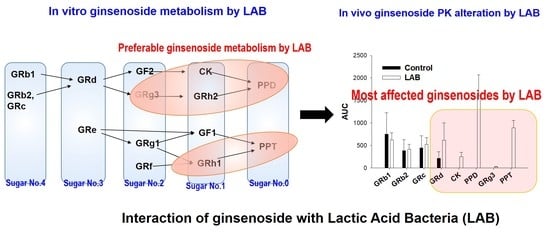
Effect of Lactic Acid Bacteria on the Pharmacokinetics and Metabolism of Ginsenosides in Mice
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials
2.2. In Vitro RGE Fermentation Study with LAB
2.3. Pharmacokinetic Study
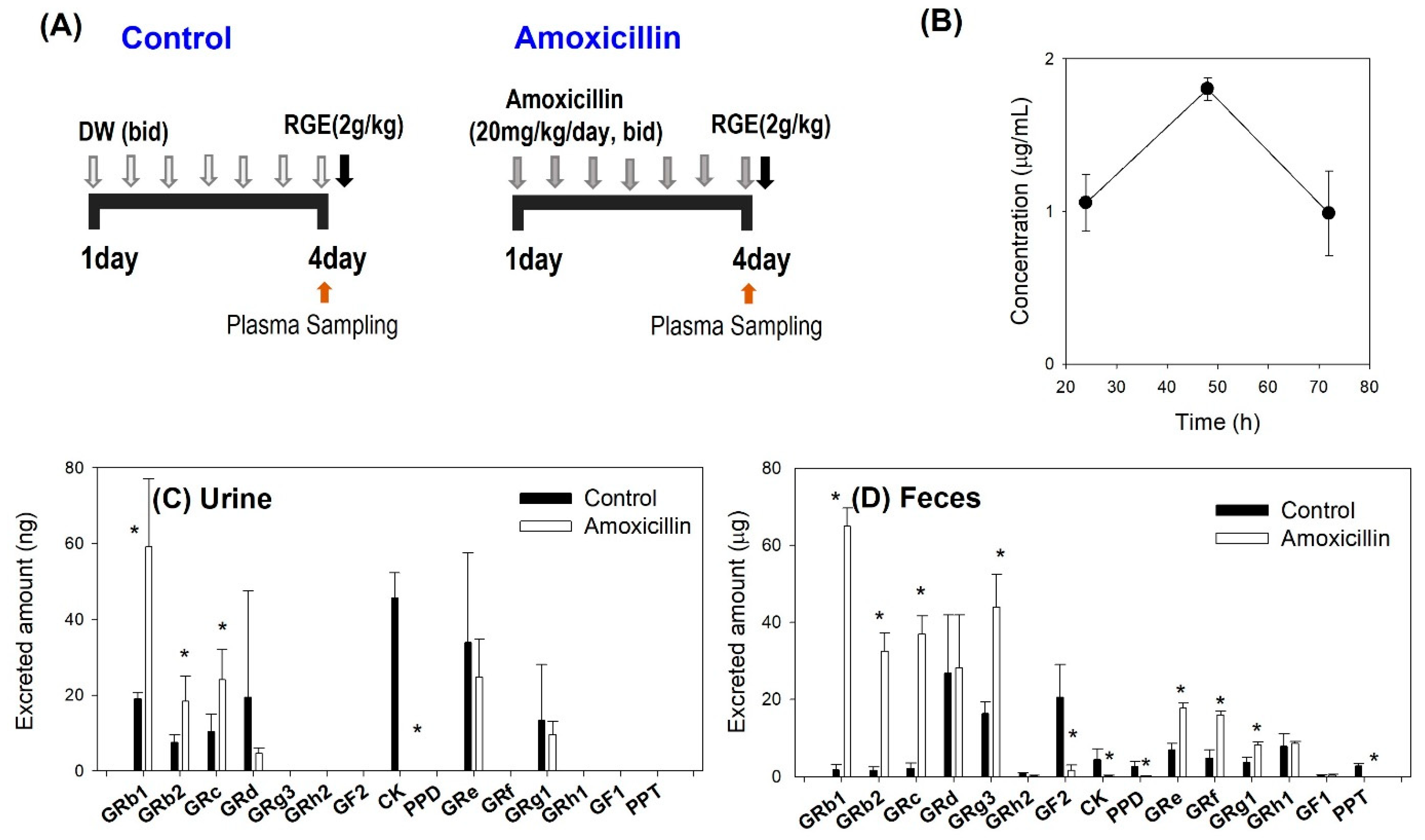
2.3.1. The Effect of Amoxicillin on the Intestinal Metabolism and the Pharmacokinetics of Ginsenosides
2.3.2. The Effect of LAB Supplementation on the Pharmacokinetics of Ginsenosides in Amoxicillin Treated Mice
2.4. Plasma Protein Binding of Ginsenosides
2.5. Plasma and Microsomal Stability of Ginsenoside
2.6. Caco-2 Permeability of Ginsenoside
2.7. LC-MS/MS Analysis
2.8. Data Analysis
3. Results
3.1. LAB-Mediated Ginsenosides Metabolism
3.2. Effect of Amoxicillin on the Metabolism of Ginsenoside
3.3. Plasma Concentrations of Ginsenosides Following Single Administration of RGE after Repeated Administration of Amoxicillin
3.4. Plasma Concentrations of Ginsenosides Following Single Oral Administration of RGE after Repeated Oral Administration of Amoxicillin with or without Repeated LAB Treatment
3.5. LAB-Mediated Metabolism of Ginsenoside
3.6. Permeability, Protein Binding, and Stability of Ginsenosides
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Won, H.-J.; Kim, H.I.; Park, T.; Kim, H.; Jo, K.; Jeon, H.; Ha, S.J.; Hyun, J.M.; Jeong, A.; Kim, J.S.; et al. Non-clinical pharmacokinetic behavior of ginsenosides. J. Ginseng Res. 2018, 43, 354–360. [Google Scholar] [CrossRef]
- Leung, K.W.; Wong, A.S. Pharmacology of ginsenosides: A literature review. Chin. Med. 2010, 5, 20–27. [Google Scholar] [CrossRef] [Green Version]
- Kim, J.H.; Yi, Y.-S.; Kim, M.-Y.; Cho, J.Y. Role of ginsenosides, the main active components of Panax ginseng, in inflammatory responses and diseases. J. Ginseng Res. 2016, 41, 435–443. [Google Scholar] [CrossRef] [Green Version]
- Gui, Q.F.; Xu, Z.R.; Xu, K.Y.; Yang, Y.M. The efficacy of ginseng-related therapies in type 2 diabetes mellitus: An updated systematic review and meta-analysis. Medicine 2016, 95, e2584. [Google Scholar] [CrossRef]
- Zhan, S.; Guo, W.; Shao, Q.; Fan, X.; Li, Z.; Cheng, Y. A pharmacokinetic and pharmacodynamic study of drug–drug inter-action between ginsenoside Rg1, ginsenoside Rb1 and schizandrin after intravenous administration to rats. J. Ethnopharmacol. 2014, 152, 333–339. [Google Scholar] [CrossRef]
- Zhan, S.-Y.; Shao, Q.; Li, Z.; Wang, Y.; Fan, X.-H. Study on PK-PD characteristics of ginsenoside Rg1 and Rb1, in rats with myocardial ischemia following intravenous administration of shengmai injection. China J. Chin. Mater. Med. 2014, 39, 1300–1305. [Google Scholar]
- Jeon, J.-H.; Kang, B.; Lee, S.; Jin, S.; Choi, M.-K.; Song, I.-S. Pharmacokinetics and intestinal metabolism of compound K in rats and mice. Pharmaceutics 2020, 12, 129. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, L.; Zhou, L.; Huang, J.; Wang, Y.; Yang, G.; Tan, Z.; Wang, Y.; Zhou, G.; Liao, J.; Ouyang, D. Single- and multiple-dose trials to determine the pharmacokinetics, safety, tolerability, and sex effect of oral ginsenoside compound K in healthy Chinese volunteers. Front. Pharmacol. 2018, 8, 965. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.L.; Zhou, L.P.; Wang, Y.Q.; Yang, G.P.; Huang, J.; Tan, Z.R.; Wang, Y.; Zhou, G.; Liao, J.; Ouyang, D. Food and sex-related impacts on the pharmacoki-netics of a single-dose of ginsenoside compound K in healthy subjects. Front. Pharmacol. 2017, 8, 636. [Google Scholar] [CrossRef] [PubMed]
- Choi, I.-D.; Ryu, J.-H.; Lee, D.-E.; Lee, M.H.; Shim, J.-J.; Ahn, Y.-T.; Sim, J.-H.; Huh, C.-S.; Shim, W.-S.; Yim, S.-V.; et al. Enhanced absorption study of ginsenoside compound K (20-O-beta-(D-glucopyranosyl)-20(S)-protopanaxadiol) after oral administration of fermented red ginseng extract (HYFRG) in healthy Korean volunteers and rats. Evid. Based. Complement. Alternat. Med. 2016, 2016, 3908142. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, H.K. Pharmacokinetics of ginsenoside Rb1 and its metabolite compound K after oral administration of Korean red ginseng extract. J. Ginseng Res. 2013, 37, 451–456. [Google Scholar] [CrossRef] [Green Version]
- Jung, J.-W.; Kang, H.-R.; Ji, G.-E.; Park, M.-S.; Song, W.-J.; Kim, M.-H.; Kwon, J.-W.; Kim, T.-W.; Park, H.-W.; Cho, S.-H.; et al. Therapeutic effects of fermented red ginseng in allergic rhinitis: A randomized, double-blind, placebo-controlled study. Allergy Asthma Immunol. Res. 2011, 3, 103–110. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Z.; Du, G.-J.; Wang, C.-Z.; Wen, X.-D.; Calway, T.; Li, Z.; He, T.-C.; Du, W.; Bissonnette, M.; Musch, M.W.; et al. Compound K, a ginsenoside metabolite, inhibits colon cancer growth via multiple pathways including p53–p21 Interactions. Int. J. Mol. Sci. 2013, 14, 2980–2995. [Google Scholar] [CrossRef] [PubMed]
- Xu, T.; Jin, Z.; Yuan, Y.; Wei, H.; Xu, X.; He, S.; Chen, S.; Hou, W.; Guo, Q.; Hua, B. Ginsenoside Rg3 serves as an adjuvant chemotherapeutic agent and vegf inhibitor in the treatment of non-small cell lung cancer: A meta-analysis and systematic review. Evid.-Based Complement. Altern. Med. 2016, 2016, 7826753. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, S.; Wang, Z.; Huang, Y.; O’Barr, S.A.; Wong, R.A.; Yeung, S.; Chow, M.S.S. Ginseng and anticancer drug combination to improve cancer chemotherapy: A critical review. Evid.-Based Complement. Altern. Med. 2014, 2014, 168940. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.-D.; Yang, Y.-Y.; Ouyang, D.-S.; Yang, G.-P. A review of biotransformation and pharmacology of ginsenoside compound K. Fitoterapia 2015, 100, 208–220. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.Y.; Sun, Y.; Fan, Q.X.; Zhang, Y.Q. Efficacy of Shenyi capsule combined with gemcitabine plus cisplatin in treatment of advanced esophageal cancer: A randomized controlled trial. Zhong Xi Yi Jie He Xue Bao 2009, 7, 1047–1051. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Wang, Y.; Niu, K.; Chen, X.; Xia, L.; Lu, D.; Kong, R.; Chen, Z.; Duan, Y.; Sun, J. Clinical benefit from EGFR-TKI plus ginsenoside Rg3 in patients with advanced non-small cell lung cancer harboring EGFR active mutation. Oncotarget 2016, 7, 70535–70545. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Peng, Z.; Wu, W.W.; Yi, P. The efficacy of ginsenoside Rg3 combined with first-line chemotherapy in the treatment of advanced non-small cell lung cancer in China: A systematic review and meta-analysis of randomized clinical trials. Front. Pharmacol. 2021, 11, 630825. [Google Scholar] [CrossRef] [PubMed]
- Choi, M.-K.; Song, I.-S. Interactions of ginseng with therapeutic drugs. Arch. Pharmacal Res. 2019, 42, 862–878. [Google Scholar] [CrossRef]
- Zhao, Q.; Li, P.; Jiang, J.; Hu, P. Pharmacokinetics of single ascending doses and multiple doses of 20(S)-ginsenoside Rg3 in Chinese healthy volunteers. Eur. J. Drug Metab. Pharmacokinet. 2015, 41, 845–853. [Google Scholar] [CrossRef]
- Yoon, S.J.; Kim, S.K.; Lee, N.Y.; Choi, Y.R.; Kim, H.S.; Gupta, H.; Youn, G.S.; Sung, H.; Shin, M.J.; Suk, K.T. Effect of Korean red ginseng on metabolic syndrome. J. Ginseng Res. 2020, 45, 380–389. [Google Scholar] [CrossRef] [PubMed]
- Yuan, H.-D.; Kim, J.T.; Kim, S.; Chung, S.-H. Ginseng and diabetes: The evidences from in vitro, animal and human studies. J. Ginseng Res. 2012, 36, 27–39. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, H.G.; Kim, K.Y.; Cha, C.J. Screening for ginseng-fermenting microorganism capable of biotransforming ginsenosides. Korean J. Microbiol. 2007, 43, 142–146. [Google Scholar]
- Jang, S.-H.; Park, J.; Kim, S.-H.; Choi, K.-M.; Ko, E.-S.; Cha, J.-D.; Lee, Y.-R.; Jang, H.; Jang, Y.-S. Red ginseng powder fermented with probiotics exerts antidiabetic effects in the streptozotocin-induced mouse diabetes model. Pharm. Biol. 2016, 55, 317–323. [Google Scholar] [CrossRef] [PubMed]
- Park, S.-H.; Oh, M.-R.; Choi, E.-K.; Kim, M.-G.; Ha, K.-C.; Lee, S.-K.; Kim, Y.-G.; Park, B.-H.; Kim, D.-S.; Chae, S.-W. An 8-wk, randomized, double-blind, placebo-controlled clinical trial for the antidiabetic effects of hydrolyzed ginseng extract. J. Ginseng Res. 2014, 38, 239–243. [Google Scholar] [CrossRef] [Green Version]
- Lee, H.-S.; Kim, M.-R.; Park, Y.; Park, H.J.; Chang, U.J.; Kim, S.Y.; Suh, H.J. Fermenting red ginseng enhances its safety and efficacy as a novel skin care anti-aging ingredient: In vitro and animal study. J. Med. Food 2012, 15, 1015–1023. [Google Scholar] [CrossRef] [Green Version]
- Nan, B.; Liu, Y.-L.; You, Y.; Li, W.-C.; Fan, J.-J.; Wang, Y.-S.; Piao, C.-H.; Hu, D.-L.; Lu, G.-J. Protective effects of enhanced minor ginsenosides in Lactobacillus fermentum KP-3-fermented ginseng in mice fed a high fat diet. Food Funct. 2018, 9, 6020–6028. [Google Scholar] [CrossRef]
- Trinh, H.-T.; Han, S.-J.; Kim, S.-W.; Lee, Y.C.; Kim, N.-H. Bifidus fermentation increases hypolipidemic and hypoglycemic effects of red ginseng. J. Microbiol. Biotechnol. 2007, 17, 1127–1133. [Google Scholar]
- Jin, S.; Lee, C.; Lim, D.; Lee, J.; Park, S.-J.; Song, I.-S.; Choi, M.-K. Improved hygroscopicity and bioavailability of solid dispersion of red ginseng extract with silicon dioxide. Pharmaceutics 2021, 13, 1022. [Google Scholar] [CrossRef]
- Jeon, J.-H.; Lee, J.; Choi, M.-K.; Song, I.-S. Pharmacokinetics of ginsenosides following repeated oral administration of red ginseng extract significantly differ between species of experimental animals. Arch. Pharmacal Res. 2020, 43, 1335–1346. [Google Scholar] [CrossRef]
- Park, S.-E.; Na, C.-S.; Yoo, S.-A.; Seo, S.-H.; Son, H.-S. Biotransformation of major ginsenosides in ginsenoside model culture by lactic acid bacteria. J. Ginseng Res. 2015, 41, 36–42. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dong, W.W.; Zhao, J.; Zhong, F.L.; Zhu, W.J.; Jiang, J.; Wu, S.; Yang, D.C.; Li, D.; Quan, L.H. Biotransformation of panax ginseng extract by rat intestinal microflora: Identification and quantification of metabolites using liquid chromatography-tandem mass spectrometry. J. Ginseng Res. 2017, 41, 540–547. [Google Scholar] [CrossRef]
- Jin, S.; Jeon, J.-H.; Lee, S.; Kang, W.Y.; Seong, S.J.; Yoon, Y.-R.; Cho, H.-J.; Song, I.-S. Detection of 13 ginsenosides (Rb1, Rb2, Rc, Rd, Re, Rf, Rg1, Rg3, Rh2, F1, compound K, 20(S)-protopanaxadiol, and 20(S)-protopanaxatriol) in human plasma and application of the analytical method to human pharmacokinetic studies following two week-repeated administration of red ginseng extract. Molecules 2019, 24, 2618. [Google Scholar] [CrossRef] [Green Version]
- Doh, E.S.; Chang, J.P.; Lee, K.H.; Seong, N.S. Ginsenoside change and antioxidation activity of fermented ginseng. Korean J. Med. Crop Sci. 2010, 18, 255–265. [Google Scholar]
- Kang, B.-H.; Lee, K.-J.; Hur, S.-S.; Lee, D.-S.; Lee, S.-H.; Shin, K.-S.; Lee, J.-M. Ginsenoside derivatives and quality characteristics of fermented ginseng using lactic acid bacteria. Korean J. Food Preserv. 2013, 20, 573–582. [Google Scholar] [CrossRef] [Green Version]
- Liu, H.; Yang, J.; Du, F.; Gao, X.; Ma, X.; Huang, Y.; Xu, F.; Niu, W.; Wang, F.; Mao, Y.; et al. Absorption and disposition of ginsenosides after oral administration of Panax notoginseng extract to rats. Drug Metab. Dispos. 2009, 37, 2290–2298. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Quan, L.H.; Kim, Y.J.; Li, G.H.; Choi, K.T.; Yang, D.C. Microbial transformation of ginsenoside rb1 to compound k by lacto-Bacillus paralimentarius. World J. Microbiol. Biotechnol. 2013, 29, 1001–1007. [Google Scholar] [CrossRef] [PubMed]
- Chi, H.; Kim, D.-H.; Ji, G.-E. Transformation of ginsenosides Rb2 and Rc from Panax ginseng by food microorganisms. Biol. Pharm. Bull. 2005, 28, 2102–2105. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bae, E.-A.; Shin, J.-E.; Kim, D.-H. Metabolism of ginsenoside re by human intestinal microflora and its estrogenic effect. Biol. Pharm. Bull. 2005, 28, 1903–1908. [Google Scholar] [CrossRef] [Green Version]
- Chi, H.L.; Lee, B.H.; You, H.J.; Park, M.S.; Ji, G.E. Differential transformation of ginsenosides from Panax ginseng by lactic acid bacteria. J. Microbiol. Biotech. 2006, 16, 1629–1633. [Google Scholar]
- Bae, E.-A.; Park, S.-Y.; Kim, D.-H. Constitutive beta-glucosidases hydrolyzing ginsenoside Rb1 and Rb2 from human intestinal bacteria. Biol. Pharm. Bull. 2000, 23, 1481–1485. [Google Scholar] [CrossRef]
- Bae, E.A.; Choo, M.K.; Park, E.K.; Park, S.Y.; Shin, H.Y.; Kim, D.H. Metabolism of ginsenoside r(c) by human intestinal bacteria and its related antiallergic activity. Biol. Pharm. Bull. 2002, 25, 743–747. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, D. Metabolism of ginsenosides to bioactive compounds by intestinal microflora and its industrial application. J. Ginseng Res. 2009, 33, 165–176. [Google Scholar] [CrossRef]
- Bae, E.-A.; Han, M.J.; Choo, M.-K.; Park, S.-Y.; Kim, D.-H. Metabolism of 20(S)- and 20(R)-ginsenoside Rg3 by human intestinal bacteria and its relation to in vitro biological activities. Biol. Pharm. Bull. 2002, 25, 58–63. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Choi, W.Y.; Choi, W.S.; Kwon, H.S.; Lee, H.Y. Enhancement of low molecular weight ginsenosides from low-quality ginseng through ultra-high-pressure and fermentation processes. Eur. Food Res. Tech. 2013, 237, 429–440. [Google Scholar] [CrossRef]
- Lee, J.H.; Hyun, Y.J.; Kim, D.H. Cloning and characterization of α-L-arabinofuranosidase and bifunctional α-L-arabinopyranosidase/β-D-galactopyranosidase from Bifidobacterium longum h-1. J. Appl. Microbiol. 2011, 111, 1097–1107. [Google Scholar] [CrossRef]
- Nicholson, J.K.; Holmes, E.; Wilson, I.D. Gut microorganisms, mammalian metabolism and personalized health care. Nat. Rev. Genet. 2005, 3, 431–438. [Google Scholar] [CrossRef]
- Park, S.J.; Kim, D.H.; Paek, N.S.; Kim, S.S. Preparation and quality characteristics of the fermentation product of ginseng by lactic acid bacteria. J. Ginseng Res. 2006, 30, 88–94. [Google Scholar]
- Delgado, S.; Florez, A.B.; Mayo, B. Antibiotic susceptibility of Lactobacillus and Bifidobacterium species from the human gastrointestinal tract. Curr. Microbiol. 2005, 50, 202–207. [Google Scholar] [CrossRef]
- Dudek-Wicher, R.K.; Junka, A.; Bartoszewicz, M. The influence of antibiotics and dietary components on gut microbiota. Gastroenterol. Rev. 2018, 13, 85–92. [Google Scholar] [CrossRef] [PubMed]
- Charteris, W.P.; Kelly, P.M.; Morelli, L.; Collins, J.K. Antibiotic susceptibility of potentially probiotic Lactobacillus species. J. Food Prot. 1998, 61, 1636–1643. [Google Scholar] [CrossRef] [PubMed]
- Nam, S.J.; Lee, T.; Choi, M.-K.; Song, I.-S. Characterization of preclinical in vitro and in vivo pharmacokinetic properties of KPLA-012, a benzopyranyl 1,2,3-triazole compound, with anti-angiogenetic and anti-tumor progressive effects. Mass Spectrom. Lett. 2018, 9, 61–65. [Google Scholar]
- Song, I.-S.; Choi, Y.A.; Choi, M.-K. Comparison of gastrointestinal permeability of caffeine, propranolol, atenolol, ofloxacin, and quinidine measured using chamber system and Caco-2 cell monolayer. Mass Spectrom. Lett. 2017, 8, 34–38. [Google Scholar]
- Choi, Y.A.; Song, I.-S.; Choi, M.-K. Pharmacokinetic drug-drug interaction and responsible mechanism between memantine and cimetidine. Pharmaceutics 2018, 10, 119. [Google Scholar] [CrossRef] [Green Version]
- Kwon, M.; Ji, H.-K.; Goo, S.H.; Nam, S.J.; Kang, Y.J.; Lee, E.; Liu, K.H.; Choi, M.-K.; Song, I.-S. Involvement of intestinal efflux and metabolic instability in the pharmacokinetics of platycodin D in rats. Drug Metab. Pharmacokinet. 2017, 32, 248–254. [Google Scholar] [CrossRef]
- Choi, M.-K.; Jin, S.; Jeon, J.-H.; Kang, W.Y.; Seong, S.J.; Yoon, Y.-R.; Han, Y.-H.; Song, I.-S. Tolerability and pharmacokinetics of ginsenosides Rb1, Rb2, Rc, Rd, and compound K after single or multiple administration of red ginseng extract in human beings. J. Ginseng Res. 2018, 44, 229–237. [Google Scholar] [CrossRef]
- Jeon, J.H.; Lee, J.; Lee, C.H.; Choi, M.K.; Song, I.-S. Correlation between the content and pharmacokinetics of ginsenosides from four different preparation of panax ginseng C.A. Meyer in rats. Mass Spectrom. Lett. 2021, 12, 16–20. [Google Scholar]
- Kwon, M.; Jeon, J.-H.; Choi, M.-K.; Song, I.-S. The development and validation of a novel “dual cocktail” probe for cytochrome P450s and transporter functions to evaluate pharmacokinetic drug-drug and herb-drug interactions. Pharmaceutics 2020, 12, 938. [Google Scholar] [CrossRef]
- Jeon, J.-H.; Lee, S.; Lee, W.; Jin, S.; Kwon, M.; Shin, C.H.; Choi, M.-K.; Song, I.-S. Herb–Drug interaction of red ginseng extract and ginsenoside Rc with valsartan in rats. Molecules 2020, 25, 622. [Google Scholar] [CrossRef] [Green Version]
- Kim, M.; Park, M.H.; Nam, G.; Lee, M.; Kang, J.; Song, I.S.; Choi, M.K.; Jin, H.K.; Bae, J.S.; Lim, M.H. A glycosylated prodrug to attenuate neuroinflammation and improve cognitive deficits in Alzheimer’s disease transgenic mice. Mol. Pharm. 2021, 18, 101–112. [Google Scholar] [CrossRef] [PubMed]
- Song, I.-S.; Jeong, H.-U.; Choi, M.-K.; Kwon, M.; Shin, Y.; Kim, J.H.; Lee, H.-S. Interactions between cyazofamid and human drug transporters. J. Biochem. Mol. Toxicol. 2020, 34, e22459. [Google Scholar] [CrossRef] [PubMed]
- Yoo, J.M.L.; Lee, J.Y.; Lee, Y.G.; Back, S.Y.; Kim, M.R. Enhanced production of compound K in fermented ginseng extracts by Lactobacillus brevis. Food Sci. Biotechnol. 2019, 28, 823–829. [Google Scholar] [CrossRef]
- Lee, N.-K.; Paik, H.-D. Bioconversion using lactic acid bacteria: Ginsenosides, GABA, and phenolic compounds. J. Microbiol. Biotechnol. 2017, 27, 869–877. [Google Scholar] [CrossRef] [PubMed]
- Renchinkhand, G.; Park, Y.W.; Song, G.-Y.; Cho, S.-H.; Urgamal, M.; Bae, H.C.; Choi, J.W.; Nam, M.S. Identification of β-glucosidase activity of Enterococcus faecalis CRNB-A3 in Airag and its potential to convert ginsenoside Rb1 from panax ginseng. J. Food Biochem. 2016, 40, 120–129. [Google Scholar] [CrossRef]
- Kim, D.-H. Gut microbiota-mediated pharmacokinetics of Ginseng saponins. J. Ginseng Res. 2017, 42, 255–263. [Google Scholar] [CrossRef]
- Westerhout, J.; van de Steeg, E.; Grossouw, D.; Zeijdner, E.E.; Krul, C.A.; Verwei, M.; Wortelboer, H.M. A new approach to predict human intestinal absorption using porcine intestinal tissue and biorelevant matrices. Eur. J. Pharm. Sci. 2014, 63, 167–177. [Google Scholar] [CrossRef]
- Volpe, D.A. Permeability classification of representative fluoroquinolones by a cell culture method. AAPS PharmSci 2004, 6, 1–6. [Google Scholar] [CrossRef] [Green Version]
- Paixão, P.; Gouveia, L.F.; Morais, J.A. Prediction of the in vitro permeability determined in Caco-2 cells by using artificial neural networks. Eur. J. Pharm. Sci. 2010, 41, 107–117. [Google Scholar] [CrossRef]
- Barber, H.; Hawksworth, G.; Kitteringham, N.; Petersen, J.; Petrie, J.; Swann, J. Protein binding of atenolol and propranolol to human serum albumin and in human plasma [proceedings]. Br. J. Clin. Pharmacol. 1978, 6, 446–447. [Google Scholar] [CrossRef]
- Van Steeg, T.; Boralli, V.; Krekels, E.; Slijkerman, P.; Freijer, J.; Danhof, M.; de Lange, E. Influence of plasma protein binding on pharmacodynamics: Estimation of in vivo receptor affinities of β blockers using a new mechanism-based PK–PD modelling approach. J. Pharm. Sci. 2009, 98, 3816–3828. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.K.; Choi, M.S.; Jeung, W.; Ra, J.; Yoo, H.H.; Kim, D.H. Effects of gut microbiota on the pharmacokinetics of protopanaxadiol ginsenosides Rd, Rg3, F2, and compound K in healthy volunteers treated orally with red ginseng. J. Ginseng Res. 2020, 44, 611–618. [Google Scholar] [CrossRef]
- Yoo, S.; Park, B.-I.; Kim, D.-H.; Lee, S.; Lee, S.-H.; Shim, W.-S.; Seo, Y.; Kang, K.; Lee, K.-T.; Yim, S.-V.; et al. Ginsenoside Absorption rate and extent enhancement of black ginseng (CJ EnerG) over red ginseng in healthy adults. Pharmaceutics 2021, 13, 487. [Google Scholar] [CrossRef] [PubMed]
- Jiang, R.; Dong, J.; Li, X.; Du, F.; Jia, W.; Xu, F.; Wang, F.; Yang, J.; Niu, W.; Li, C. Molecular mechanisms governing different pharmacokinetics of ginsenosides and potential for ginsenoside-perpetrated herb–drug interactions on OATP1B3. Br. J. Pharmacol. 2015, 172, 1059–1073. [Google Scholar] [CrossRef] [Green Version]
- Chiu, N.T.C.; Guns, E.S.T.; Adomat, H.; Jia, W.; Deb, S. Identification of human cytochrome P450 enzymes involved in the hepatic and intestinal biotransformation of 20(S)-protopanaxadiol. Biopharm. Drug Dispos. 2013, 35, 104–118. [Google Scholar] [CrossRef] [PubMed]








| PPD-Type | Content (mg/g RGE) | PPT-Type | Content (mg/g RGE) |
|---|---|---|---|
| GRb1 | 4.7 | GRg3 | 3.5 |
| GRb2 | 2.3 | GRh2 | ND |
| GRc | 2.5 | CK | ND |
| GRd | 1.3 | PPD | ND |
| F2 | ND | Total content of PPD-type 14.3 mg/g RGE | |
| PPT-type | Content (mg/g RGE) | PPT-type | Content (mg/g RGE) |
| GRe | 1.3 | GRh1 | 1.6 |
| GRg1 | 0.6 | F1 | ND |
| GRf | 1.1 | PPT | ND |
| Total content of PPD-type 4.6 mg/g RGE | |||
| Species | Species | Species |
|---|---|---|
| L. helveticus | L. rhamnosus | B. lactis |
| L. bulgaricus | L. casei | Enterococcus faecium |
| L. fermentum | L. reuteri | Enterococcus faecalis |
| L. gasseri | L. plantarum | Lactococcus lactis |
| L. paracasei | L. salivarius | Streptococcus thermophiles |
| L. acidophilus | B. longum | |
| B. breve | B. bifidum | Total 1 billion CFU/2 g |
| Group | Amoxicillin | Control | |||||
|---|---|---|---|---|---|---|---|
| Sampling Time (h) | Group 1 (n = 3) | Group 2 (n = 3) | Group 3 (n = 3) | Blood Volume (μL) | Group 4 (n = 3) | Group 5 (n = 3) | Blood Volume (μL) |
| 0 | RO–right | 80 | RO–right | 80 | |||
| 2 | RO–right | 80 | RO–right | 80 | |||
| 4 | RO–left | 80 | RO–left | 80 | |||
| 8 | RO–left | 80 | RO–left | 80 | |||
| 24 | RO–right | AA | 100 | AA | 100 | ||
| 48 | RO–left | AA | 100 | AA | 100 | ||
| 72 | AA | 100 | |||||
| Group | Amoxicillin + LAB | Amoxicillin | ||||
|---|---|---|---|---|---|---|
| Sampling Time (h) | Group 1 (n = 3) | Group 2 (n = 3) | Blood Volume (μL) | Group 3 (n = 3) | Group 4 (n = 3) | Blood Volume (μL) |
| 0 | RO–right | 80 | RO–right | 80 | ||
| 2 | RO–right | 80 | RO–right | 80 | ||
| 4 | RO–left | 80 | RO–left | 80 | ||
| 8 | RO–left | 80 | RO–left | 80 | ||
| 24 | AA | 100 | AA | 100 | ||
| 48 | AA | 100 | AA | 100 | ||
| Ginsenosides | Control | ||||
|---|---|---|---|---|---|
| AUC (ng/mL·h) | Cmax (ng/mL) | Tmax (h) | MRT (h) | Half-Life (h) | |
| GRb1 | 895.9 ± 81.3 | 41.3 ± 14.3 | 8.0 ± 0.0 | 16.3 ± 2.7 | 25.9 ± 4.7 |
| GRb2 | 361.4 ± 74.3 | 12.5 ± 2.1 | 8.0 ± 0.0 | 19.4 ± 1.6 | 24.6 ± 6.7 |
| GRc | 508.6 ± 109 | 17.2 ± 3.1 | 8.0 ± 0.0 | 19.9 ± 1.7 | 22.8 ± 6.7 |
| GRd | 278.8 ± 117 | 8.6 ± 1.8 * | 13.3 ± 9.2 | 21.3 ± 3.0 | 24.2 ± 3.1 |
| CK | 69.94 ± 37.8 | 3.6 ± 1.4 | 8.0 ± 0.0 | 13.9 ± 5.3 | 13.2 ± 3.2 |
| PPD | 608.7 ± 133.8 | 29.3 ± 7.0 | 8.0 ± 0.0 | 12.6 ± 4.7 | 7.9 ± 1.8 |
| PPT | 547.9 ± 77.6 | 20.3 ± 8.9 | 2.0 ± 0.0 | 20.2 ± 2.2 | 20.4 ± 2.3 |
| Ginsenosides | Amoxicillin | ||||
| AUC (ng/mL·h) | Cmax (ng/mL) | Tmax (h) | MRT (h) | Half-life (h) | |
| GRb1 | 926.0 ± 439 | 28.2 ± 12.9 | 8.0 ± 0.0 | 19.3 ± 1.4 | 21.7 ± 3.6 |
| GRb2 | 391.5 ± 192 | 10.3 ± 4.6 | 13.3 ± 9.2 | 21.4 ± 1.6 | 25.1 ± 2.7 |
| GRc | 571.7 ± 276 | 14.8 ± 6.6 | 13.3 ± 9.2 | 21.9 ± 1.5 | 25.1 ± 9.9 |
| GRd | 132.4 ± 77 | 3.2 ± 1.6 | 24.7 ± 23.0 | 24.0 ± 2.1 | 29.1 ± 8.6 |
| CK | NC | NC | NC | NC | NC |
| PPD | NC | NC | NC | NC | NC |
| PPT | NC | NC | NC | NC | NC |
| Ginsenosides | Amoxicillin | ||||
|---|---|---|---|---|---|
| AUC (ng/mL·h) | Cmax (ng/mL) | Tmax (h) | MRT (h) | Half-Life (h) | |
| GRb1 | 752.3 ± 477.6 | 23.7 ± 11.6 | 3.3 ± 1.2 | 18.6 ± 1.6 | 20.1 ± 2.2 |
| GRb2 | 385.9 ± 243.6 | 10.9 ± 5.2 | 4.0 ± 0.0 | 20.2 ± 1.7 | 19.2 ± 2.3 |
| GRc | 448.2 ± 268.1 | 13.2 ± 6.6 | 4.7 ± 3.1 | 20.6 ± 1.2 | 20.2 ± 1.8 |
| GRd | 213.1 ± 151.3 | 6.5 ± 5.1 | 5.3 ± 2.3 | 24.1 ± 3.9 | 29.2 ± 2.7 |
| CK | NC | NC | NC | NC | NC |
| PPD | NC | NC | NC | NC | NC |
| GRg3 | NC | NC | NC | NC | NC |
| PPT | NC | NC | NC | NC | NC |
| Ginsenosides | Amoxicillin + LAB | ||||
| AUC (ng/mL·h) | Cmax (ng/mL) | Tmax (h) | MRT (h) | Half-life (h) | |
| GRb1 | 622.1 ± 163.6 | 26.2 ± 4.6 | 4.7 ± 3.1 | 16.9 ± 1.0 | 16.2 ± 0.9 |
| GRb2 | 419.2 ± 108.6 | 15.1 ± 3.4 | 8.0 ± 0.0 | 18.1 ± 0.9 | 21.6 ± 1.7 |
| GRc | 524.4 ± 146.6 | 18.3 ± 4.3 | 8.0 ± 0.0 | 19.0 ± 1.3 | 24.1 ± 2.4 |
| GRd | 618.1 ± 385.6 * | 18.4 ± 6.9 * | 16.0 ± 9.2 | 20.6 ± 3.7 | 25.5 ± 3.8 |
| CK | 249.8 ± 103.0 | 9.3 ± 2.9 | 8.0 ± 0.0 | 13.7 ± 6.2 | 12.0 ± 5.0 |
| PPD | 1607.3 ± 463.7 | 73.0 ± 14.6 | 6.7 ± 2.3 | 16.0 ± 1.2 | 18.4 ± 2.2 |
| GRg3 | 25.3 ± 10.6 | 4.8 ± 1.9 | 2.5 ± 1.0 | 3.5 ± 0.9 | 5.3 ± 1.1 |
| PPT | 894.4 ± 161.3 | 63.4 ± 0.6 | 4.0 ± 0.0 | 13.7 ± 2.9 | 14.4 ± 3.0 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jeon, J.-H.; Lee, J.; Park, J.-H.; Lee, C.-H.; Choi, M.-K.; Song, I.-S. Effect of Lactic Acid Bacteria on the Pharmacokinetics and Metabolism of Ginsenosides in Mice. Pharmaceutics 2021, 13, 1496. https://doi.org/10.3390/pharmaceutics13091496
Jeon J-H, Lee J, Park J-H, Lee C-H, Choi M-K, Song I-S. Effect of Lactic Acid Bacteria on the Pharmacokinetics and Metabolism of Ginsenosides in Mice. Pharmaceutics. 2021; 13(9):1496. https://doi.org/10.3390/pharmaceutics13091496
Chicago/Turabian StyleJeon, Ji-Hyeon, Jaehyeok Lee, Jin-Hyang Park, Chul-Haeng Lee, Min-Koo Choi, and Im-Sook Song. 2021. "Effect of Lactic Acid Bacteria on the Pharmacokinetics and Metabolism of Ginsenosides in Mice" Pharmaceutics 13, no. 9: 1496. https://doi.org/10.3390/pharmaceutics13091496
APA StyleJeon, J.-H., Lee, J., Park, J.-H., Lee, C.-H., Choi, M.-K., & Song, I.-S. (2021). Effect of Lactic Acid Bacteria on the Pharmacokinetics and Metabolism of Ginsenosides in Mice. Pharmaceutics, 13(9), 1496. https://doi.org/10.3390/pharmaceutics13091496