; Roberts, L.R. Induction of Lysosome-associated Protein Transmembrane 4 Beta via Sulfatase 2 Enhances Autophagic Flux in Liver Cancer Cells. Hepatol. Commun. 2019, 3, 1520–1543. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Koh, S.P.; Wickremesekera, A.C.; Brasch, H.D.; Marsh, R.; Tan, S.T.; Itinteang, T. Expression of Cathepsins B, D, and G in Isocitrate Dehydrogenase-Wildtype Glioblastoma. Front. Surg. 2017, 4, 28. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Basu, S.; Cheriyamundath, S.; Gavert, N.; Brabletz, T.; Haase, G.; Ben-Ze’ev, A. Increased expression of cathepsin D is required for L1-mediated colon cancer progression. Oncotarget 2019, 10, 5217–5228. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liaudet-Coopman, E.; Beaujouin, M.; Derocq, D.; Garcia, M.; Glondu-Lassis, M.; Laurent-Matha, V.; Prébois, C.; Rochefort, H.; Vignon, F. Cathepsin D: Newly discovered functions of a long-standing aspartic protease in cancer and apoptosis. Cancer Lett. 2006, 237, 167–179. [Google Scholar] [CrossRef] [Green Version]
- Baldwin, E.T.; Bhat, T.N.; Gulnik, S.; Hosur, M.V.; Sowder, R.C.; Cachau, R.E.; Collins, J.; Silva, A.M.; Erickson, J.W. Crystal structures of native and inhibited forms of human cathepsin D: Implications for lysosomal targeting and drug design. Proc. Natl. Acad. Sci. USA 1993, 90, 6796–6800. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lowry, J.R.; Klegeris, A. Emerging roles of microglial cathepsins in neurodegenerative disease. Brain Res. Bull. 2018, 139, 144–156. [Google Scholar] [CrossRef]
- Arodola, O.A.; Soliman, M.E. Hybrid 2D/3D-quantitative structure-activity relationship and modeling studies perspectives of pepstatin A analogs as cathepsin D inhibitors. Future Med. Chem. 2018, 10, 5–26. [Google Scholar] [CrossRef] [PubMed]
- Gan, P.; Ding, L.; Hang, G.; Xia, Q.; Huang, Z.; Ye, X.; Qian, X. Oxymatrine Attenuates Dopaminergic Neuronal Damage and Microglia-Mediated Neuroinflammation Through Cathepsin D-Dependent HMGB1/TLR4/NF-κB Pathway in Parkinson’s Disease. Front. Pharmacol. 2020, 11, 776. [Google Scholar] [CrossRef]
- Liang, Z.; Liu, Z.; Sun, X.; Tao, M.; Xiao, X.; Yu, G.; Wang, X. The effect of fucoidan on cellular oxidative stress and the CATD-Bax signaling axis in MN9D cells damaged by 1-methyl-4-phenypyridinium. Front. Aging Neurosci. 2018, 10, 429. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Singh, M.P.; Sharma, C.; Kang, S.C. Morin hydrate attenuates adenine-induced renal fibrosis via targeting cathepsin D signaling. Int. Immunopharmacol. 2021, 90, 107234. [Google Scholar] [CrossRef] [PubMed]
- Ellis, C.R.; Tsai, C.C.; Lin, F.Y.; Shen, J. Conformational dynamics of cathepsin D and binding to a small-molecule BACE1 inhibitor. J. Comput. Chem. 2017, 38, 1260–1269. [Google Scholar] [CrossRef]
- Miyazaki, Y.; Ono, N.; Huang, M.; Altaf-Ul-Amin, M.; Kanaya, S. Comprehensive Exploration of Target-specific Ligands Using a Graph Convolution Neural Network. Mol. Inform. 2020, 39, e1900095. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jantas, D.; Chwastek, J.; Grygier, B.; Lasoń, W. Neuroprotective Effects of Necrostatin-1 Against Oxidative Stress–Induced Cell Damage: An Involvement of Cathepsin D Inhibition. Neurotox. Res. 2020, 37, 525–542. [Google Scholar] [CrossRef] [Green Version]
- Grädler, U.; Czodrowski, P.; Tsaklakidis, C.; Klein, M.; Werkmann, D.; Lindemann, S.; Maskos, K.; Leuthner, B. Structure-based optimization of non-peptidic Cathepsin D inhibitors. Bioorg. Med. Chem. Lett. 2014, 24, 4141–4150. [Google Scholar] [CrossRef] [PubMed]
- Goyal, S.; Patel, K.V.; Nagare, Y.; Raykar, D.B.; Raikar, S.S.; Dolas, A.; Khurana, P.; Cyriac, R.; Sarak, S.; Gangar, M.; et al. Identification and structure-activity relationship studies of small molecule inhibitors of the human cathepsin D. Bioorg. Med. Chem. 2021, 29, 115879. [Google Scholar] [CrossRef]
- Liu, C.M.; Shen, H.T.; Lin, Y.A.; Yu, Y.L.; Chen, Y.S.; Liu, C.J.; Hsieh, Y.H. Antiproliferative and antimetastatic effects of praeruptorin C on human non–small cell lung cancer through inactivating ERK/CTSD signalling pathways. Molecules 2020, 25, 1625. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bagamanshina, A.V.; Troitskaya, O.S.; Nushtaeva, A.A.; Yunusova, A.Y.; Starykovych, M.O.; Kuligina, E.V.; Kit, Y.Y.; Richter, M.; Wohlfromm, F.; Kähne, T.; et al. Cytotoxic and antitumor activity of lactaptin in combination with autophagy inducers and inhibitors. Biomed Res. Int. 2019, 2019. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gan, P.; Xia, Q.; Hang, G.; Zhou, Y.; Qian, X.; Wang, X.; Ding, L. Knockdown of cathepsin D protects dopaminergic neurons against neuroinflammation-mediated neurotoxicity through inhibition of NF-κB signalling pathway in Parkinson’s disease model. Clin. Exp. Pharmacol. Physiol. 2019, 46, 337–349. [Google Scholar] [CrossRef]
- Donia, T.; Jyoti, B.; Suizu, F.; Hirata, N.; Tanaka, T.; Ishigaki, S.; Pranzatelli, T.J.F.; Nio-Kobayashi, J.; Iwanaga, T.; Chiorini, J.A.; et al. Identification of RNA aptamer which specifically interacts with PtdIns(3)P. Biochem. Biophys. Res. Commun. 2019, 517, 146–154. [Google Scholar] [CrossRef] [PubMed]
- Cox, J. Cystatins as regulators of cancer. Med. Res. Arch. 2017, 5. [Google Scholar] [CrossRef] [Green Version]
- Završnik, J.; Butinar, M.; Prebanda, M.T.; Krajnc, A.; Vidmar, R.; Fonović, M.; Grubb, A.; Turk, V.; Turk, B.; Vasiljeva, O. Cystatin C deficiency suppresses tumor growth in a breast cancer model through decreased proliferation of tumor cells. Oncotarget 2017, 8, 73793–73809. [Google Scholar] [CrossRef]
- Oh, B.M.; Lee, S.-J.; Cho, H.J.; Park, Y.S.; Kim, J.-T.; Yoon, S.R.; Lee, S.C.; Lim, J.-S.; Kim, B.-Y.; Choe, Y.-K.; et al. Cystatin SN inhibits auranofin-induced cell death by autophagic induction and ROS regulation via glutathione reductase activity in colorectal cancer. Cell Death Dis. 2017, 8, e2682. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Komura, T.; Takabatake, H.; Harada, K.; Yamato, M.; Miyazawa, M.; Yoshida, K.; Honda, M.; Wada, T.; Kitagawa, H.; Ohta, T.; et al. Clinical features of cystatin A expression in patients with pancreatic ductal adenocarcinoma. Cancer Sci. 2017, 108, 2122–2129. [Google Scholar] [CrossRef] [Green Version]
- McAdoo, M.H.; Dannenberg, A.M.; Hayes, C.J.; James, S.P.; Sanner, J.H. Inhibition of cathepsin D-type proteinase of macrophages by pepstatin, a specific pepsin inhibitor, and other substances. Infect. Immun. 1973, 7, 655–665. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Musi, M.; Tessitore, L.; Bonelli, G.; Kazakova, O.V.; Baccino, F.M. Changes in rat liver immunoreactive cathepsin D after cycloheximide. Biochem. Int. 1985, 10, 283–290. [Google Scholar]
- Hannapel, D.J. Nucleotide and deduced amino acid sequence of the 22-kilodalton cathepsin D inhibitor protein of potato (Solanum tuberosum L.). Plant Physiol. 1993, 101, 703–704. [Google Scholar] [CrossRef] [Green Version]
- Galesa, K.; Pain, R.; Jongsma, M.A.; Turk, V.; Lenarcic, B. Structural characterization of thyroglobulin type-1 domains of equistatin. FEBS Lett. 2003, 539, 120–124. [Google Scholar] [CrossRef] [Green Version]
- Rakitzis, E.T.; Malliopoulou, T.B. Inactivation of cathepsin D by dithiophosgene and by 2,2-dichloro-1,3-dithiacyclobutanone. Biochem. J. 1976, 153, 737–739. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kregar, I.; Stanovnik, B.; Tisler, M.; Nisi, C.; Gubensek, F.; Turk, V. Inactivation studies of cathepsin D with diazo compounds. Acta Biol. Med. Ger. 1977, 36, 1927–1930. [Google Scholar]
- Lin, T.Y.; Williams, H.R. Inhibition of cathepsin D by synthetic oligopeptides. J. Biol. Chem. 1979, 254, 11875–11883. [Google Scholar] [CrossRef]
- Gunn, J.M.; Owens, R.A.; Liu, W.S.; Glover, G.I. Biological activity of aspartic proteinase inhibitors related to pepstatin. Acta Biol. Med. Ger. 1981, 40, 1547–1553. [Google Scholar] [PubMed]
- Jupp, R.A.; Dunn, B.M.; Jacobs, J.W.; Vlasuk, G.; Arcuri, K.E.; Veber, D.F.; Perlow, D.S.; Payne, L.S.; Boger, J.; de Laszlo, S. The selectivity of statine-based inhibitors against various human aspartic proteinases. Biochem. J. 1990, 265, 871–878. [Google Scholar] [CrossRef] [Green Version]
- Rao, C.M.; Scarborough, P.E.; Kay, J.; Batley, B.; Rapundalo, S.; Klutchko, S.; Taylor, M.D.; Lunney, E.A.; Humblet, C.C.; Dunn, B.M. Specificity in the binding of inhibitors to the active site of human/primate aspartic proteinases: Analysis of P2-P1-P1’-P2’ variation. J. Med. Chem. 1993, 36, 2614–2620. [Google Scholar] [CrossRef]
- Schulze, H.; Kolter, T.; Sandhoff, K. Principles of lysosomal membrane degradation. Cellular topology and biochemistry of lysosomal lipid degradation. Biochim. Biophys. Acta Mol. Cell Res. 2009, 1793, 674–683. [Google Scholar] [CrossRef] [Green Version]
- Rudzińska, M.; Daglioglu, C.; Savvateeva, L.V.; Kaci, F.N.; Antoine, R.; Zamyatnin, A.A. Current Status and Perspectives of Protease Inhibitors and Their Combination with Nanosized Drug Delivery Systems for Targeted Cancer Therapy. Drug Des. Devel. Ther. 2021, 15, 9–20. [Google Scholar] [CrossRef] [PubMed]

| DNA 1 /Protein Change | Type of Mutation | Effect on Protease | NCL Type | Ref. |
|---|---|---|---|---|
| c.392A>G/p.Tyr131Cys | Missense | Reduced enzymatic activity | Late infantile NCL (LINCL) | [65] |
| c.446G>T/p.Gly149Val | Missense | Reduced enzymatic activity | Juvenile NCL (JNCL) | [66] |
| c.1196G>A/p.Arg399His | ||||
| c.299C>T/p.Ser100Phe | Missense | Reduced enzymatic activity | Congenital CLN 2 (CLN10) | [67] |
| c.764dupA/p.Tyr255 | Nonsense | Absence of protein | CLN10 | [56] |
| c.6517T>A/p.Phe229Ile | Missense | Reduced protein amount and enzymatic activity | NCL-like disorder | [63] |
| c.10267G>C/p.Trp383Cys |
| Inhibitor | Mechanism | Ref. |
|---|---|---|
| Natural Compounds | ||
| Pepstatin A from Actinomycetes | Non-competitive inhibitor | [141] |
| Cycloheximide from Streptomyces griseus | Protein synthesis inhibitor | [142] |
| The 22-kDa cathepsin D inhibitor protein of potatoes (PDI) from Solanum tuberosum | Reversible inhibitor | [143] |
| Equistatin from Actinia equina | Reversible inhibitor | [144] |
| Fucoidan from brown seaweeds and algae | Down-regulator of the expression | [126] |
| Oxymatrine from Sophora flavescens | Down-regulator of the expression | [125] |
| Morin hydrate from Maclura pomifera, Maclura tinctoria, Psidium guajava | Reversible inhibitor | [127] |
| Synthetic compounds | ||
| Dithiophosgene | Irreversible covalent inhibitor | [145] |
| 2,2-Dichloro-1,3-dithiacyclobutanone | ||
| Diazo compounds | Irreversible covalent inhibitor | [146] |
| Pro-Pro-Phe-Phe-Val-D-Leu | Reversible inhibitor | [147] |
| Cbz-Val-Val-(3S4S)-statine | Reversible inhibitor | [148] |
| Ibu-His-Pro-Phe-HCys-Sta-Leu-NH-[CH2]2-S-Acm | Reversible inhibitor | [149] |
| Derivatives of 4-(morpholinylsulphonyl)-L-Phe-P2-(cyclohexyl)Ala psi[isostere]-P1′-P2′ | Irreversible covalent inhibitor | [150] |
| Lentiviral shRNA constructs | RNA interference inhibitor | [76] |
| Acylguanidines | Reversible inhibitor | [131,132] |
| LY2811376 | Reversible inhibitor | [128] |
| PtdIns(3)P 1 RNA aptamer | Inhibitor of PtdIns(3)P | [136] |
| Macrocyclic inhibitors | Competitive inhibitor | [36] |
| Necrostatin-1 | Suppressor of activity | [130] |
| Polytherapy | ||
| RL2 2, with chloroquine, Ku55933, and rapamycin from Streptomyces hygroscopicus | Suppressor of activity | [134] |
| Praeruptorin C from Peucedanum praeruptorum and U0126 | Inhibitor through ERK1/2 3 signaling pathway | [133] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mijanovic, O.; Petushkova, A.I.; Brankovic, A.; Turk, B.; Solovieva, A.B.; Nikitkina, A.I.; Bolevich, S.; Timashev, P.S.; Parodi, A.; Zamyatnin, A.A., Jr. Cathepsin D—Managing the Delicate Balance. Pharmaceutics 2021, 13, 837. https://doi.org/10.3390/pharmaceutics13060837
Mijanovic O, Petushkova AI, Brankovic A, Turk B, Solovieva AB, Nikitkina AI, Bolevich S, Timashev PS, Parodi A, Zamyatnin AA Jr. Cathepsin D—Managing the Delicate Balance. Pharmaceutics. 2021; 13(6):837. https://doi.org/10.3390/pharmaceutics13060837
Chicago/Turabian StyleMijanovic, Olja, Anastasiia I. Petushkova, Ana Brankovic, Boris Turk, Anna B. Solovieva, Angelina I. Nikitkina, Sergey Bolevich, Peter S. Timashev, Alessandro Parodi, and Andrey A. Zamyatnin, Jr. 2021. "Cathepsin D—Managing the Delicate Balance" Pharmaceutics 13, no. 6: 837. https://doi.org/10.3390/pharmaceutics13060837
APA StyleMijanovic, O., Petushkova, A. I., Brankovic, A., Turk, B., Solovieva, A. B., Nikitkina, A. I., Bolevich, S., Timashev, P. S., Parodi, A., & Zamyatnin, A. A., Jr. (2021). Cathepsin D—Managing the Delicate Balance. Pharmaceutics, 13(6), 837. https://doi.org/10.3390/pharmaceutics13060837









