Antibacterial Activity of Co(III) Complexes with Diamine Chelate Ligands against a Broad Spectrum of Bacteria with a DNA Interaction Mechanism
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials and Growth Conditions
2.2. Synthesis of [CoCl2(N,N)2]Cl
2.3. Synthesis of [Co(dap)2FLU]Cl2
2.4. Physicochemical Measurements
2.5. Determination of the Minimum Inhibitory Concentration (MIC) and Minimum Bactericidal Concentration (MBC)
2.6. Serial Passages Assay
2.7. Synergy Assay
2.8. Microscopic Analysis
2.8.1. Transmission Electron Microscopy (TEM)
2.8.2. Scanning Electron Microscopy (SEM)
2.8.3. Confocal Microscopy Assay
2.9. Interactions of Compounds (1) and (2) with Bacterial DNA
2.9.1. DNA Binding Studies of Diamine Co(III) Complexes
2.9.2. DNA Cleavage Study
2.10. Statistical Analysis
3. Results
3.1. The Minimum Inhibitory Concentration (MIC) and Minimum Bactericidal Concentration (MBC)
| Strains | [CoCl2(dap)2]Cl (1) | [CoCl2(en)2]Cl (2) | Ampicillin | Ciprofloxacin | ||||
|---|---|---|---|---|---|---|---|---|
| MIC | MBC | MIC | MBC | MIC | MBC | MIC | MBC | |
| P. aeruginosa ATCC 9027 | 1667 ± 516 | 2333 ± 817 | 1500 ± 548 | 2333 ± 817 | >2333 | >2333 | 0.67 ± 0.3 | 0.83 ± 0.3 |
| P. aeruginosa 12274 | 667 ± 258 | 1500 ± 548 | 1333 ± 516 | 1500 ± 548 | >2333 | >2333 | 107 ± 33 | 341 ± 132 |
| P. mirabilis | 750 ± 246 | 833 ± 258 | 1167 ± 408 | 1333 ± 204 | >2333 | >2333 | 21 ± 8.3 | 19 ± 6.5 |
| P. mirabilis 1268 | 667 ± 258 | 917 ± 204 | 1333 ± 516 | 1333 ± 516 | >2333 | >2333 | 17 ± 6.5 | 21 ± 8.3 |
| E. hirae ATCC 1052 | 2167 ± 983 | 2333 ± 817 | 2333 ± 817 | 2333 ± 817 | 117 ± 26 | 192 ± 70 | 107 ± 33 | 149 ± 52 |
| E. faecium 38344825 | 6833 ± 2858 | 7333 ± 1633 | 9333 ± 3266 | >9333 | >2333 | >2333 | 85 ± 33 | 171 ± 66 |
| E. faecalis ATCC 51299 | 2333 ± 817 | 4667 ± 1633 | 2667 ± 1033 | 7333 ± 1633 | 171 ± 66 | 171 ± 52 | 1.33 ± 0.5 | 1.5 ± 0.6 |
| E. faecalis 3937158 | 1333 ± 516 | 1733 ± 653 | 1333 ± 516 | 2667 ± 1033 | >2333 | >2333 | 53 ± 17 | 171 ± 66 |
| E. faecalis 16274 | 458 ± 102 | 833 ± 258 | 1200 ± 408 | 1600 ± 516 | 235 ± 52 | 1195 ± 418 | 149 ± 52 | 234 ± 52 |
| S. aureus ATCC 6538 | 375 ± 137 | 750 ± 433 | 667 ± 258 | 1333 ± 516 | 27 ± 8.3 | 27 ± 8.3 | 0.38 ± 0.1 | 0.58 ± 0.3 |
| S. aureus MSSA 56/AS | 333 ± 129 | 458 ± 102 | 583 ± 204 | 583 ± 204 | 1365 ± 529 | >2333 | 0.67 ± 0.3 | 0.67 ± 0.3 |
| S. aureus MRSA 12673 | 667 ± 258 | 750 ± 418 | 833 ± 258 | 916 ± 204 | >2333 | >2333 | 149 ± 52 | 149 ± 52 |
| S. aureus MRSA (hetero-VISA) 6347 | 458 ± 102 | 667 ± 258 | 667 ± 516 | 667 ± 204 | >2333 | >2333 | 0.42 ± 0.1 | 0.67 ± 0.3 |
| S. aureus MRSA N315 (ref.) | 667 ± 258 | 750 ± 204 | 583 ± 204 | 583 ± 204 | >2333 | >2333 | 1.33 ± 0.5 | 1.33 ± 0.5 |
| S. aureus MRSA 13251 | 542 ± 225 | 583 ± 204 | 833 ± 258 | 833 ± 258 | >2333 | >2333 | 107 ± 33 | 213 ± 66 |
| S. aureus MRSA 15732 | 458 ± 102 | 833 ± 258 | 917 ± 204 | 917 ± 204 | >2333 | >2333 | 171 ± 66 | 171 ± 52 |
| S. aureus MRSA 13318 | 667 ± 258 | 833 ± 258 | 458 ± 102 | 833 ± 258 | >2333 | >2333 | 149 ± 52 | 149 ± 33 |
| K. pneumoniae ATCC 700603 | 1333 ± 516 | 1667 ± 516 | 1667 ± 516 | 3000 ± 1095 | 853 ± 264 | 1365 ± 529 | 43 ± 16 | 107 ± 66 |
| K. pneumoniae 16205 | 750 ± 274 | 667 ± 258 | 750 ± 274 | 833 ± 258 | 1877 ± 418 | >2333 | 85 ± 33 | 149 ± 52 |
| K. pneumoniae 12828 | 667 ± 274 | 1167 ± 408 | 1333 ± 516 | 1333 ± 204 | 1877 ± 418 | >2333 | 43 ± 17 | 171 ± 66 |
| E. coli ATCC 8739 | 833 ± 258 | 1083 ± 491 | 1500 ± 548 | 1833 ± 408 | 107 ± 33 | 384 ± 140 | 0.23 ± 0.08 | 0.84 ± 0.3 |
| E. coli 12519 | 667 ± 258 | 833 ± 258 | 1167 ± 408 | 1333 ± 516 | >2333 | >2333 | 13 ± 4.1 | 171 ± 66 |
| E. coli 12293 | 833 ± 258 | 1667 ± 516 | 1667 ± 516 | 2333 ± 817 | >2333 | >2333 | 21 ± 8.3 | 107 ± 33 |
| S. marcescens 12795 | 583 ± 333 | 1333 ± 516 | 917 ± 258 | 917 ± 204 | 149 ± 52 | 213 ± 66 | 0.19 ± 0.07 | 0.21 ± 0.07 |
| S. marcescens 13148/2 | 667 ± 258 | 1833 ± 408 | 917 ± 204 | 1333 ± 516 | 341 ± 132 | 427 ± 132 | 0.33 ± 0.1 | 0.33 ± 01 |
| C. sporogenes | 208 ± 65 | 1667 ± 516 | 115 ± 25 | 1833 ± 408 | 0.833 ± 0.3 | 1.17 ± 0.4 | 0.83 ± 0.3 | 1.17 ± 0.4 |
| Propionibacterium acnes | 208 ± 65 | 208 ± 65 | 229 ± 51 | 229 ± 51 | 171 ± 66 | 171 ± 66 | 0.75 ± 0.3 | 1.13 ± 0.5 |
| S. epidermidis ATCC 1499 | 667 ± 258 | 1083 ± 492 | 1667 ± 516 | 1667 ± 516 | 21 ± 8.3 | 21 ± 6.5 | 0.21 ± 0.06 | 0.42 ± 0.1 |
| S. epidermidis MRSE 13199 | 917 ± 204 | 1833 ± 1602 | 1833 ± 408 | 1833 ± 408 | 683 ± 264 | >2333 | 0.19 ± 0.07 | 0.19 ± 0.07 |
| Salmonella enterica | 1333 ± 516 | 1667 ± 516 | 1833 ± 408 | 2333 ± 817 | 341 ± 132 | >2333 | 48 ± 18 | 170 ± 66.1 |
| Helicobacter pylori | 167 ± 65 | 1833 ± 408 | 208 ± 65 | 3000 ± 1095 | 3.33 ± 1.03 | 5.3 ± 2.1 | 0.67 ± 0.3 | 1.17 ± 0.4 |
| S. pyogenes | 229 ± 51 | 521 ± 26 | 208 ± 65 | 229 ± 51 | 1.7 ± 0.52 | 4.7 ± 1.6 | 6 ± 1.9 | 24 ± 8.8 |
| S. pneumoniae | 208 ± 65 | 208 ± 65 | 229 ± 51 | 292 ± 102 | 1.8 ± 0.41 | 1.8 ± 0.5 | 11 ± 4.1 | - |
3.2. Serial Passages Assay
3.3. Synergy Assay
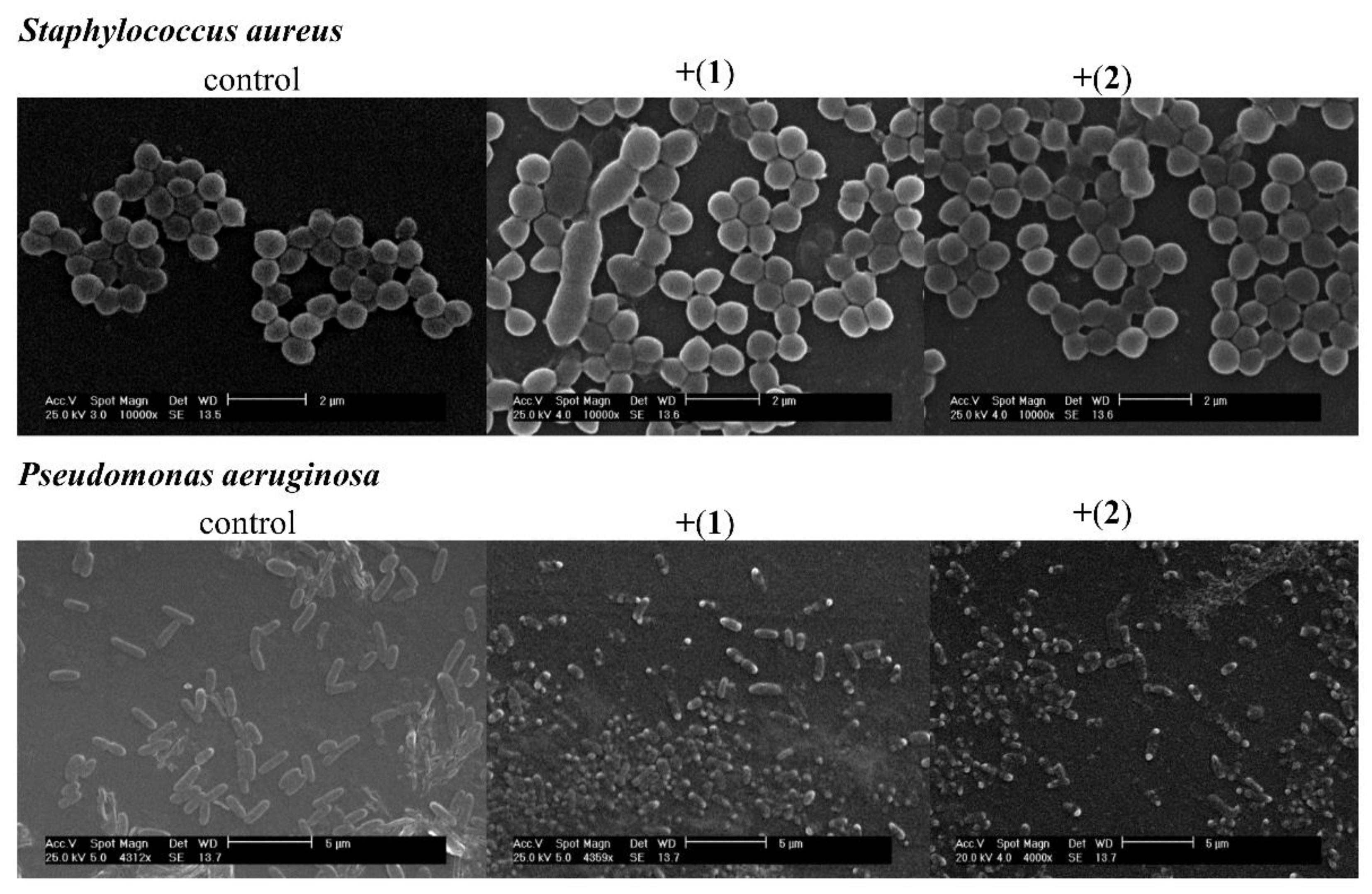
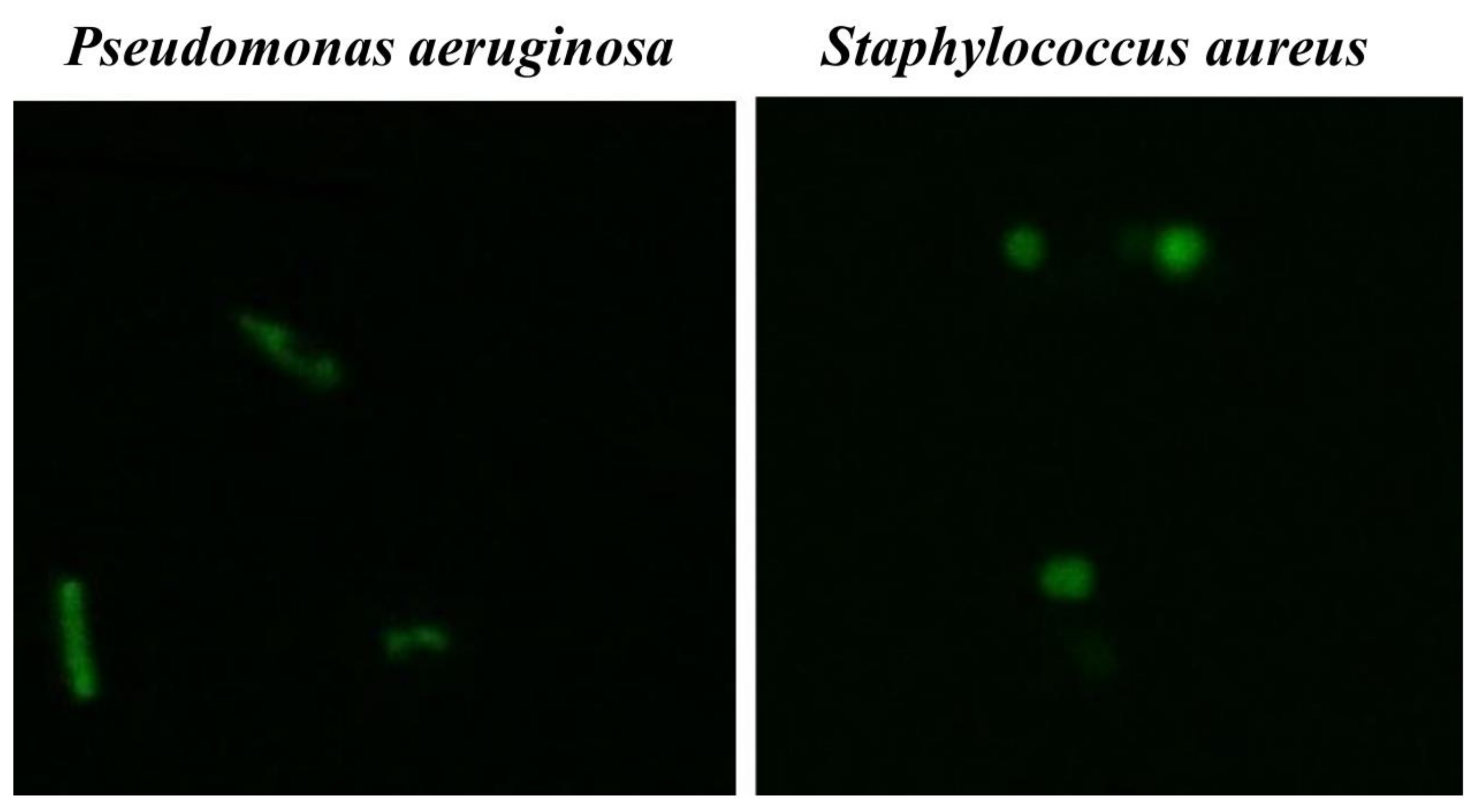
3.4. Microscopic Analysis
3.4.1. Transmission Electron Microscopy
3.4.2. Scanning Electron Microscopy
3.4.3. Fluorescent Microscopy Assay
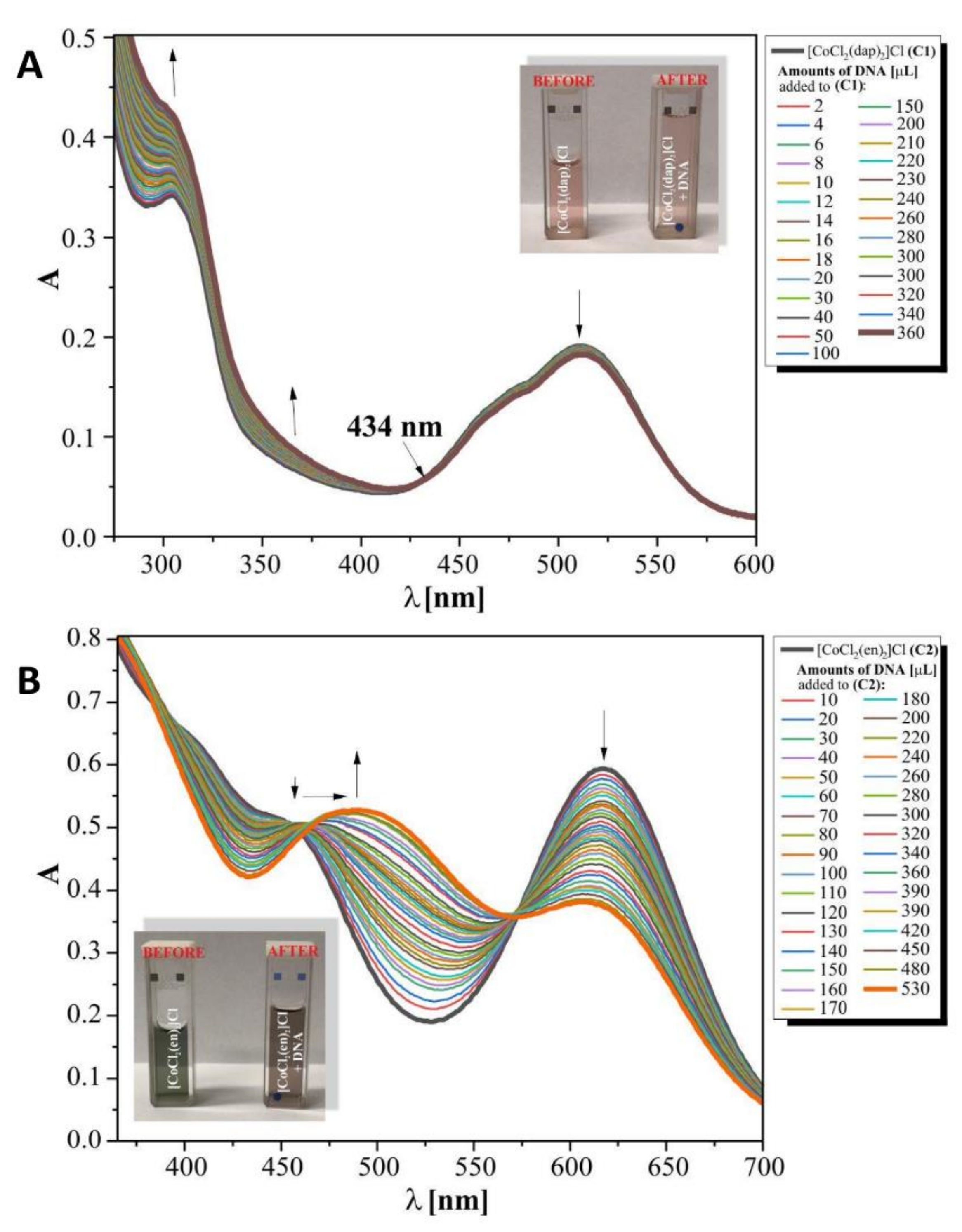
3.5. Interactions of Compounds (1) and (2) with DNA
3.5.1. Binding Studies in the Presence of E. coli DNA
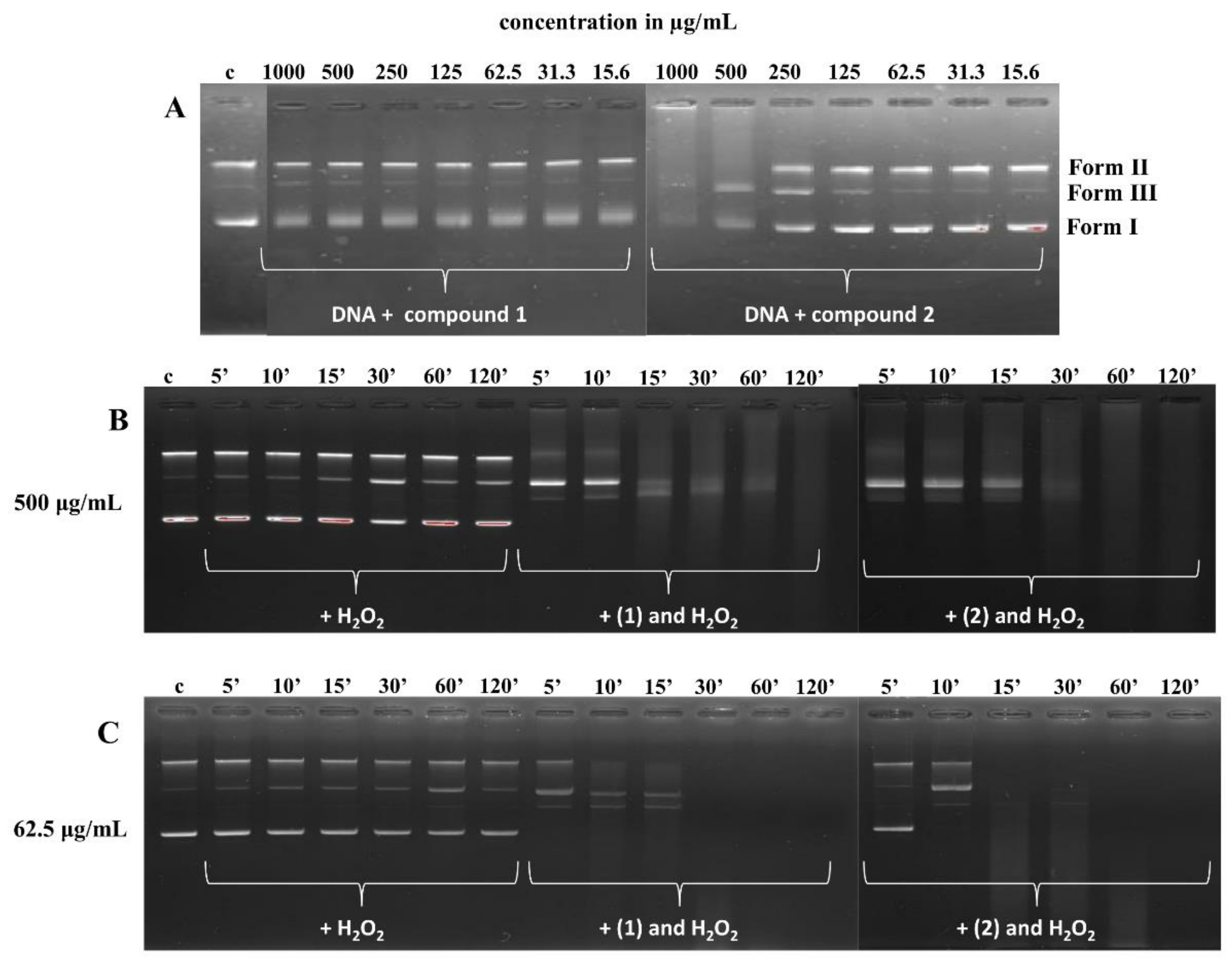
3.5.2. DNA Cleavage Study
4. Discussion
4.1. The Minimum Inhibitory Concentration (MIC) and Minimum Bactericidal Concentration (MBC)
4.2. Serial Passages Assay
4.3. Synergy Assay
4.4. Microscopic Analysis
4.5. Interactions of Compounds (1) and (2) with DNA
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Patrick, G.L. An introduction to Medicinal Chemistry; Oxford University Press: Oxford, UK, 2001; p. 264. [Google Scholar]
- Giordano, T.J.; Palenik, G.J.; Palenik, R.C.; Sullivan, D.A. Preparation and x-ray crystal structure of a seven-coordinate cobalt(II) complex with 2,6-diacetylpyridine bis(2-picolinoylhydrazone. Inorg. Chem. 1979, 18, 2445. [Google Scholar] [CrossRef]
- Rekha, T.H.; Ibrahim, K.M.; Abdallah, A.M.; Hassanian, M.M. Synthesis, spectral and antimicrobial activity studies of o-aminoacetophenone o-hydroxybenzoylhydrazone complexes. Synth. React. Inorg. Met. Org. Chem. 1996, 26, 1113. [Google Scholar]
- Nawar, N.; Hosny, N.M. Mono- and Bi-nuclerar Schiff base complexes derived from glycine, 3-acetylpyridine and transition metal ions. Transit. Met. Chem. 2000, 25, 1–8. [Google Scholar] [CrossRef]
- Turecka, K.; Chylewska, A.; Kawiak, A.; Waleron, K. Antifungal activity and mechanism of action of the Co(III) coordination complexes with diamine chelate ligands against reference and clinical strains of Candida spp. Front. Microbiol. 2018, 9, 1594. [Google Scholar] [CrossRef]
- Da Silva Dantas, F.G.; de Almeida-Apolonio, A.A.; de Araújo, R.P.; Favarin, L.R.V.; de Castilho, P.F.; de Oliveira Galvão, F.; Svidzinski, T.I.E.; Casagrande, G.A.; de Oliveira, K.M.P. A promising copper(II) complex as antifungal and anibiofilm drug against yeast infection. Molecules 2018, 23, 1856. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Favarin, L.R.V.; Oliveira, L.B.; Silva, H.; Micheletti, A.C.; Pizzuti, L.; Machulek-Júnior, A.; Caires, A.R.L.; Back, D.F.; Lima, S.M.; Andrade, L.H.C.; et al. Sonochemical synthesis of highly luminescent silver complexes: Photophysical properties and preliminary in vitro antitumor and antibacterial assays. Inorg. Chim. Acta 2018, 492, 235–242. [Google Scholar] [CrossRef]
- Favarin, L.R.V.; Laranjeira, G.B.; Teixeira, C.F.A.; Silva, H.; Micheletti, A.C.; Pizzuti, L.; Machulek-Júnior, A.; Caires, A.R.L.; Deflon, V.M.; Pesci, R.R.B.P.; et al. Harvesting greenish blue luminescence in gold(I) complexes and their application as promising bioactive molecules and cellular bioimaging agents. New J. Chem. 2020, 44, 6862–6871. [Google Scholar] [CrossRef]
- Rosenberg, B. Platinum complexes for the treatment of cancer. Interdiscipl. Sci. Rev. 1978, 3, 134–147. [Google Scholar] [CrossRef]
- Sadler, P.J. The biological chemistry of gold. Struct. Bond. 1984, 29, 171–214. [Google Scholar] [CrossRef] [Green Version]
- Clark, M.J.E. Ruthenium and Other Metal Complexes in Cancer Chemotherapy; Springer: Heidelberg, Germany, 1989. [Google Scholar]
- Cowan, J.A. Inorganic Biochemistry, 2nd ed.; Wiley-VCH: New York, NY, USA, 1997. [Google Scholar]
- Merchant, B. Gold, the noble metal and the paradoxes of its toxicology. Biologicals 1998, 26, 49–59. [Google Scholar] [CrossRef]
- Bertini, L.; Gray, H.B.; Stiefel, E.I.; Valentine, J.S. Biological Inorganic Chemistry: Structure and Rectivity; University Science Books: Sausalito, CA, USA, 2007. [Google Scholar]
- Walker, G.W.; Gene, R.J.; Sargeson, A.M.; Behm, C.A. Surface-active cobalt cage complexes: Synthesis, surface chemistry, biological activity, and redox properties. Dalton Trans. 2003, 2992–3001. [Google Scholar] [CrossRef]
- Yilmaz, I.; Cukurovali, A. Characterization and Antimicrobial Activity of the Schiff Bases Derived from 2,4-Disubstituted Thiazole and 3-Methoxy Salicylaldehyde and Their Cobalt(II), Copper(II), Nickel(II) and Zinc(II) Complexes. Trans Met. Chem. 2003, 28, 399–404. [Google Scholar] [CrossRef]
- Liang, F.; Wang, P.; Zhou, X.; Li, T.; Li, Z.; Lin, H.; Gao, D.; Zheng, C.; Wu, C. Nickle(II) and cobalt(II) complexes of hydroxyl-substituted triazamacrocyclic ligand as potential antitumor agents. Bioorg Med. Chem Lett. 2004, 14, 1901–1904. [Google Scholar] [CrossRef] [PubMed]
- Belicchi-Ferrari, M.; Bisceglie, F.; Casoli, C.; Durot, S.; Morgerstern-Badarau, I.; Pelosi, G.; Pilloti, E.; Pinelli, S.; Tarasconi, P. Copper(II) and Cobalt(III) Pyridoxal Thiosemicarbazone Complexes with Nitroprusside as Counterion: Syntheses, Electronic Properties, and Antileukemic Activity. J. Med. Chem. 2005, 48, 1671–1679. [Google Scholar] [CrossRef] [PubMed]
- Lv, J.; Liu, T.; Cai, S.; Wang, X.; Liu, L.; Wang, Y. Synthesis, structure and biological activity of cobalt(II) and copper(II) complexes of valine-derived schiff bases. J. Inorg. Biochem. 2006, 100, 1888–1896. [Google Scholar] [CrossRef]
- Zhong, X.; Yi, J.; Sun, J.; Wei, H.L.; Liu, W.S.; Yu, K.B. Synthesis and crystal structure of some transition metal complexes with a novel bis-Schiff base ligand and their antitumor activities. Eur. J. Med. Chem. 2006, 41, 1090–1092. [Google Scholar] [CrossRef] [PubMed]
- Bisceglie, F.; Baldini, M.; Belicchi-Ferrari, M.; Buluggiu, E.; Careri, M.; Pelosi, G.; Pinelli, S.; Tarasconi, P. Metal complexes of retinoid derivatives with antiproliferative activity: Synthesis, characterization and DNA interaction studies. Eur. J. Med. Chem. 2007, 42, 627–634. [Google Scholar] [CrossRef]
- Penumaka, N.; Satyanarayana, S. DNA Binding and Photocleavage Studies of Cobalt(III) Polypyridine Complexes: [Co(en)2PIP]3+, [Co(en)2IP]3+, and [Co(en)2phen-dione]3+. Bioinorg. Chem. Appl. 2007, 8, 1–8. [Google Scholar] [CrossRef] [Green Version]
- Konidaris, K.F.; Raptopoulou, C.P.; Psycharis, V.; Perlepes, S.P.; Manessi-Zoupa, E.; Stamatatos, T.C. Use of the 2-Pyridinealdoxime/N,N′-Donor Ligand Combination in Cobalt (III) Chemistry: Synthesis and Characterization of Two Cationic Mononuclear Cobalt (III) Complexes. Bioinorg. Chem. Appl. 2010, 7, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Dias, B.B.; da Silva Dantas, F.G.; Galvão, F.; Cupozk-Pinheiro, W.J.; Wender, H.; Pizzuti, L.; Rosa, P.P.; Tenório, K.V.; Gatto, C.C.; Negri, M.; et al. Synthesis, structural characteryzation, and prospects for new cobalt (II) complexes with tiocarbamoyl-pyrazoline ligands as promising antifungal agents. J. Ion. Bioch. 2020, 213, 111277. [Google Scholar] [CrossRef] [PubMed]
- Carter, M.T.; Rodriguez, M.; Bard, A.J. Voltammetric studies of the interaction of metal chelates with DNA. 2. Tris-chelated complexes of co(III) and iron(III) with 1,10-phenantroline and 2,2′-bipyridine. J. Am. Chem. Soc. 1989, 111, 8901–8911. [Google Scholar] [CrossRef]
- Jungwirth, U.; Kowol, C.R.; Keppler, B.K.; Hartinger, C.G.; Berger, W.; Heffeter, P. Anticancer Activity of Metal Complexes: Involvement of Redox Processes. Europe PubMed Central. Antioxid Redox Signal. 2011, 15, 1085–1127. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dwyer, F.P.; Sargeson, A.M. Stereospecific Influences in Metal Complexes Containing Optically Active Ligands. III. The Reaction of Dichlorobis-(ethylenediamine)-cobalt(III) Chloride with levoprophylenediamine. J. Am. Chem. Soc. 1959, 81, 5269–5272. [Google Scholar] [CrossRef]
- Mishra, A.; Kaushik, N.K.; Verma, A.K.; Gupta, R. Synthesis, characterization and antibacterial activity of cobalt(III) complexes with pyridine–amide ligands. Eur. J. Med. Chem. 2008, 43, 2189–2196. [Google Scholar] [CrossRef] [PubMed]
- Asbell, P.A.; Epstein, S.P.; Wallace, J.A.; Epstein, D.; Stewart, C.C.; Burger, R.M. Efficacy of kobalt chelates in the Rabbit eye model for epithelial herpetic keratitis. Cornea 1998, 17, 550–557. [Google Scholar] [PubMed]
- Brown, J.M. The Hypoxic Cell: A Target for Selective Cancer Theraphy—Eighteenth Bruce F. Cain Memorial Award Lecture. Cancer Res. 1999, 59, 5863–5870. [Google Scholar] [PubMed]
- Dachs, G.U.; Tozer, G.M. Hypoxia modulated gene expression: Angiogenesis, metatesis and therapeutic exploitation. Eur. J. Cancer 2000, 36, 1649–1660. [Google Scholar] [CrossRef]
- Kumar, R.S.; Arunachalam, S. Synthesis, micellar properties, DNA binding and antimicrobial studies of some surfactant-cobalt(III) complexes. Biophys. Chem. 2008, 136, 136–144. [Google Scholar] [CrossRef]
- El-Ayaan, U.; Abdel-Aziz, A.A.M. Synthesis, antimicrobial activity and molecular modeling of cobalt and nickel complexes containing the bulky ligand: Bis[N-(2,6-diisopropylphenyl)imino]acenaphthene. Eur. J. Med. Chem. 2005, 40, 1214–1221. [Google Scholar] [CrossRef]
- Chylewska, A.; Biedulsak, M.; Sumczyński, P.; Makowski, M. Metallopharmaceuticals in therapy—A new horizon for scientific research. Cur. Med. Chem. 2018, 25, 1729–1791. [Google Scholar] [CrossRef]
- Chylewska, A.; Turecka, K.; Dąbrowska, A.; Werel, W.; Chmurzyński, L. Synthesis, physicochemical characterization and antimicrobial activity of Co (III) complexes with diamine chelate ligands. IJAPBC 2013, 2, 454–464. [Google Scholar]
- Meletiadis, J.; Verweji, P.E.; TeDorsthorst, T.A.; Meis, J.F.G.M.; Mouton, J.W. Assessing in vitro combinations of antifungal drugs against yeasts and filamentous fungi: Comparison of different drug interaction models. Med. Mycol. 2005, 43, 133–152. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Odds, F.C. Synergy, antagonism, and what the chequerboard puts between them. J Ant. Chem. 2003, 52, 1. [Google Scholar] [CrossRef]
- Banasiuk, R.; Frackowiak, J.E.; Krychowiak, M.; Matuszewska, M.; Kawiak, A.; Ziabka, M.; Lendzion-Bielun, Z.; Narajczyk, M.; Królicka, A. Synthesis of antimicrobial silver nanoparticles through a photomediated reaction in an aqueous environment. Inter. J. Nanomed. 2016, 11, 315–324. [Google Scholar] [CrossRef] [Green Version]
- Khan, I.; Bahuguna, A.; Kumar, P.; Bajpal, V.K.; Kang, S.C. Antimicrobial Potential of Carvacrol against Uropathogenic Escherichia coli via Membrane Disruption, Depolarization, and Reactive Oxygen Species Generation. Front. Microbiol. 2017, 8, 2421. [Google Scholar] [CrossRef] [Green Version]
- Reichmann, M.E.; Rice, S.A.; Thomas, C.A.; Doty, P.A. Further Examination of the Molecular Weight and Size of Desoxypentose Nucleic Acid. J. Am. Chem. Soc. 1954, 76, 3047–3053. [Google Scholar] [CrossRef]
- Bouwman, E.; Day, R.; Driessen, W.L.; Tremel, W.; Krebs, B.; Wood, J.S.; Reedijk, J. Two different copper(II) coordination geometries imposed by two closely related chelating imidazole-thioether (N2S2) ligands. Crystal structures of (1,6-bis(4-imidazolyl)-2,5-dithiahexane)chlorocopper(II) tetrafluoroborate, (1,6-bis(5-methyl-4-imidazolyl)-2,5-dithiahexane)chloro(tetrafluoroborato)copper(II), and (1,6-bis(5-methyl-4-imidazolyl)-2,5-dithiahexane)nitrato(thiocyanato-N)copper(II). Inorg. Chem. 1988, 27, 4614–4618. [Google Scholar] [CrossRef]
- Vijayalakshmi, R.; Kanthimathi, M.; Subramanian, V.; Nair, B.U. Interaction of DNA with [Cr(Schiff base)(H2O)2]ClO4. Biochim. Biophys. Acta 2000, 1475, 157–162. [Google Scholar] [CrossRef]
- Suller, M.T.E.; Lloyd, D. The antibacterial activity of vancomycin towards Staphylococcus aureus under aerobic and anaerobic conditions. J Appl. Microbiol. 2001, 92, 866–872. [Google Scholar] [CrossRef] [Green Version]
- Oliveira, A.P.A.; Ferreira, J.F.G.; Farias, L.M.; Magalhães, P.P.; Teixeira, L.|R.; Beraldo, H. Antimicrobial Effects of Silver(I) and Bismuth(III) Complexes with Secnidazole-Derived Schiff Base Ligands: The Role of the Nitro Group Reduction. J. Braz. Chem. Soc. 2019, 30, 2299–2307. [Google Scholar] [CrossRef]
- Martinez, J.L.; Baquero, F.; Andersson, D.I. Beyond serial passages: New methods for predicting the emergence of resistance to novel antibiotics. Curr. Opin. Pharmacol. 2011, 11, 439–445. [Google Scholar] [CrossRef] [PubMed]
- Davies, T.A.; Pankuch, G.A.; Dewasse, B.E.; Jacobs, M.R.; Appelbaum, P.C. In vitro development of resistance to five quinolones and amoxicillin-claulanate in Streptococcus pneumoniae. Antimicrob. Agents Chemother. 1999, 42, 1177–1182. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Clark, C.; McGhee, P.; Appelbaum, P.C.; Kosowska-Shick, K. Multistep resistance development studies of ceftaroline in gram-positive and -negative bacteria. Antimicrob. Agents Chemother. 2011, 55, 2344–2351. [Google Scholar] [CrossRef] [Green Version]
- Mohammad, H.; Younis, W.; Ezzat, H.G.; Peters, C.E.; Abdel-Khalek, A.; Cooper, B.; Pogliano, K.; Pogliano, J.; Mayhoub, A.S.; Seleem, M.N. Bacteriological profiling of diphenylureas as a novel class of antibiotics against methicillin-resistant Staphylococcus aureus. PLoS ONE 2017, 12, 6. [Google Scholar] [CrossRef] [Green Version]
- D’Lima, L.; Friedman, L.; Wang, L.; Xu, P.; Anderson, M.; Debabov, D. No Decrease in Susceptibility to NVC-422 in Multiple-Passage Studies with Methicillin-Resistant Staphylococcus aureus, S. aureus, Pseudomonas aeruginosa, and Escherichia coli. Antimicrob Agents Chemother. 2012, 56, 2753–2755. [Google Scholar] [CrossRef] [Green Version]
- Pommier, Y.; Leo, E.; Zhang, H.; Marchand, C. DNA topoisomerases and their poisoning by anticancer and antibacterial drugs. Chem. Biol. 2010, 17, 421–433. [Google Scholar] [CrossRef] [Green Version]
- Shahabadi, N.; Kashanian, S.; Purfoulad, M. DNA interaction studies of a platinum(II) complex, PtCl2(NN) (NN = 4,7-dimethyl-1,10-phenanthroline), using different instrumental methods. Spectrochim. Acta Part A 2009, 72, 757–761. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Zhang, T.; Lu, T.; Qu, L.; Zhou, H.; Zhang, Q.; Ji, L. DNA-binding and cleavage studies of macrocyclic copper(II) complexes. J. Inorg. Biochem. 2002, 91, 269–276. [Google Scholar] [CrossRef]
- Liu, J.; Zhang, C.; Chen, H.; Deng, T.; Lu, T.; Ji, L. Interaction of macrocyclic copper(II) complexes with calf thymus DNA: Effects of the side chains of the ligands on the DNA-binding behaviors. Dalton Trans. 2003, 114–119. [Google Scholar] [CrossRef]
- Zhang, S.; Zhu, Y.; Tu, C.; Wei, H.; Yang, Z.; Lin, L.; Ding, J.; Zhang, Z.; Guo, A. A novel cytotoxic ternary copper(II) complex of 1,10-phenantroline and L-threonine with DNA nuclease activity. J. Inorg Biochem. 2004, 98, 2099–2106. [Google Scholar] [CrossRef]
- Thamilarasan, V.; Sengottuvelan, N.; Sudha, A.; Srinivasan, P.; Chakkaravarthi, G. Cobalt(III) complexes as potential anticancer agents: Physicochemical, structural, cytotoxic activity and DNA/protein interactions. J. Photochem. Photobiol. B Biol. 2016, 162, 558–569. [Google Scholar] [CrossRef]
- Haribabu, J.; Jeyallakshmi, K.; Arun, Y.; Bhuvanesh, N.S.P.; Perumal, P.T.; Karvembu, R. Synthesis, DNA/protein binding, molecular docking, DNA cleavage and in vitro anticancer activity of nickel(II) bis(thiosemicarbazone) complexes. RSC Adv. 2015, 5, 46031–46049. [Google Scholar] [CrossRef]
- Kumar, M.P.; Tejaswi, S.; Rambabu, A.; Kalalbandi, V.K.A. Synthesis, crystal structure, DNA binding and cleavage studies of cooper(II) complexes with isoxazole Schiff bases. Polyhedron 2015, 102, 111–120. [Google Scholar] [CrossRef]
- Mahalakshmi, R.; Raman, N. Enthused research on DNA-binding and DNA-cleavage aptitude of mixed ligand metal complexes. Spectrochim. Acta A Mol. Biomol. Spectrosc. 2013, 112, 198–205. [Google Scholar] [CrossRef] [PubMed]
- Van Eldik, R.; Reedijk, J. Advances in Inorganic Chemistry: Homogeneous Biomimetic Oxidation Catalysis; Elsevier: Amsterdam, The Netherlands, 2006. [Google Scholar]
- Qian, L.; Browne, W.R.; Roelfes, G. DNA cleavage activity of Fe(II)N4Py under photo irradiation in the presence of 1,8-naphthalimide and 9-aminocridine: Unexpected effects of reactive oxygen species scavengers. Inorg. Chem. 2011, 50, 8318–8325. [Google Scholar] [CrossRef]
- Qian, L.; Browne, W.R.; Roelfes, G. Photoenhanced oxidative DNA cleavage with non-heme iron(II) complexes. Inorg. Chem. 2010, 49, 11009–11017. [Google Scholar] [CrossRef] [Green Version]
- Zhao, Y.; Zhu, J.; He, W.; Yang, Z.; Zhu, Z.; Li, Y.; Zhang, J.; Guo, Z. Oxidative DNA cleavage promoted by multinuclear copper complexes: Activity dependence on the complex structure. Chem.-Eur. J. 2006, 12, 6621–6629. [Google Scholar] [CrossRef]
- Uma, V.; Kanthimathi, M.; Nair, B.U. Oxidative DNA cleavage mediated by a new cooper(II) terpyridine complex: Crystal structure and DNA binding studies. J. Inorg. Biochem. 2005, 99, 2299–2307. [Google Scholar] [CrossRef]
- Li, D.D.; Tian, J.L.; Gu, W.; Liu, X.; Zeng, H.H.; Yan, S.P. DNA binding, oxidative DNA cleavage, cytotoxicity and apoptosis-inducing activity of cooper(II) complexes with 1,4-Tpbd(N,N,N’N’-tetrakis(2-yridylme- thyl)benzene-1,4-diamine) ligand. J. Inorg. Bioch. 2011, 105, 894–901. [Google Scholar] [CrossRef]
- Li, D.D.; Huang, F.P.; Chen, G.J.; Gao, C.Y.; Tian, J.L.; Gu, W.; Liu, X.; Yan, S.P. Four new copper(II) complexes with 1,3-tpbd ligand: Synthesis, crystal structures, magnetism, oxidative and hydrolytic cleavage of pBR322 DNA. J. Inorg. Bioch. 2009, 104, 431–441. [Google Scholar] [CrossRef] [PubMed]
- Prakash, H.; Shodai, A.; Yasui, H.; Sakurai, H.; Hirota, S. Photocontrol of Spatial Orientation and DNA Cleavage Activity of Copper(II)-Bound Dipeptides Linked by an Azobenzene Derivative. Inorg. Chem. 2008, 47, 5045–5047. [Google Scholar] [CrossRef]
- Li, H.; Le, X.Y.; Pang, D.W.; Deng, H.; Xu, Z.H.; Lin, Z.H. DNA-binding and cleavage studies of novel copper(II) complex with L-phenylalaninate and 1,4,8,9-tetra-aza-triphenylene ligands. J. Inorg. Biochem. 2005, 99, 2240–2247. [Google Scholar] [CrossRef]
- Xu, W.; Louka, F.; Doulain, P.E.; Landry, C.A.; Mautner, F.A.; Massoud, S.S. Hydrolytic cleavage of DNA promoted by cobalt(III)–tetraamine complexes: Synthesis and characterization of carbonatobis [2-(2-pyridylethyl)]-(2-pyridylmethyl)aminecobalt(III) perchlorate. Polyhedron 2009, 28, 1221–1228. [Google Scholar] [CrossRef]
- Massoud, S.S.; Perkins, R.S.; Louka, F.R.; Xu, W.; LeRoux, A.; Dutercq, Q.; Fischer, R.C.; Mautner, F.A.; Handa, M.; Hiraoka, Y.; et al. Efficient hydrolytic cleavage of plasmid DNA by chloro-cobalt(II) complexes based on sterically hindered pyridyl tripod tetraamine ligands: Synthesis, crystal structure and DNA cleavage. Dalton. Trans. 2014, 43, 10086–10103. [Google Scholar] [CrossRef] [Green Version]
- Molenveld, P.; Engbersen, J.F.J.; Reinhoudt, D.N. Dinuclear metallo-phosphodiesterase models: Application of calix [4]arenes as molecular scaffolds. Chem. Soc. Rev. 2000, 29, 75–86. [Google Scholar] [CrossRef]
- Jin, Y.; Lewis, M.A.; Gokhale, N.H.; Long, E.C.; Cowan, J.A. Influence of Stereochemistry and Redox Potentials on the Single- and Double-Strand DNA Cleavage Efficiency of Cu(II)·and Ni(II)·Lys-Gly-His-Derived ATCUN Metallopeptides. J. Am. Chem. Soc. 2007, 129, 8353–8361. [Google Scholar] [CrossRef] [PubMed]
- Jin, Y.; Cowan, A. DNA Cleavage by Copper−ATCUN Complexes. Factors Influencing Cleavage Mechanism and Linearization of dsDNA. J. Am. Chem. Soc. 2005, 127, 8408–8415. [Google Scholar] [CrossRef]
- Jiang, Q.; Xiao, N.; Shi, P.; Zhu, Y.; Guo, Z. Design of artificial metallonucleases with oxidative mechanism. Coord. Chem. Rev. 2007, 251, 1951–1972. [Google Scholar] [CrossRef]
- Turel, I.; Kljun, J. Interactions of Metal Ions with DNA, Its Constituents and Derivatives, which may be Relevant for Anticancer Research. Curr. Top. Med. Chem. 2011, 11, 2661–2687. [Google Scholar] [CrossRef]
- Ahmadi, R.A.; Amani, S.S. Synthesis, Spectroscopy, Thermal Analysis, Magnetic Properties and Biological Activity Studies of Cu(II) and Co(II) Complexes with Schiff Base Dye Ligands. Molecules 2012, 17, 6434–6448. [Google Scholar] [CrossRef]
- Muhammad, N.; Guo, Z. Metal-based anticancer chemotherapeutic agents. Curr. Opin. Chem. Biol. 2014, 19, 144–153. [Google Scholar] [CrossRef]
- Tümer, M.; Köksal, H.; Serin, S.; Dig, M. Antimicrobial activity studies of mononuclear and binuclear mixed-ligand copper (II) complexes derived from Schiff base ligands and 1,10-phenantroline. Transit. Met. Chem. 1999, 24, 13–17. [Google Scholar] [CrossRef]
- Costa, S.S.; Viveiros, M.; Amaral, L.; Couto, I. Multidrug efflux pumps in Staphylococcus aureus: An uptade. Open Microbiol. J. 2013, 7 (Suppl. 1-M5), 59–71. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Anes, J.; McCusker, M.P.; Fanning, S.; Martins, M. The ins and outs of RND efflux pumps in Escherichia coli. Front. Microbiol. 2015, 6, 587. [Google Scholar] [CrossRef] [PubMed] [Green Version]

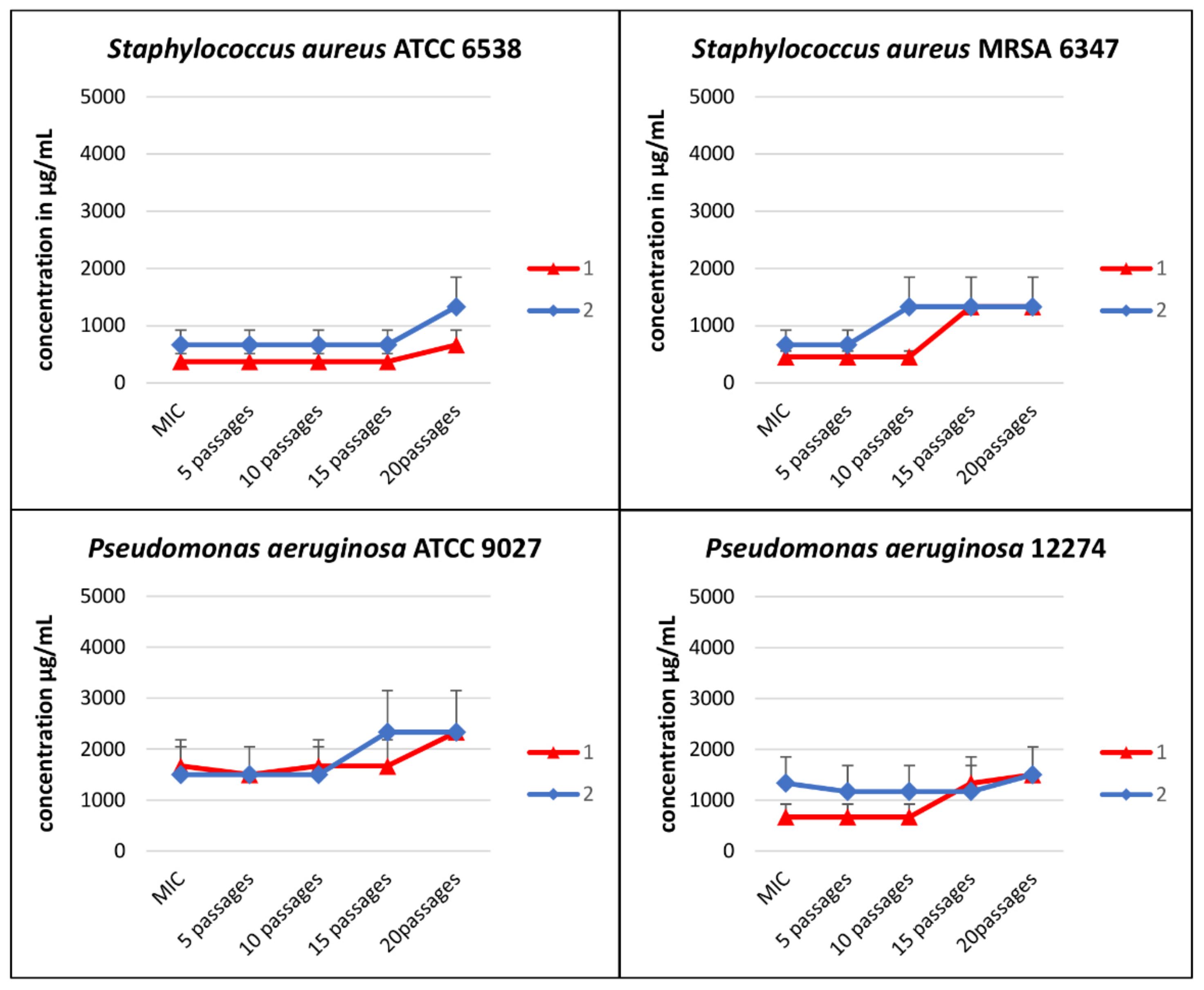


| Strains | Compound | Initial MIC (µg/mL) | MIC after 20 Passages (µg/mL) |
|---|---|---|---|
| S. aureus ATCC 6538 | [CoCl2(dap)2]Cl (1) | 375 ± 137 | 667 ± 258 |
| [CoCl2(en)2]Cl (2) | 667 ± 258 | 1333 ± 516 | |
| S. aureus MRSA 6347 | [CoCl2(dap)2]Cl (1) | 458 ± 102 | 1333 ± 516 |
| [CoCl2(en)2]Cl (2) | 667 ± 516 | 2333 ± 817 | |
| P. aeruginosa ATCC 9027 | [CoCl2(dap)2]Cl (1) | 1667 ± 516 | 2333 ± 817 |
| [CoCl2(en)2]Cl (2) | 1500 ± 548 | 2333 ± 817 | |
| P. aeruginosa 12274 | [CoCl2(dap)2]Cl (1) | 667 ± 258 | 1500 ± 548 |
| [CoCl2(en)2]Cl (2) | 1333 ± 516 | 1500 ± 548 |
| (a) | |||||||||
| The Bacterial Strain | Antibiotic | MIC of Antibiotics (µg/mL) | MIC of (1) (µg/mL) | Fold Reduction of MIC of Antibiotic | Fold Reduction of MIC of (1) | FIC | Interpretation | ||
| Alone | Com. | Alone | Com | ||||||
| S. aureus ATCC 6530 | Ampicillin | 27 ± 8.3 | 27 ± 8.3 | 458 ± 102 | 417 ± 144 | - | 0.9 | 2.1 | indifference |
| Gentamicin | 0.438 ± 0.13 | 0.438 ± 0.13 | 375 ± 137 | 375 ± 137 | - | - | 2 | indifference | |
| Ciprofloxacin | 0.38 ± 0.1 | 0.38 ± 0.1 | 375 ± 137 | 375 ± 137 | - | - | 2 | indifference | |
| Nalidixic acid | 53.3 ± 18.5 | 53.3 ± 18.5 | 375 ± 137 | 375 ± 137 | - | - | 2 | indifference | |
| Polymyxin B | 85.3 ± 37 | 85.3 ± 37 | 417 ± 144 | 208 ± 72 | - | 2 | 1.5 | indifference | |
| S. aureus MRSA 12673 | Ampicillin | >4096 | >4096 | 667 ± 258 | 667 ± 258 | - | - | - | - |
| Gentamicin | 0.23 ± 0.08 | 0.12 ± 0.03 | 667 ± 258 | 667 ± 258 | 1.9 | - | 1.5 | indifference | |
| Ciprofloxacin | 149 ± 52 | 85.3 ± 37 | 416.7 ± 144 | 208 ± 72 | 1.8 | 2 | 1 | indifference | |
| Nalidixic acid | 448 ± 128 | 56 ± 16 | 667 ± 258 | 667 ± 258 | 8 | - | 1.1 | indifference | |
| Polymyxin B | 224 ± 64 | 224 ± 64 | 667 ± 258 | 667 ± 258 | - | - | 2 | indifference | |
| E. coli ATCC 8739 | Ampicillin | 107 ± 37 | 53.5 ± 28.5 | 833 ± 258 | 417 ± 144 | 2 | 2 | 0.7 | indifference |
| Gentamicin | 0.42 ± 0.14 | 0.22 ± 0.06 | 437 ± 125 | 219 ± 63 | 2 | 2 | 1 | indifference | |
| Ciprofloxacin | 0.23 ± 0.08 | 0.12 ± 0.03 | 900 ± 223 | 313 ± 125 | 1.9 | 4 | 0.7 | indifference | |
| Nalidixic acid | 7.0 ± 2.0 | 3.6 ± 0.9 | 900 ± 223 | 437 ± 125 | 1.9 | 2 | 1 | indifference | |
| Polymyxin B | 0.438 ± 0.13 | 0.89 ± 0.25 | 875 ± 250 | 875 ± 250 | - | - | 3 | indifference | |
| E. coli 12519 | Ampicillin | 3584 ± 1024 | 3584 ± 1024 | 667 ± 258 | 667 ± 258 | - | - | 2 | indifference |
| Gentamicin | 112 ± 32 | 56 ± 16 | 667 ± 258 | 667 ± 258 | 2 | - | 1.5 | indifference | |
| Ciprofloxacin | 13 ± 4.1 | 13 ± 4.1 | 667 ± 258 | 667 ± 258 | - | - | 2 | indifference | |
| Nalidixic acid | 1792 ± 512 | 448 ± 128 | 667 ± 258 | 219 ± 63 | 4 | 3 | 0.6 | indifference | |
| Polymyxin B | 0.112 ± 0.03 | 0.438 ± 0.13 | 667 ± 258 | 667 ± 258 | - | - | 5 | antagonistic | |
| (b) | |||||||||
| The Bacterial Strain | Antibiotic | MIC of Antibiotics (µg/mL) | MIC of (2) (µg/mL) | Fold Reduction of MIC of Antibiotic | Fold Reduction of MIC of (2) | FIC | Interpretation | ||
| Alone | Com. | Alone | Com. | ||||||
| S. aureus ATCC 6530 | Ampicillin | 27 ± 8.3 | 27 ± 8.3 | 667 ± 258 | 667 ± 258 | - | - | 2 | indifference |
| Gentamicin | 0.438 ± 0.13 | 0.23 ± 0.08 | 667 ± 258 | 667 ± 258 | 2 | - | 1.5 | indifference | |
| Ciprofloxacin | 0.38 ± 0.1 | 0.17 ± 0.06 | 667 ± 258 | 667 ± 258 | 2.2 | - | 1.5 | indifference | |
| Nalidixic acid | 56 ± 16 | 7.0 ± 2.0 | 667 ± 258 | 667 ± 258 | 8 | - | 1.1 | indifference | |
| Polymyxin B | 80 ± 32 | 80 ± 32 | 667 ± 258 | 333 ± 63 | - | 2 | 1 | indifference | |
| S. aureus MRSA 12673 | Ampicillin | >4096 | >4096 | 883 ± 258 | 883 ± 258 | - | - | 1.2 | indifference |
| Gentamicin | 0.23 ± 0.08 | 0.12 ± 0.03 | 438 ± 125 | 438 ± 125 | 2 | - | 1.5 | indifference | |
| Ciprofloxacin | 149 ± 52 | 74.5 ± 29 | 438 ± 125 | 225 ± 56 | 2 | 1.9 | 1 | indifference | |
| Nalidixic acid | 461 ± 114 | 115 ± 33 | 438 ± 125 | 225 ± 56 | 4 | 1.9 | 0.7 | indifference | |
| Polymyxin B | 224 ± 64 | 224 ± 64 | 438 ± 125 | 438 ± 125 | - | - | 2 | indifference | |
| E. coli ATCC 8739 | Ampicillin | 107 ± 33 | 53.5 ± 19 | 1500 ± 548 | 750 ± 274 | 2 | 2 | 1 | indifference |
| Gentamicin | 0.438 ± 0.13 | 0.12 ± 0.03 | 1750 ± 500 | 875 ± 250 | 4 | 2 | 0.7 | indifference | |
| Ciprofloxacin | 0.23 ± 0.08 | 0.23 ± 0.08 | 1750 ± 500 | 875 ± 250 | - | 2 | 1.5 | indifference | |
| Nalidixic acid | 7.2 ± 1.8 | 7.2 ± 1.8 | 1750 ± 500 | 875 ± 250 | - | 2 | 1.5 | indifference | |
| Polymyxin B | 0.438 ± 0.13 | 0.22 ± 0.06 | 1750 ± 500 | 875 ± 250 | 2 | 2 | 1 | indifference | |
| E. coli 12519 | Ampicillin | 3584 ± 1024 | 3584 ± 1024 | 1167 ± 408 | 1167 ± 408 | - | - | 2 | indifference |
| Gentamicin | 115 ± 29 | 115 ± 29 | 1200 ± 447 | 1200 ± 447 | - | - | 2 | indifference | |
| Ciprofloxacin | 13 ± 4.1 | 13 ± 4.1 | 1167 ± 408 | 600 ± 224 | - | 1.9 | 1.5 | indifference | |
| Nalidixic acid | 1792 ± 512 | 320 ± 128 | 1200 ± 447 | 1200 ± 447 | 5.6 | - | 1.1 | indifference | |
| Polymyxin B | 0.109 ± 0.03 | 0.22 ± 0.06 | 1167 ± 408 | 1167 ± 408 | - | - | 3 | indifference | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Turecka, K.; Chylewska, A.; Rychłowski, M.; Zakrzewska, J.; Waleron, K. Antibacterial Activity of Co(III) Complexes with Diamine Chelate Ligands against a Broad Spectrum of Bacteria with a DNA Interaction Mechanism. Pharmaceutics 2021, 13, 946. https://doi.org/10.3390/pharmaceutics13070946
Turecka K, Chylewska A, Rychłowski M, Zakrzewska J, Waleron K. Antibacterial Activity of Co(III) Complexes with Diamine Chelate Ligands against a Broad Spectrum of Bacteria with a DNA Interaction Mechanism. Pharmaceutics. 2021; 13(7):946. https://doi.org/10.3390/pharmaceutics13070946
Chicago/Turabian StyleTurecka, Katarzyna, Agnieszka Chylewska, Michał Rychłowski, Joanna Zakrzewska, and Krzysztof Waleron. 2021. "Antibacterial Activity of Co(III) Complexes with Diamine Chelate Ligands against a Broad Spectrum of Bacteria with a DNA Interaction Mechanism" Pharmaceutics 13, no. 7: 946. https://doi.org/10.3390/pharmaceutics13070946
APA StyleTurecka, K., Chylewska, A., Rychłowski, M., Zakrzewska, J., & Waleron, K. (2021). Antibacterial Activity of Co(III) Complexes with Diamine Chelate Ligands against a Broad Spectrum of Bacteria with a DNA Interaction Mechanism. Pharmaceutics, 13(7), 946. https://doi.org/10.3390/pharmaceutics13070946