Characterization of the Secretome of Pathogenic Candida glabrata and Their Effectiveness against Systemic Candidiasis in BALB/c Mice for Vaccine Development
Abstract
1. Introduction
2. Materials and Methods
2.1. The Strains and Growth Conditions
2.2. Antifungal Susceptibility Profiling
2.3. Screening of Pathogenicity Markers in Clinical Strains of C. glabrata
2.3.1. Adherence Assay
2.3.2. Biofilm Formation
2.3.3. Hydrolytic Enzymes Production
2.4. Secretory Protein Extraction and LC–MS Analysis
2.4.1. Extraction of Secreted Proteins from Growth Media
2.4.2. Sample Preparation for LC–MS Analysis
2.4.3. LC–MS Data Acquisition
2.4.4. LC–MS Database Searching
2.4.5. Bioinformatic and Functional Analyses
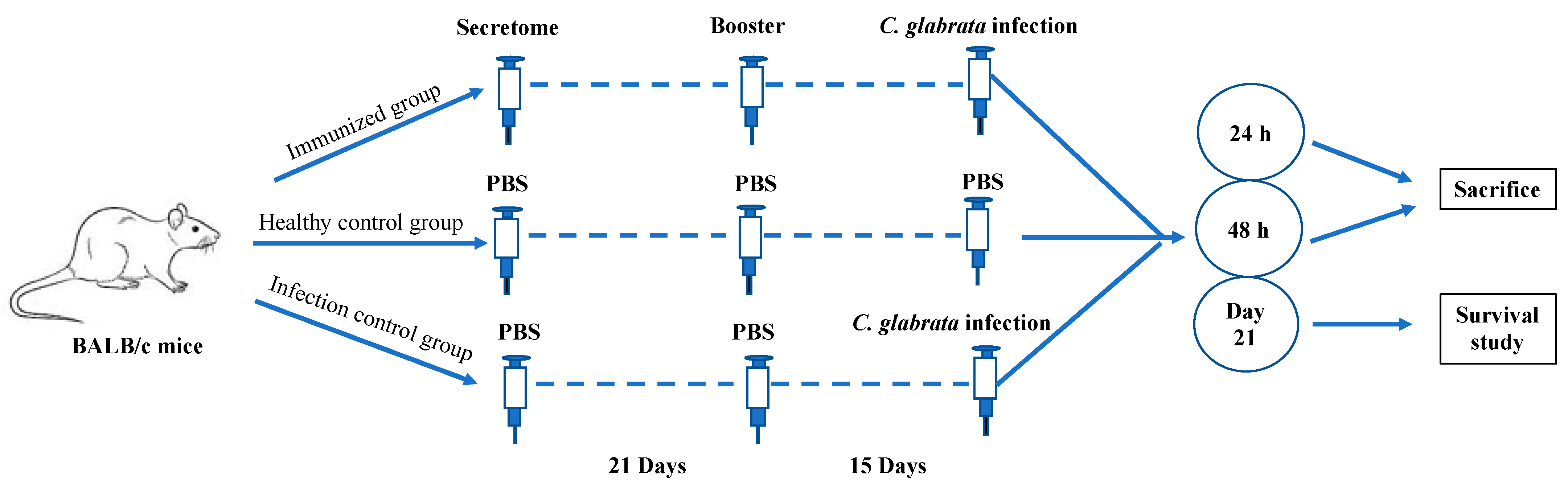
2.5. In Vivo Studies
2.6. Immunization Protocols
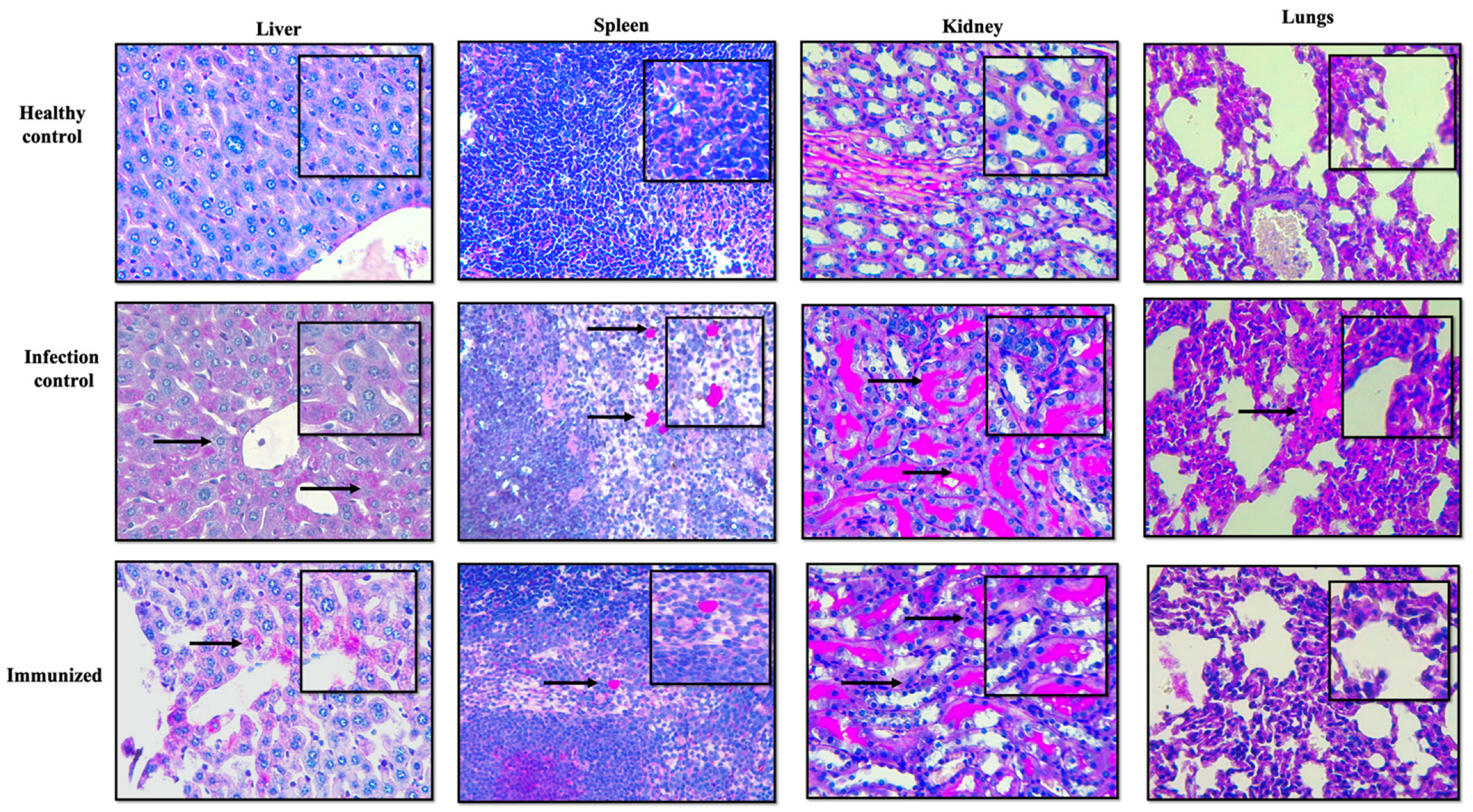
2.7. Histopathologic Examination
2.8. Statistical Analysis
3. Results and Discussion
3.1. Antifungal Susceptibility Profiling
3.2. Pathogenicity Markers
3.2.1. Adherence
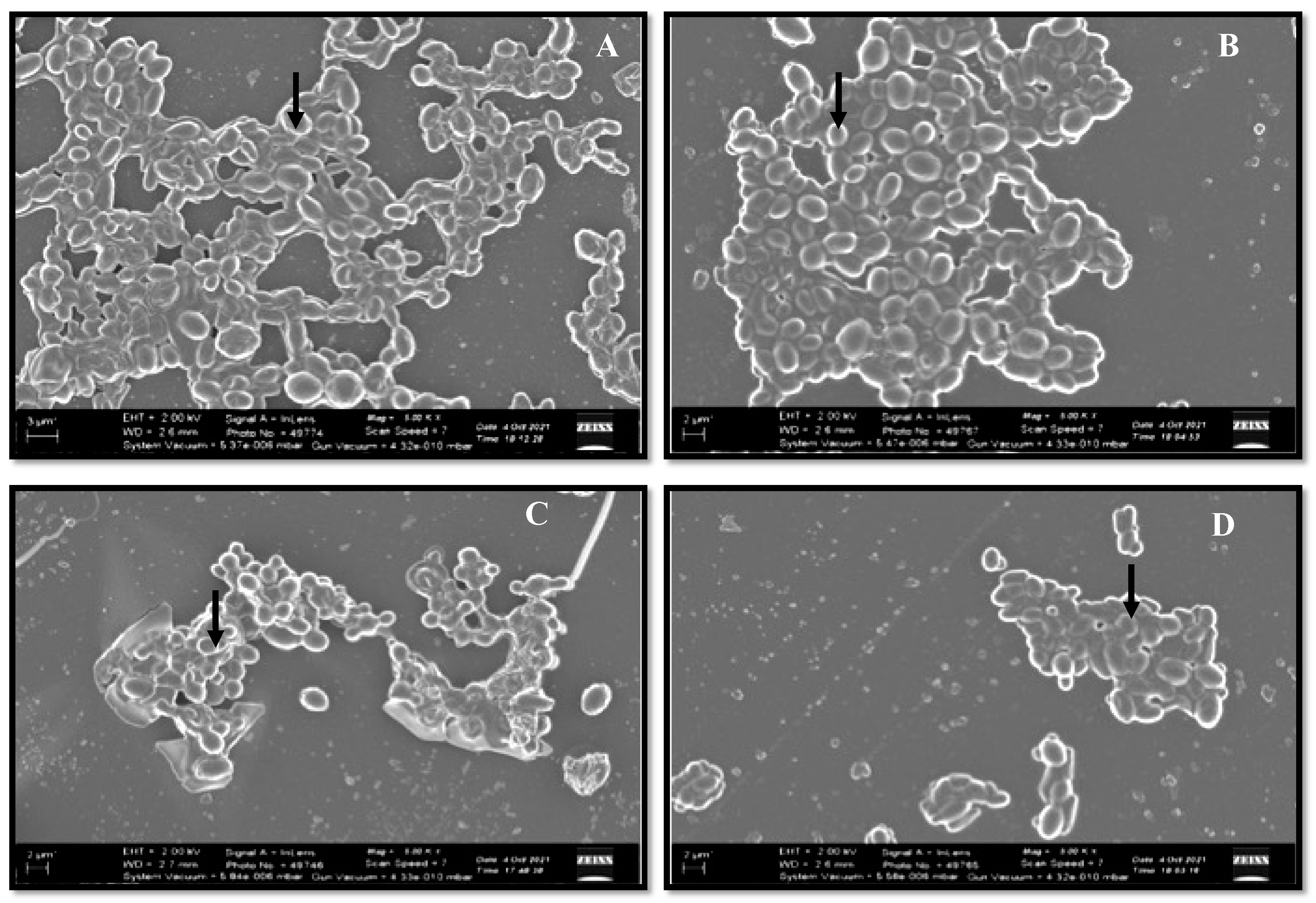
3.2.2. Biofilm Formation
3.2.3. Hydrolytic Enzymes
3.3. Proteomic Analysis of C. glabrata Secretome
3.4. Functional Analysis of Predicted Antigens of C. glabrata Secretome
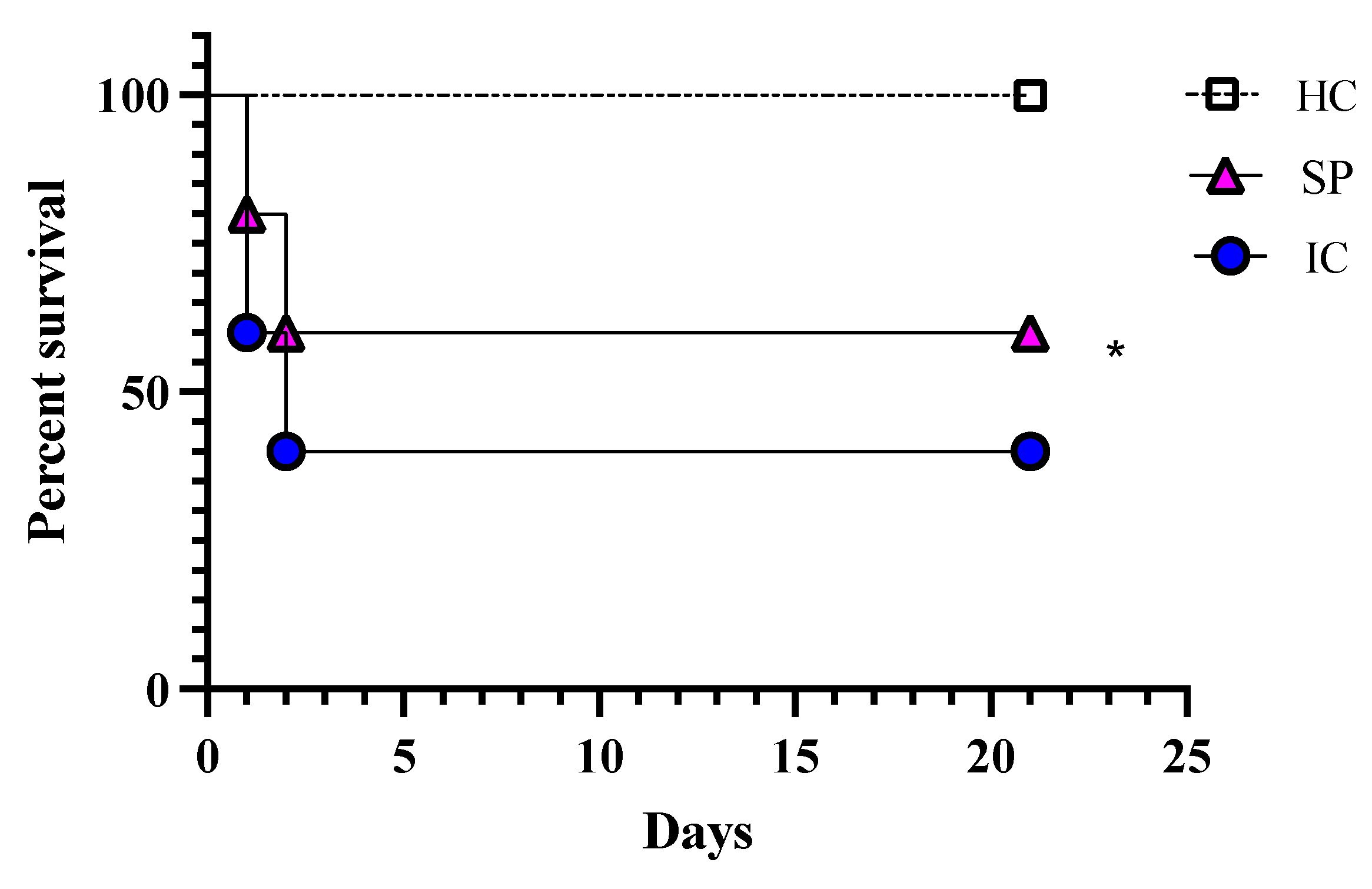
3.5. In Vivo Studies
Effectiveness of Secreted Proteins against C. glabrata Infection
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Brown, G.D.; Denning, D.W.; Gow, N.A.; Levitz, S.M.; Netea, M.G.; White, T.C. Hidden killers: Human fungal infections. Sci. Transl. Med. 2010, 4, 165rv13. [Google Scholar] [CrossRef] [PubMed]
- Pappas, P.; Lionakis, M.; Arendrup, M.; Ostrosky-Zeichner, L.; Jan Kullberg, B. Invasive candidiasis. Nat. Rev. Dis. Primers 2018, 4, 18026. [Google Scholar] [CrossRef] [PubMed]
- Saijo, S.; Ikeda, S.; Yamabe, K.; Kakuta, S.; Ishigame, H.; Akitsu, A.; Fujikado, N.; Kusaka, T.; Kubo, S.; Chung, S.H.; et al. Dectin-2 recognition of α-mannans and induction of Th17 cell differentiation is essential for host defense against Candida albicans. Immunity 2010, 32, 681–691. [Google Scholar] [CrossRef] [PubMed]
- Guinea, J. Global trends in the distribution of Candida species causing candidemia. Clin. Microbiol. Infect. 2014, 20, 5–10. [Google Scholar] [CrossRef] [PubMed]
- Khatib, R.; Johnson, L.B.; Fakih, M.G.; Riederer, K.; Briski, L. Current trends in candidemia and species distribution among adults: Candida glabrata surpasses C. albicans in diabetic patients and abdominal sources. Mycoses 2016, 59, 781–786. [Google Scholar] [CrossRef]
- Castanheira, M.; Messer, S.A.; Rhomberg, P.R.; Pfaller, M.A. Antifungal susceptibility patterns of a global collection of fungal isolates: Results of the SENTRY Antifungal Surveillance Program (2013). Diagn. Microbiol. Infect. Dis. 2016, 85, 200–204. [Google Scholar] [CrossRef]
- Dujon, B.; Sherman, D.; Fischer, G.; Durrens, P.; Casaregola, S.; Lafontaine, I.; De Montigny, J.; Marck, C.; Neuvéglise, C.; Talla, E.; et al. Genome evolution in yeasts. Nature 2004, 430, 35–44. [Google Scholar] [CrossRef]
- Rodrigues, C.F.; Silva, S.; Henriques, M. Candida glabrata: A review of its features and resistance. Eur. J. Clin. Microbiol. Infect. Dis. 2014, 33, 673–688. [Google Scholar] [CrossRef]
- Gabaldon, T.; Carrete, L. The birth of a deadly yeast: Tracing the evolutionary emergence of virulence traits in Candida glabrata. FEMS Yeast Res. 2016, 16, fov110. [Google Scholar] [CrossRef]
- Kumar, K.; Askari, F.; Sahu, M.; Kaur, R. Candida glabrata: A lot more than meets the eye. Microorganisms 2019, 7, 39. [Google Scholar] [CrossRef]
- Silva, S.; Negri, M.; Henriques, M.; Oliveira, R.; Williams, D.W.; Azeredo, J. Candida glabrata, Candida parapsilosis and Candida tropicalis: Biology, epidemiology, pathogenicity and antifungal resistance. FEMS Microbiol. Rev. 2012, 36, 288–305. [Google Scholar] [CrossRef] [PubMed]
- Pfaller, M.A.; Pappas, P.G.; Wingard, J.R. Invasive fungal Pathogens: Current epidemiological. Trends Clin. Infect. Dis. 2006, 43, S3–S14. [Google Scholar] [CrossRef]
- Kaur, R.; Ma, B.; Cormack, B.P. A family of glycosylphosphatidylinositol-linked aspartyl proteases is required for virulence of Candida glabrata. Proc. Natl. Acad. Sci. USA 2007, 104, 2638–7633. [Google Scholar] [CrossRef] [PubMed]
- Seider, K.; Brunke, S.; Schild, L.; Jablonowski, N.; Wilson, D.; Majer, O.; Barz, D.; Haas, A.; Kuchler, K.; Schaller, M.; et al. The facultative intracellular pathogen Candida glabrata subverts macrophage cytokine production and phagolysosome maturation. J. Immunol. 2011, 187, 3072–3086. [Google Scholar] [CrossRef] [PubMed]
- Rasheed, M.; Battu, A.; Kaur, R. Aspartyl proteases in Candida glabrata are required for suppression of the host innate immune response. J. Biol. Chem. 2018, 293, 6410–6433. [Google Scholar] [CrossRef] [PubMed]
- Sorgo, A.G.; Heilmann, C.J.; Dekker, H.L.; Brul, S.; de Koster, C.G.; Klis, F.M. Mass spectrometric analysis of the secretome of Candida albicans. Yeast 2010, 27, 661–672. [Google Scholar] [CrossRef] [PubMed]
- Lee, P.Y.; Gam, L.H.; Yong, V.C.; Rosli, R.; Ng, K.P.; Chong, P.P. Immunoproteomic analysis of antibody response to cell wall-associated proteins of Candida tropicalis. J. Appl. Microbiol. 2014, 117, 854–865. [Google Scholar] [CrossRef]
- El Khoury, P.; Awad, A.; Wex, B.; Khalaf, R.A. Proteomic analysis of a Candida albicans pir32 null strain reveals proteins involved in adhesion, filamentation and virulence. PLoS ONE 2018, 13, e0194403. [Google Scholar] [CrossRef]
- Awad, A.; El Khoury, P.; Geukgeuzian, G.; Khalaf, R.A. Cell wall proteome profiling of a Candida albicans fluconazole-resistant strain from a Lebanese hospital patient using Tandem Mass Spectrometry—A pilot study. Microorganisms 2021, 9, 1161. [Google Scholar] [CrossRef]
- De Groot, P.W.; Kraneveld, E.A.; Yin, Q.Y.; Dekker, H.L.; Groß, U.; Crielaard, W.; de Koster, C.G.; Bader, O.; Klis, F.M.; Weig, M. The cell wall of the human pathogen Candida glabrata: Differential incorporation of novel adhesin-like wall proteins. Eukar Cell 2008, 7, 1951–1964. [Google Scholar] [CrossRef]
- Rasheed, M.; Kumar, N.; Kaur, R. Global secretome characterization of the pathogenic yeast Candida glabrata. J. Proteome Res. 2020, 19, 49–63. [Google Scholar] [CrossRef] [PubMed]
- Thomas, D.P.; Viudes, A.; Monteagudo, C.; Lazzell, A.L.; Saville, S.P.; Lopez-Ribot, J.L. A proteomic-based approach for the identification of Candida albicans protein components present in a subunit vaccine that protects against disseminated candidiasis. Proteomics 2006, 6, 6033–6041. [Google Scholar] [CrossRef] [PubMed]
- Klis, F.M.; Sosinska, G.J.; de Groot, P.W.; Brul, S. Covalently linked cell wall proteins of Candida albicans and their role in fitness and virulence. FEMS Yeast Res. 2009, 9, 1013–1028. [Google Scholar] [CrossRef] [PubMed]
- Luo, G.; Ibrahim, A.S.; Spellberg, B.; Nobile, C.J.; Mitchell, A.P.; Fu, Y. Candida albicans Hyr1p confers resistance to neutrophil killing and is a potential vaccine target. J. Infect. Dis. 2010, 201, 1718–1728. [Google Scholar] [CrossRef]
- Singh, S.; Uppuluri, P.; Mamouei, Z.; Alqarihi, A.; Elhassan, H.; French, S.; Lockhart, S.R.; Chiller, T.; Edwards, J.E., Jr.; Ibrahim, A.S. The NDV-3A vaccine protects mice from multidrug resistant Candida auris infection. PLoS Pathog. 2019, 15, e1007460. [Google Scholar] [CrossRef]
- Tarang, S.; Kesherwani, V.; LaTendresse, B.; Lindgren, L.; Rocha-Sanchez, S.M.; Weston, M.D. In silico design of a multivalent vaccine against Candida albicans. Sci. Rep. 2020, 10, 1066. [Google Scholar] [CrossRef]
- CLSI. Reference Method for Broth Dilution Antifungal Susceptibility Testing of Yeast, 4th ed.; CLSI Standard M27; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2017. [Google Scholar]
- Srivastava, V.; Wani, M.Y.; Al-Bogami, A.S.; Ahmad, A. Piperidine based 1,2,3-triazolylacetamide derivatives induce cell cycle arrest and apoptotic cell death in Candida auris. J. Adv. Res. 2020, 29, 121–135. [Google Scholar] [CrossRef]
- Patel, M.; Gulube, Z.; Dutton, M. The effect of Dodonaea viscosavar. Angusti folia on Candida albicans proteinase and phospholipase production and adherence to oral epithelial cells. J. Ethnopharmacol. 2009, 24, 562–565. [Google Scholar] [CrossRef]
- Staniszewska, M.; Bondaryk, M.; Swoboda-Kopec, E.; Siennicka, K.; Sygitowicz, G.; Kurzatkowski, W. Candida albicans morphologies revealed by scanning electron microscopy analysis. Braz. J. Microbiol. 2013, 44, 813–821. [Google Scholar] [CrossRef] [PubMed]
- Yousuf, S.; Ahmad, A.; Khan, A.; Manzoor, N.; Khan, L.A. Effect of garlic-derived allyl sulphides on morphogenesis and hydrolytic enzyme secretion in Candida albicans. Med. Mycol. 2011, 49, 444–448. [Google Scholar] [CrossRef]
- Wang, J.; Shi, L.; He, X.; Lu, L.; Li, X.; Chen, B. Comparative secretome analysis reveals perturbation of host secretion pathways by a hypovirus. Sci. Rep. 2016, 6, 34308. [Google Scholar] [CrossRef] [PubMed]
- Shevchenko, A.; Tomas, H.; Havli, J.; Olsen, J.V.; Mann, M. In-gel digestion for mass spectrometric characterization of proteins and proteomes. Nat. Protoc. 2006, 1, 2856–2860. [Google Scholar] [CrossRef] [PubMed]
- Skrzypek, M.S.; Binkley, J.; Binkley, G.; Miyasato, S.R.; Simison, M.; Sherlock, G. The Candida Genome Database (CGD): Incorporation of assembly 22, systematic identifiers and visualization of high throughput sequencing data. Nucleic Acids Res. 2017, 45, D592–D596. [Google Scholar] [CrossRef]
- Doytchinova, I.A.; Flower, D.R. VaxiJen: A server for prediction of protective antigens, tumour antigens and subunit vaccines. BMC Bioinform. 2007, 8, 4. [Google Scholar] [CrossRef] [PubMed]
- Almagro Armenteros, J.J.; Tsirigos, K.D.; Sønderby, C.K.; Petersen, T.N.; Winther, O.; Brunak, S.; von Heijne, G.; Nielsen, H. Signal P5.0 improves signal peptide predictions using deep neural networks. Nat. Biotechnol. 2019, 37, 420–423. [Google Scholar] [CrossRef]
- Almagro Armenteros, J.J.; Sonderby, C.K.; Sonderby, S.K.; Nielsen, H.; Winther, O. DeepLoc: Prediction of protein subcellular localization using deep learning. Bioinformatics 2017, 33, 3387–3395. [Google Scholar] [CrossRef] [PubMed]
- Erum, R.; Samad, F.; Khan, A.; Kazmi, S.U. A comparative study on production of extracellular hydrolytic enzymes of Candida species isolated from patients with surgical site infection and from healthy individuals and their co-relation with antifungal drug resistance. BMC Microbiol. 2020, 20, 368. [Google Scholar] [CrossRef]
- Hruskova-Heidingsfeldova, O. Secreted proteins of Candida albicans. Front. Biosci. 2008, 13, 7227–7242. [Google Scholar] [CrossRef]
- Ene, I.V.; Heilmann, C.J.; Sorgo, A.G.; Walker, L.A.; de Koster, C.G.; Munro, C.A. Carbon source-induced reprogramming of the cell wall proteome and secretome modulates the adherence and drug resistance of the fungal pathogen Candida albicans. Proteomics 2012, 12, 3164–3179. [Google Scholar] [CrossRef]
- Sorgo, A.G.; Heilmann, C.J.; Brul, S.; de Koster, C.G.; Klis, F.M. Beyond the wall: Candida albicans secret(e)s to survive. FEMS Microbiol. Lett. 2013, 338, 10–17. [Google Scholar] [CrossRef]
- Gil-Bona, A.; Llama-Palacios, A.; Parra, C.M.; Vivanco, F.; Nombela, C.; Monteoliva, L.; Gil, C. Proteomics unravels extracellular vesicles as carriers of classical cytoplasmic proteins in Candida albicans. J. Proteome Res. 2015, 14, 142–153. [Google Scholar] [CrossRef] [PubMed]
- Gil-Bona, A.; Amador-Garcia, A.; Gil, C.; Monteoliva, L. The external face of Candida albicans: A proteomic view of the cell surface and the extracellular environment. J. Proteomics 2018, 30, 70–79. [Google Scholar] [CrossRef] [PubMed]
- Klis, F.M.; Brul, S. Adaptations of the secretome of Candida albicans in response to host-related environmental conditions. Eukaryot Cell 2015, 14, 1165–1172. [Google Scholar] [CrossRef]
- Nombela, C.; Gil, C.; Chaffin, W.L. Non-conventional protein secretion in yeast. Trends Microbiol. 2006, 14, 15–21. [Google Scholar] [CrossRef] [PubMed]
- Satala, D.; Satala, G.; Karkowska-Kuleta, J.; Bukowski, M.; Kluza, A.; Rapala-Kozik, M. Structural insights into the interactions of candidal enolase with human vitronectin, fibronectin and plasminogen. Int. J. Mol. Sci. 2020, 21, 7843. [Google Scholar] [CrossRef] [PubMed]
- Parnanen, P.; Sorsa, T.; Tervahartiala, T.; Nikula-Ijas, P. Isolation, characterization and regulation of moonlighting proteases from Candida glabrata cell wall. Microb. Pathog. 2020, 149, 104547. [Google Scholar] [CrossRef] [PubMed]
- Pitarch, A.; Abian, J.; Carrascal, M.; Sanchez, M.; Nombela, C.; Gil, C. Proteomics-based identification of novel Candida albicans antigens for diagnosis of systemic candidiasis in patients with underlying hematological malignancies. Proteomics 2004, 4, 3084–3106. [Google Scholar] [CrossRef]
- Pitarch, A.; Jiménez, A.; Nombela, C.; Gil, C. Decoding serological response to Candida cell wall immunome into novel diagnostic, prognostic, and therapeutic candidates for systemic candidiasis by proteomic and bioinformatic analyses. Mol. Cell Proteomics 2006, 5, 79–96. [Google Scholar] [CrossRef]
- Huertas, B.; Prieto, D.; Pitarch, A.; Gil, C.; Pla, J.; Diez-Orejas, R. Serum antibody profile during colonization of the mouse gut by Candida albicans: Relevance for protection during systemic infection. J. Proteome Res. 2017, 16, 335–345. [Google Scholar] [CrossRef]
- Vaz, C.; Pitarch, A.; Gómez-Molero, E.; Amador-García, A.; Weig, M.; Bader, O.; Monteoliva, L.; Gil, C. Mass spectrometry-based proteomic and immunoproteomic analyses of the Candida albicans hyphal secretome reveal diagnostic biomarker candidates for invasive candidiasis. J. Fungi 2021, 7, 501. [Google Scholar] [CrossRef]
- Kim, S.W.; Joo, Y.J.; Kim, J. Asc1p, a ribosomal protein, plays a pivotal role in cellular adhesion and virulence in Candida albicans. J. Microbiol. 2010, 48, 842–848. [Google Scholar] [CrossRef] [PubMed]
- Juchimiuk, M.; Kruszewska, J.; Palamarczyk, G. Dolichol phosphate mannose synthase from the pathogenic yeast Candida albicans is a multimeric enzyme. Biochim. Biophys. Acta 2015, 1850, 2265–2275. [Google Scholar] [CrossRef] [PubMed]
- Anderson, R.M.; Bitterman, K.J.; Wood, J.G.; Medvedik, O.; Sinclair, D.A. Nicotinamide and PNC1 govern lifespan extension by calorie restriction in Saccharomyces cerevisiae. Nature 2003, 423, 181–185. [Google Scholar] [CrossRef] [PubMed]
- Mendelsohn Endelsohn, B.A.; Li, A.M.; Vargas, C.A.; Riehman, K.; Watson, A.; Fridovich-Keil, J.L. Genetic and biochemical interactions between SCP160 and EAP1 in yeast. Nucleic Acids Res. 2003, 31, 5838–5847. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Gil-Bona, A.; Parra-Giraldo, C.M.; Hernaez, M.L.; Reales-Calderon, J.A.; Solis, N.V.; Filler, S.G.; Monteoliva, L.; Gil, C. Candida albicans cell shaving uncovers new proteins involved in cell wall integrity, yeast to hypha transition, stress response and host-pathogen interaction. J. Proteomics 2015, 127, 340–351. [Google Scholar] [CrossRef]
- Hirayama, T.; Miyazaki, T.; Ito, Y.; Wakayama, M.; Shibuya, K.; Yamashita, K.; Takazono, T.; Saijo, T.; Shimamura, S.; Yamamoto, K.; et al. Virulence assessment of six major pathogenic Candida species in the mouse model of invasive candidiasis caused by fungal translocation. Sci. Rep. 2020, 10, 3814. [Google Scholar] [CrossRef] [PubMed]
- Kasper, L.; Seider, K.; Hube, B. Intracellular survival of Candida glabrata in macrophages: Immune evasion and persistence. FEMS Yeast Res. 2015, fov042. [Google Scholar] [CrossRef]
- Duggan, S.; Essig, F.; Hünniger, K.; Mokhtari, Z.; Bauer, L.; Lehnert, T.; Brandes, S.; Häder, A.; Jacobsen, I.D.; Martin, R.; et al. Neutrophil activation by Candida glabrata but not Candida albicans promotes fungal uptake by monocytes. Cell Microbiol. 2015, 17, 1259–1276. [Google Scholar] [CrossRef]
- Hassan, Y.; Chew, S.Y.; Than, L.T.L. Candida glabrata: Pathogenicity and resistance mechanisms for adaptation and survival. J. Fungi 2021, 7, 667. [Google Scholar] [CrossRef]
- Srikantha, T.; Daniels, K.J.; Wu, W.; Lockhart, S.R.; Yi, S.; Sahni, N.; Ma, N.; Soll, D.R. Dark brown is the more virulent of the switch phenotypes of Candida glabrata. Microbiology 2008, 154, 3309–3318. [Google Scholar] [CrossRef]
- Jacobsen, I.D.; Brunke, S.; Seider, K.; Schwarzmuller, T.; Firon, A.; d’Enfert, C.; Kuchler, K.; Hube, B. Candida glabrata persistence in mice does not depend on host immunosuppression and is unaffected by fungal amino acid auxotrophy. Infect. Immun. 2010, 78, 1066–1077. [Google Scholar] [CrossRef] [PubMed]




| Species | MIC/MFC (µg/mL) | |||
|---|---|---|---|---|
| Am B | CAS | |||
| MIC | MFC | MIC | MFC | |
| C. glabrata ATCC2950 | 1 | 2.0 | 0.5 | 2.0 |
| C. glabrata 3411 | 0.5 | 1.0 | 0.25 | 1.0 |
| C. glabrata 3469 | 0.125 | 0.5 | 0.25 | 1.0 |
| C. glabrata 3471 | 0.125 | 0.25 | 0.25 | 1.0 |
| Sr. No. | C. glabrata ID | Common Name | Subcellular Localization | Soluble/Membrane | Secretory Peptide |
|---|---|---|---|---|---|
| 1. | CAGL0A01716g | Pnc1 | Cyto | Sol | No |
| 2. | CAGL0A04521g | Rps8A | Cyto | Sol | No |
| 3. | CAGL0D00858g | Rps29B | Cyto | Sol | No |
| 4. | CAGL0D02090g | Asc1 | Nuc | Sol | No |
| 5. | CAGL0E02739g | SGTA_dimer domain-containing protein | Cyto | Sol | No |
| 6. | CAGL0F02937g | Rpl12B | Cyto | Sol | No |
| 7. | CAGL0F04477g | Proteasome subunit beta type-6 | Cyto | Sol | No |
| 8. | CAGL0G00220g | Bgl2 | ExC | Sol | Yes |
| 9. | CAGL0G01078g | Rpl26A | Cyto | Sol | No |
| 10. | CAGL0G05357g | NAD(P)H-hydrate epimerase | Cyto | Sol | No |
| 11. | CAGL0G09955g | Dolichol-phosphate mannosyltransferase subunit 1 (DPMS) | ER | Memb | No |
| 12. | CAGL0H01705g | Fpr2 | ER | Sol | Yes |
| 13. | CAGL0H02057g | Gar1 | Nuc | Sol | No |
| 14. | CAGL0H02321g | Coupling of ubiquitin conjugation to ER degradation protein 1 | ER | Memb | No |
| 15. | CAGL0H04521g | Rpl32 | Cyto | Sol | No |
| 16. | CAGL0H07023g | ATP synthase subunit e, mitochondrial | Mito | Memb | No |
| 17. | CAGL0H08541g | Nhp6B | Nuc | Sol | No |
| 18. | CAGL0I00594g | Gsp1 | Cyto | Sol | No |
| 19. | CAGL0I07969g | ATP synthase subunit K, mitochondrial | Mito | Memb | No |
| 20. | CAGL0J01463g | Cwp1 | ExC | Sol | Yes |
| 21. | CAGL0J02508g | Awp1 | ExC | Memb | Yes |
| 22. | CAGL0J03234g | Rps24A | Cyto | Sol | No |
| 23. | CAGL0J07678g | Sui1 | Cyto | Sol | No |
| 24. | CAGL0K06281g | Guk1 | Cyto | Sol | No |
| 25. | CAGL0K07744g | Nudix hydrolase domain-containing protein | Mito | Sol | No |
| 26. | CAGL0K08382g | Rps21A | Cyto | Sol | No |
| 27. | CAGL0K11572g | Sba1 | Nuc | Sol | No |
| 28. | CAGL0K12826g | Rpl10 | Cyto | Sol | No |
| 29. | CAGL0M02497g | Rpl33A | Cyto | Sol | No |
| 30. | CAGL0M03223g | Protein SCP160 | Cyto | Sol | No |
| 31. | CAGL0M04983g | Mbf1 | Nuc | Sol | No |
| 32. | CAGL0M12595g | Actin-related protein 2/3 complex subunit 5 | Cyto | Sol | No |
| 33. | CAGL0M13277g | Ynk1 | Cyto | Sol | No |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kamli, M.R.; Sabir, J.S.M.; Malik, M.A.; Ahmad, A. Characterization of the Secretome of Pathogenic Candida glabrata and Their Effectiveness against Systemic Candidiasis in BALB/c Mice for Vaccine Development. Pharmaceutics 2022, 14, 1989. https://doi.org/10.3390/pharmaceutics14101989
Kamli MR, Sabir JSM, Malik MA, Ahmad A. Characterization of the Secretome of Pathogenic Candida glabrata and Their Effectiveness against Systemic Candidiasis in BALB/c Mice for Vaccine Development. Pharmaceutics. 2022; 14(10):1989. https://doi.org/10.3390/pharmaceutics14101989
Chicago/Turabian StyleKamli, Majid Rasool, Jamal S. M. Sabir, Maqsood Ahmad Malik, and Aijaz Ahmad. 2022. "Characterization of the Secretome of Pathogenic Candida glabrata and Their Effectiveness against Systemic Candidiasis in BALB/c Mice for Vaccine Development" Pharmaceutics 14, no. 10: 1989. https://doi.org/10.3390/pharmaceutics14101989
APA StyleKamli, M. R., Sabir, J. S. M., Malik, M. A., & Ahmad, A. (2022). Characterization of the Secretome of Pathogenic Candida glabrata and Their Effectiveness against Systemic Candidiasis in BALB/c Mice for Vaccine Development. Pharmaceutics, 14(10), 1989. https://doi.org/10.3390/pharmaceutics14101989







