Efficient Peptide-Mediated In Vitro Delivery of Cas9 RNP
Abstract
:1. Introduction
2. Materials & Methods
2.1. Cell Culture
2.2. Cas9 RNP Preparation
2.3. Protein to Peptide Complexation
2.4. RNAiMAX and CRISPRMax RNP Transfection
2.5. Flow Cytometry
2.6. DLS
2.7. Microscopy
2.8. Storage Testing
2.9. DNA Extraction
2.10. PCR and DNA Analysis Methods
2.11. Statistical Analysis
2.12. Next-Generation Sequencing (NGS)
3. Results
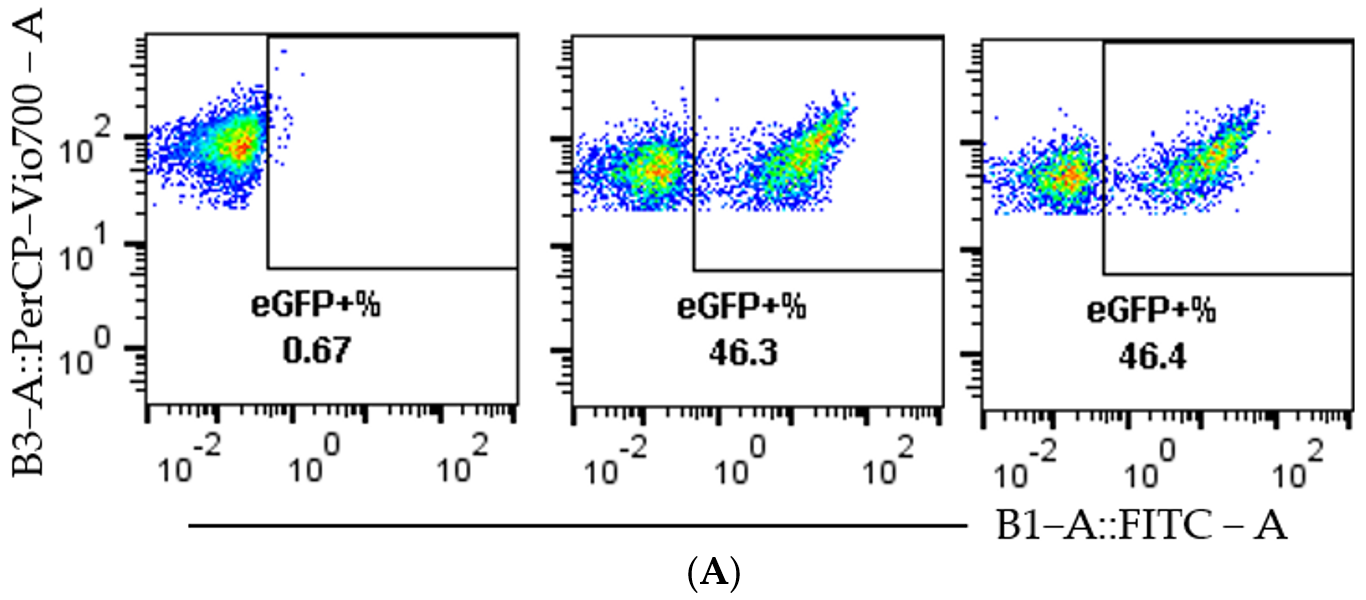
3.1. Cas9 RNP Delivery
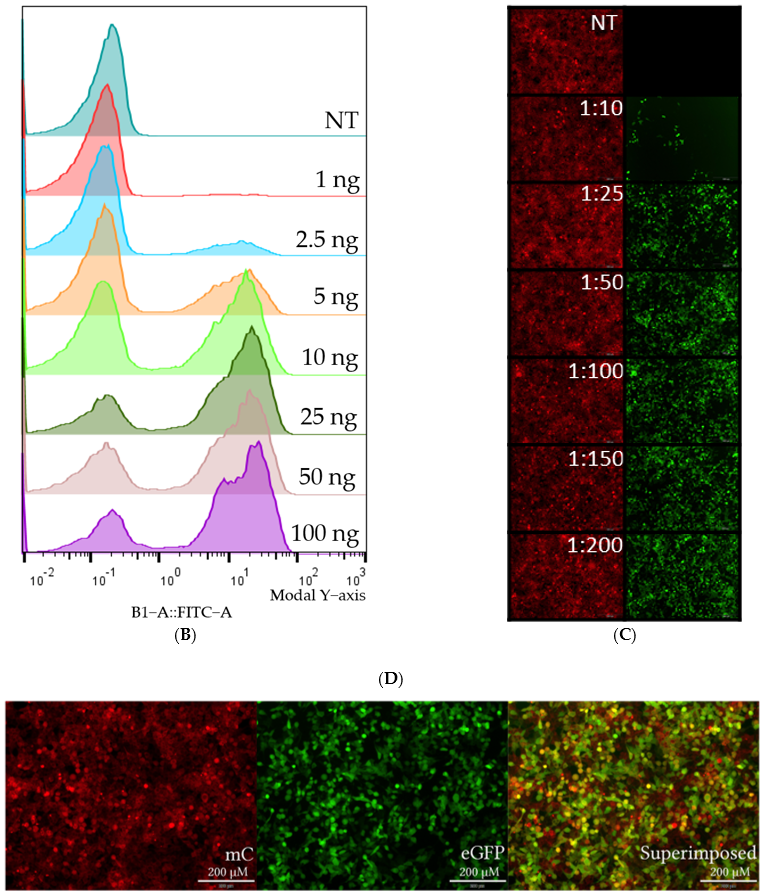
3.2. PVA-PEG Addition to Complexation Increases Editing Rates
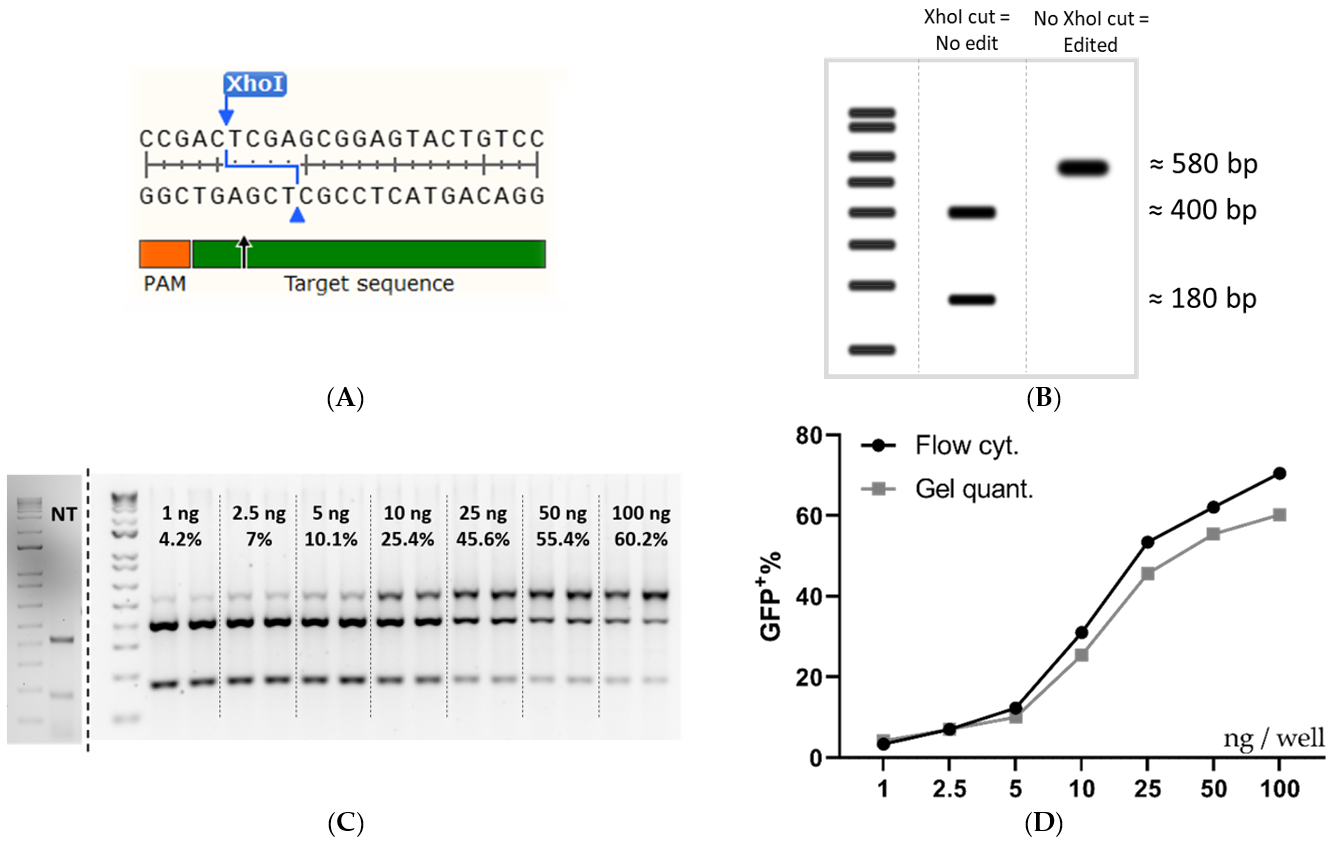
3.3. Validation of the Editing Efficiency
3.4. Storage Conditions Effect on Transfection Efficiency
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Shim, G.; Kim, D.; Park, G.T.; Jin, H.; Suh, S.K.; Oh, Y.K. Therapeutic Gene Editing: Delivery and Regulatory Perspectives. Acta Pharmacol. Sin. 2017, 38, 738–753. [Google Scholar] [CrossRef] [Green Version]
- Qi, L.S.S.; Larson, M.H.H.; Gilbert, L.A.A.; Doudna, J.A.A.; Weissman, J.S.S.; Arkin, A.P.P.; Lim, W.A.A. Repurposing CRISPR as an RNA-Guided Platform for Sequence-Specific Control of Gene Expression. Cell 2013, 152, 1173–1183. [Google Scholar] [CrossRef] [Green Version]
- Anzalone, A.; Randolph, P.B.; Davis, J.R.; Sousa, A.A.; Koblan, L.W.; Levy, J.M.; Chen, P.J.; Wilson, C.; Newby, G.A.; Raguram, A.; et al. Search-and-Replace Genome Editing without Double-Strand Breaks or Donor DNA. Nature 2019, 576, 149–157. [Google Scholar] [CrossRef]
- Gaudelli, N.M.; Komor, A.C.; Rees, H.A.; Packer, M.S.; Badran, A.H.; Bryson, D.I.; Liu, D.R. Programmable Base Editing of T to G C in Genomic DNA without DNA Cleavage. Nature 2017, 551, 464–471. [Google Scholar] [CrossRef] [PubMed]
- Komor, A.C.; Kim, Y.B.; Packer, M.S.; Zuris, J.A.; Liu, D.R. Programmable Editing of a Target Base in Genomic DNA without Double-Stranded DNA Cleavage. Nature 2016, 533, 420–424. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Frangoul, H.; Altshuler, D.; Cappellini, M.D.; Chen, Y.-S.; Domm, J.; Eustace, B.K.; Foell, J.; de la Fuente, J.; Grupp, S.; Handgretinger, R.; et al. CRISPR-Cas9 Gene Editing for Sickle Cell Disease and β-Thalassemia. N. Engl. J. Med. 2021, 384, 252–260. [Google Scholar] [CrossRef] [PubMed]
- Doudna, J.A. The Promise and Challenge of Therapeutic Genome Editing. Nature 2020, 578, 229–236. [Google Scholar] [CrossRef]
- Schumann, K.; Lin, S.; Boyer, E.; Simeonov, D.R.; Subramaniam, M.; Gate, R.E.; Haliburton, G.E.; Ye, C.J.; Bluestone, J.A.; Doudna, J.A.; et al. Generation of Knock-in Primary Human T Cells Using Cas9 Ribonucleoproteins. Proc. Natl. Acad. Sci. USA 2015, 112, 10437–10442. [Google Scholar] [CrossRef] [Green Version]
- Hultquist, J.F.; Schumann, K.; Woo, J.M.; Manganaro, L.; McGregor, M.J.; Doudna, J.; Simon, V.; Krogan, N.J.; Marson, A. A Cas9 Ribonucleoprotein Platform for Functional Genetic Studies of HIV-Host Interactions in Primary Human T Cells. Cell Rep. 2016, 17, 1438–1452. [Google Scholar] [CrossRef] [Green Version]
- Staahl, B.T.; Benekareddy, M.; Coulon-Bainier, C.; Banfal, A.A.; Floor, S.N.; Sabo, J.K.; Urnes, C.; Munares, G.A.; Ghosh, A.; Doudna, J.A. Efficient Genome Editing in the Mouse Brain by Local Delivery of Engineered Cas9 Ribonucleoprotein Complexes. Nat. Biotechnol. 2017, 35, 431–434. [Google Scholar] [CrossRef]
- Wilson, J.M. Lessons learned from the gene therapy trial for ornithine transcarbamylase deficiency. In Getting to Good: Research Integrity in the Biomedical Sciences; Springer International Publishing: New York, NY, USA, 2018; Volume 96, pp. 490–497. ISBN 9783319513584. [Google Scholar]
- Platt, R.J.; Chen, S.; Zhou, Y.; Yim, M.J.; Swiech, L.; Kempton, H.R.; Dahlman, J.E.; Parnas, O.; Eisenhaure, T.M.; Jovanovic, M.; et al. CRISPR-Cas9 Knockin Mice for Genome Editing and Cancer Modeling. Cell 2014, 159, 440–455. [Google Scholar] [CrossRef] [Green Version]
- Colella, P.; Ronzitti, G.; Mingozzi, F. Emerging Issues in AAV-Mediated In Vivo Gene Therapy. Mol. Ther. Methods Clin. Dev. 2018, 8, 87–104. [Google Scholar] [CrossRef] [Green Version]
- Nelson, C.E.; Wu, Y.; Gemberling, M.P.; Oliver, M.L.; Waller, M.A.; Bohning, J.D.; Robinson-Hamm, J.N.; Bulaklak, K.; Castellanos Rivera, R.M.; Collier, J.H.; et al. Long-Term Evaluation of AAV-CRISPR Genome Editing for Duchenne Muscular Dystrophy. Nat. Med. 2019, 25, 427–432. [Google Scholar] [CrossRef]
- Kelton, W.J.; Pesch, T.; Matile, S.; Reddy, S.T. Surveying the Delivery Methods of CRISPR/Cas9 for Ex Vivo Mammalian Cell Engineering. Chimia 2016, 70, 439–442. [Google Scholar] [CrossRef] [Green Version]
- Zhang, S.; Shen, J.; Li, D.; Cheng, Y. Strategies in the Delivery of Cas9 Ribonucleoprotein for CRISPR/Cas9 Genome Editing. Theranostics 2020, 11, 614–648. [Google Scholar] [CrossRef]
- Finn, J.D.; Smith, A.R.; Patel, M.C.; Shaw, L.; Youniss, M.R.; van Heteren, J.; Dirstine, T.; Ciullo, C.; Lescarbeau, R.; Seitzer, J.; et al. A Single Administration of CRISPR/Cas9 Lipid Nanoparticles Achieves Robust and Persistent In Vivo Genome Editing. Cell Rep. 2018, 22, 2227–2235. [Google Scholar] [CrossRef] [Green Version]
- Glass, Z.; Li, Y.; Xu, Q. Nanoparticles for CRISPR-Cas9 Delivery. Nat. Biomed. Eng. 2017, 1, 854–855. [Google Scholar] [CrossRef]
- Sun, W.; Ji, W.; Hall, J.M.; Hu, Q.; Wang, C.; Beisel, C.L.; Gu, Z. Self-Assembled DNA Nanoclews for the Efficient Delivery of CRISPR-Cas9 for Genome Editing. Angew. Chem.-Int. Ed. 2015, 54, 12029–12033. [Google Scholar] [CrossRef]
- D’Astolfo, D.S.; Pagliero, R.J.; Pras, A.; Karthaus, W.R.; Clevers, H.; Prasad, V.; Lebbink, R.J.; Rehmann, H.; Geijsen, N. Efficient Intracellular Delivery of Native Proteins. Cell 2015, 161, 674–690. [Google Scholar] [CrossRef] [Green Version]
- Gee, P.; Lung, M.S.Y.; Okuzaki, Y.; Sasakawa, N.; Iguchi, T.; Makita, Y.; Hozumi, H.; Miura, Y.; Yang, L.F.; Iwasaki, M.; et al. Extracellular Nanovesicles for Packaging of CRISPR-Cas9 Protein and SgRNA to Induce Therapeutic Exon Skipping. Nat. Commun. 2020, 11, 1334. [Google Scholar] [CrossRef] [Green Version]
- Patel, S.G.; Sayers, E.J.; He, L.; Narayan, R.; Williams, T.L.; Mills, E.M.; Allemann, R.K.; Luk, L.Y.P.; Jones, A.T.; Tsai, Y.H. Cell-Penetrating Peptide Sequence and Modification Dependent Uptake and Subcellular Distribution of Green Florescent Protein in Different Cell Lines. Sci. Rep. 2019, 9, 6298. [Google Scholar] [CrossRef]
- Hoffmann, K.; Milech, N.; Juraja, S.M.; Cunningham, P.T.; Stone, S.R.; Francis, R.W.; Anastasas, M.; Hall, C.M.; Heinrich, T.; Bogdawa, H.M.; et al. A Platform for Discovery of Functional Cell-Penetrating Peptides for Efficient Multi-Cargo Intracellular Delivery. Sci. Rep. 2018, 8, 12538. [Google Scholar] [CrossRef]
- Lostalé-Seijo, I.; Louzao, I.; Juanes, M.; Montenegro, J. Peptide/Cas9 Nanostructures for Ribonucleoprotein Cell Membrane Transport and Gene Edition. Chem. Sci. 2017, 8, 7923–7931. [Google Scholar] [CrossRef] [Green Version]
- Rouet, R.; Thuma, B.A.; Roy, M.D.; Lintner, N.G.; Rubitski, D.M.; Finley, J.E.; Wisniewska, H.M.; Mendonsa, R.; Hirsh, A.; de Oñate, L.; et al. Receptor-Mediated Delivery of CRISPR-Cas9 Endonuclease for Cell-Type-Specific Gene Editing. J. Am. Chem. Soc. 2018, 140, 6596–6603. [Google Scholar] [CrossRef]
- Ramakrishna, S.; Kwaku Dad, A.B.; Beloor, J.; Gopalappa, R.; Lee, S.K.; Kim, H. Gene Disruption by Cell-Penetrating Peptide-Mediated Delivery of Cas9 Protein and Guide RNA. Genome Res. 2014, 24, 1020–1027. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Morris, M.C.; Depollier, J.; Mery, J.; Heitz, F.; Divita, G. A Peptide Carrier for the Delivery of Biologically Active Proteins into Mammalian Cells. Nat. Biotechnol. 2001, 19, 1173–1176. [Google Scholar] [CrossRef]
- Chakrabarti, A.M.; Henser-Brownhill, T.; Monserrat, J.; Poetsch, A.R.; Luscombe, N.M.; Scaffidi, P. Target-Specific Precision of CRISPR-Mediated Genome Editing. Mol. Cell 2019, 73, 699–713.e6. [Google Scholar] [CrossRef] [Green Version]
- Ezzat, K.; Andaloussi, S.E.L.; Zaghloul, E.M.; Lehto, T.; Lindberg, S.; Moreno, P.M.D.; Viola, J.R.; Magdy, T.; Abdo, R.; Guterstam, P.; et al. PepFect 14, a Novel Cell-Penetrating Peptide for Oligonucleotide Delivery in Solution and as Solid Formulation. Nucleic Acids Res. 2011, 39, 5284–5298. [Google Scholar] [CrossRef] [PubMed]
- Mäe, M.; el Andaloussi, S.; Lundin, P.; Oskolkov, N.; Johansson, H.J.; Guterstam, P.; Langel, Ü. A Stearylated CPP for Delivery of Splice Correcting Oligonucleotides Using a Non-Covalent Co-Incubation Strategy. J. Control. Release 2009, 134, 221–227. [Google Scholar] [CrossRef]
- De Jong, O.G.; Murphy, D.E.; Mäger, I.; Willms, E.; Garcia-Guerra, A.; Gitz-Francois, J.J.; Lefferts, J.; Gupta, D.; Steenbeek, S.C.; van Rheenen, J.; et al. A CRISPR-Cas9-Based Reporter System for Single-Cell Detection of Extracellular Vesicle-Mediated Functional Transfer of RNA. Nat. Commun. 2020, 11, 1113. [Google Scholar] [CrossRef]
- Liang, X.; Potter, J.; Kumar, S.; Zou, Y.; Quintanilla, R.; Sridharan, M.; Carte, J.; Chen, W.; Roark, N.; Ranganathan, S.; et al. Rapid and Highly Efficient Mammalian Cell Engineering via Cas9 Protein Transfection. J. Biotechnol. 2015, 208, 44–53. [Google Scholar] [CrossRef] [Green Version]
- Saher, O.; Lehto, T.; Gissberg, O.; Gupta, D.; Gustafsson, O.; el Andaloussi, S.; Darbre, T.; Lundin, K.E.; Edvard Smith, C.I.; Zain, R. Sugar and Polymer Excipients Enhance Uptake and Splice-Switching Activity of Peptide-Dendrimer/Lipid/Oligonucleotide Formulations. Pharmaceutics 2019, 11, 666. [Google Scholar] [CrossRef]
- Green, M.; Ishino, M.; Loewenstein, P.M. Mutational Analysis of HIV-1 Tat Minimal Domain Peptides: Identification of Trans-Dominant Mutants That Suppress HIV-LTR-Driven Gene Expression. Cell 1989, 58, 215–223. [Google Scholar] [CrossRef]
- Ezzat, K.; Helmfors, H.; Tudoran, O.; Juks, C.; Lindberg, S.; Padari, K.; El-Andaloussi, S.; Pooga, M.; Langel, Ü. Scavenger Receptor-mediated Uptake of Cell-penetrating Peptide Nanocomplexes with Oligonucleotides. FASEB J. 2012, 26, 1172–1180. [Google Scholar] [CrossRef]
- Juks, C.; Padari, K.; Margus, H.; Kriiska, A.; Etverk, I.; Arukuusk, P.; Koppel, K.; Ezzat, K.; Langel, Ü.; Pooga, M. The Role of Endocytosis in the Uptake and Intracellular Trafficking of PepFect14-Nucleic Acid Nanocomplexes via Class A Scavenger Receptors. Biochim. Biophys. Acta-Biomembr. 2015, 1848, 3205–3216. [Google Scholar] [CrossRef] [Green Version]
- El-Andaloussi, S.; Järver, P.; Johansson, H.J.; Langel, Ü. Cargo-Dependent Cytotoxicity and Delivery Efficacy of Cell-Penetrating Peptides: A Comparative Study. Biochem. J. 2007, 407, 285–292. [Google Scholar] [CrossRef] [Green Version]
- Vives, E.; Richard, J.-; Rispal, C.; Lebleu, B. TAT Peptide Internalization: Seeking the Mechanism of Entry. Curr. Protein Pept. Sci. 2005, 4, 125–132. [Google Scholar] [CrossRef]
- Nakase, I.; Niwa, M.; Takeuchi, T.; Sonomura, K.; Kawabata, N.; Koike, Y.; Takehashi, M.; Tanaka, S.; Ueda, K.; Simpson, J.C.; et al. Cellular Uptake of Arginine-Rich Peptides: Roles for Macropinocytosis and Actin Rearrangement. Mol. Ther. 2004, 10, 1011–1022. [Google Scholar] [CrossRef]
- El-Andaloussi, S.; Johansson, H.J.; Lundberg, P.; Langel, Ü. Induction of Splice Correction by Cell-Penetrating Peptide Nucleic Acids. J. Gene Med. 2006, 8, 1262–1273. [Google Scholar] [CrossRef]






| sgRNAs | Target Sequence |
| Stop-Light guide RNA (gRNA) | GGACAGTACTCCGCTCGAGT |
| HPRT (Hypoxanthine-guanine phosphoribosyltransferase) Human gRNA | AATTATGGGGATTACTAGGA |
| PCR primers | Sequence |
| PCR primer-Stop-Light FWD | ACATCACCTCCCACAACGAG |
| PCR primer-Stop-Light REV | GGTCTTGTAGTTGCCGTCGT |
| PCR primer-HPRT FWD | AAGAATGTTGTGATAAAAGGTGATGCT |
| PCR primer-HPRT REV | ACACATCCATGGGACTTCTGCCTC |
| Target | Tag | Tag Weight (kDa) | Tag Isoelectric Point | Purchased from Biosciences. |
|---|---|---|---|---|
| Rat IgG2a kappa Isotype Control (eBR2a) | FITC | 0.39 | 4.70 | cat# 554680 |
| CD45.1 Monoclonal Antibody | APC | 105 | 5.00 | cat# 17045382 |
| Mouse IgG1, κ Isotype Control | PE | 240 | 4.15 | cat# 554680 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gustafsson, O.; Rädler, J.; Roudi, S.; Lehto, T.; Hällbrink, M.; Lehto, T.; Gupta, D.; Andaloussi, S.E.; Nordin, J.Z. Efficient Peptide-Mediated In Vitro Delivery of Cas9 RNP. Pharmaceutics 2021, 13, 878. https://doi.org/10.3390/pharmaceutics13060878
Gustafsson O, Rädler J, Roudi S, Lehto T, Hällbrink M, Lehto T, Gupta D, Andaloussi SE, Nordin JZ. Efficient Peptide-Mediated In Vitro Delivery of Cas9 RNP. Pharmaceutics. 2021; 13(6):878. https://doi.org/10.3390/pharmaceutics13060878
Chicago/Turabian StyleGustafsson, Oskar, Julia Rädler, Samantha Roudi, Tõnis Lehto, Mattias Hällbrink, Taavi Lehto, Dhanu Gupta, Samir EL Andaloussi, and Joel Z. Nordin. 2021. "Efficient Peptide-Mediated In Vitro Delivery of Cas9 RNP" Pharmaceutics 13, no. 6: 878. https://doi.org/10.3390/pharmaceutics13060878
APA StyleGustafsson, O., Rädler, J., Roudi, S., Lehto, T., Hällbrink, M., Lehto, T., Gupta, D., Andaloussi, S. E., & Nordin, J. Z. (2021). Efficient Peptide-Mediated In Vitro Delivery of Cas9 RNP. Pharmaceutics, 13(6), 878. https://doi.org/10.3390/pharmaceutics13060878







