Multiplex Analysis of CircRNAs from Plasma Extracellular Vesicle-Enriched Samples for the Detection of Early-Stage Non-Small Cell Lung Cancer
Abstract
1. Introduction
2. Materials and Methods
2.1. Patient Samples
2.2. Plasma Processing
2.3. Enrichment of EVs
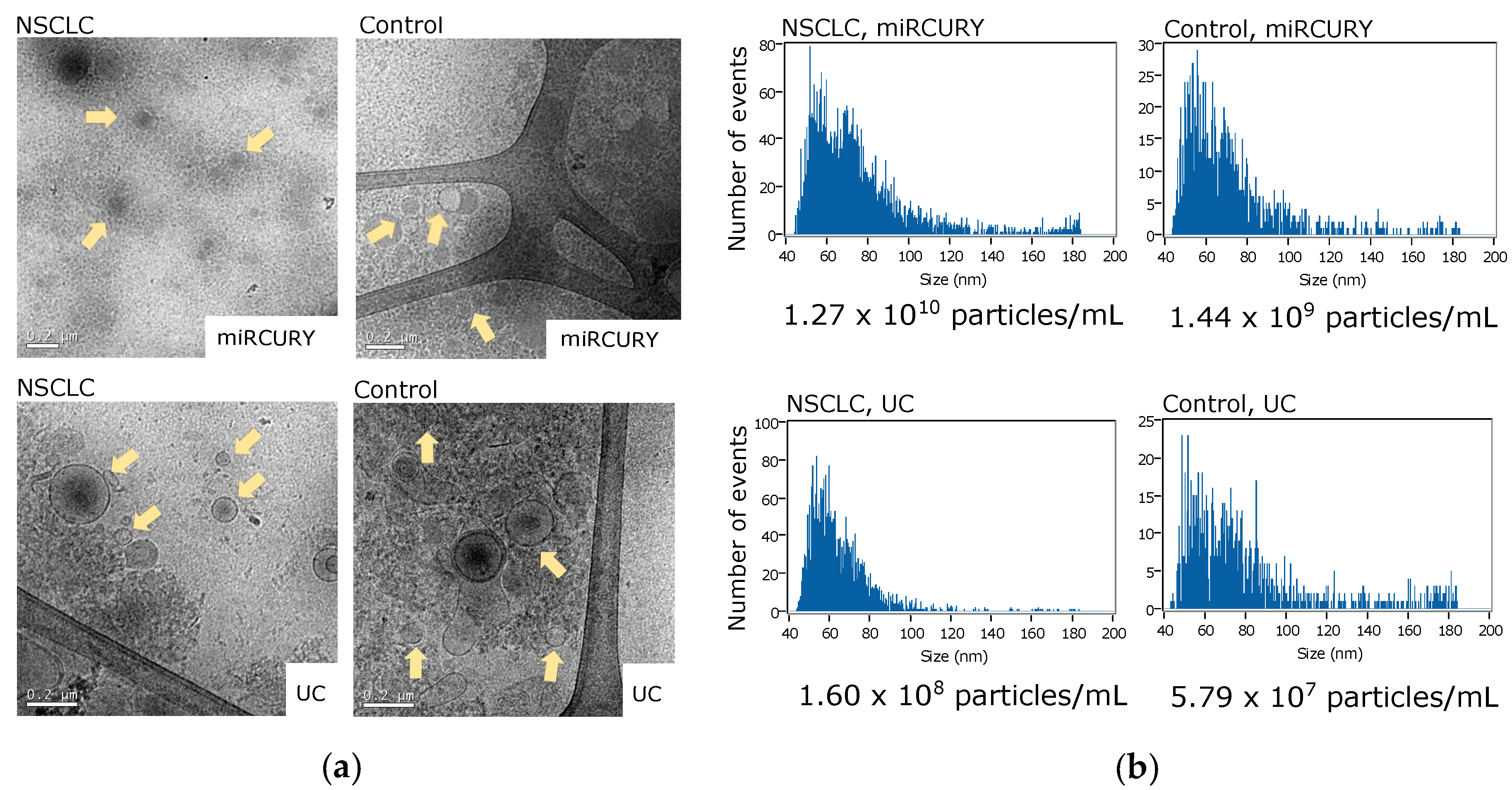
2.4. Transmission Electron Microscopy (TEM)
2.5. Nano-Flow Cytometry Measurements
2.6. RNA Isolation and DNase Treatment
2.7. RT-qPCR and Sanger Sequencing Analysis
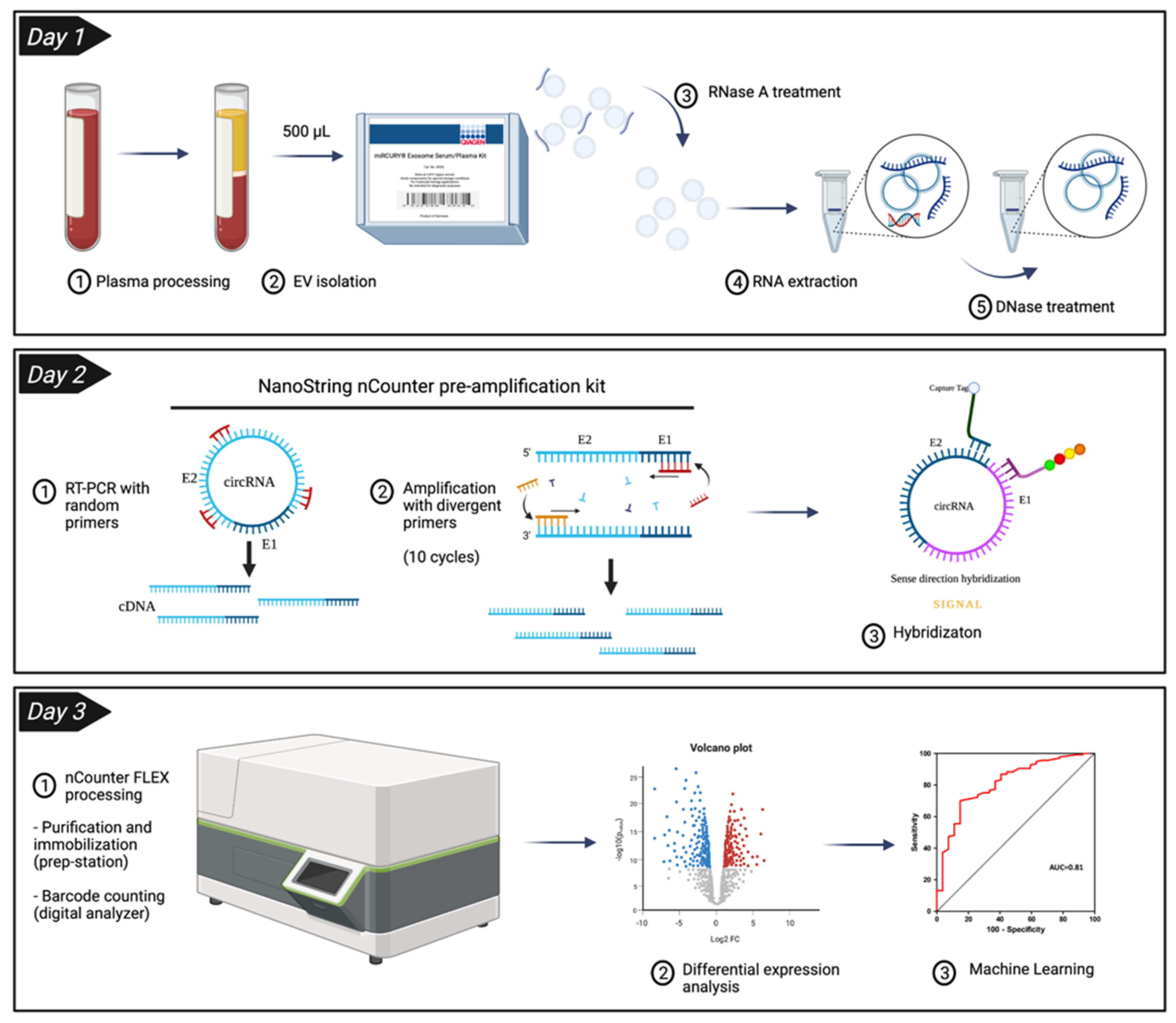
2.8. nCounter Processing
2.9. Differential Expression Analysis
2.10. Data Pre-Processing and Normalization for Signature Development
2.11. Machine Learning (ML) for Signature Development
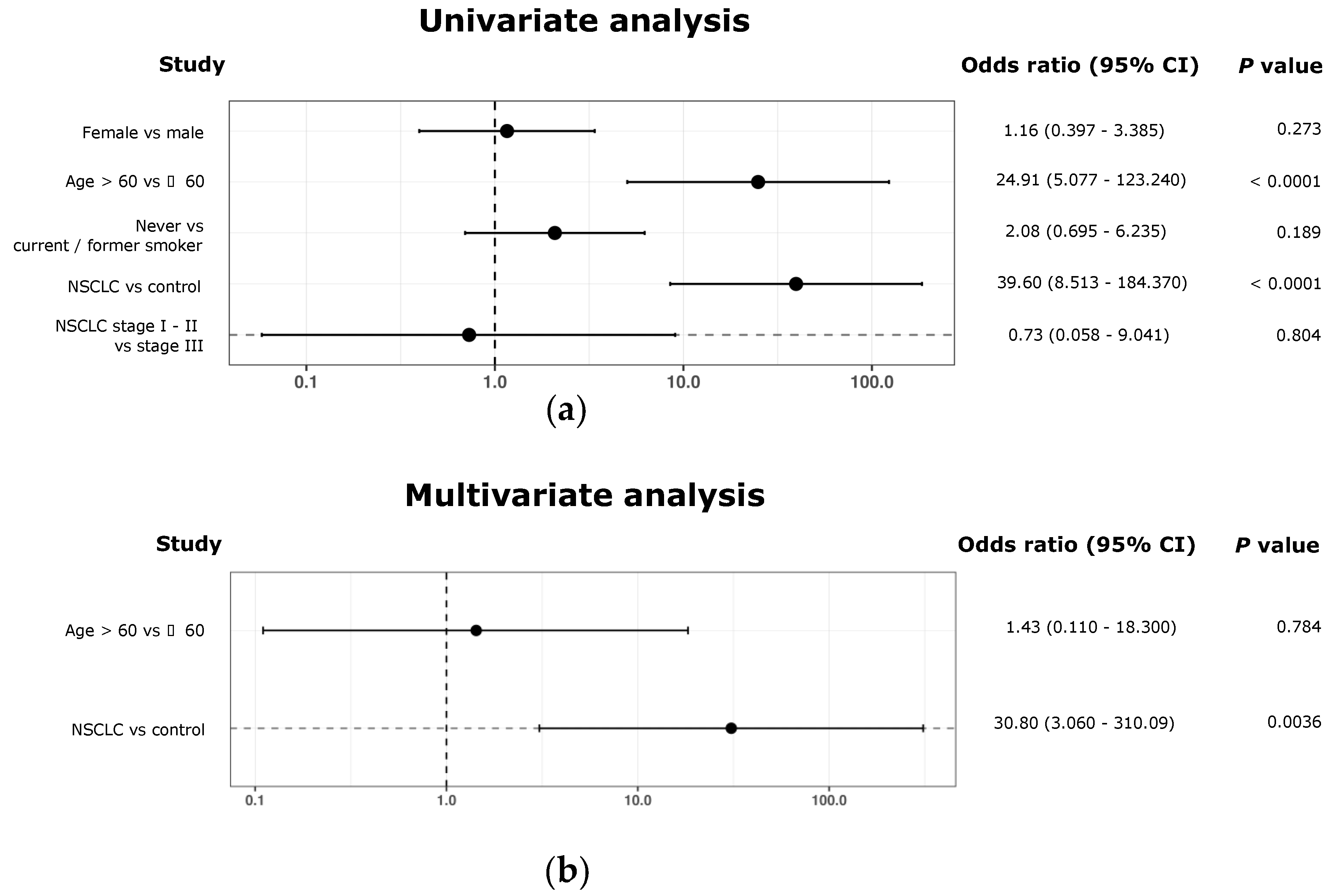
2.12. Univariant and Multivariant Analyses
3. Results
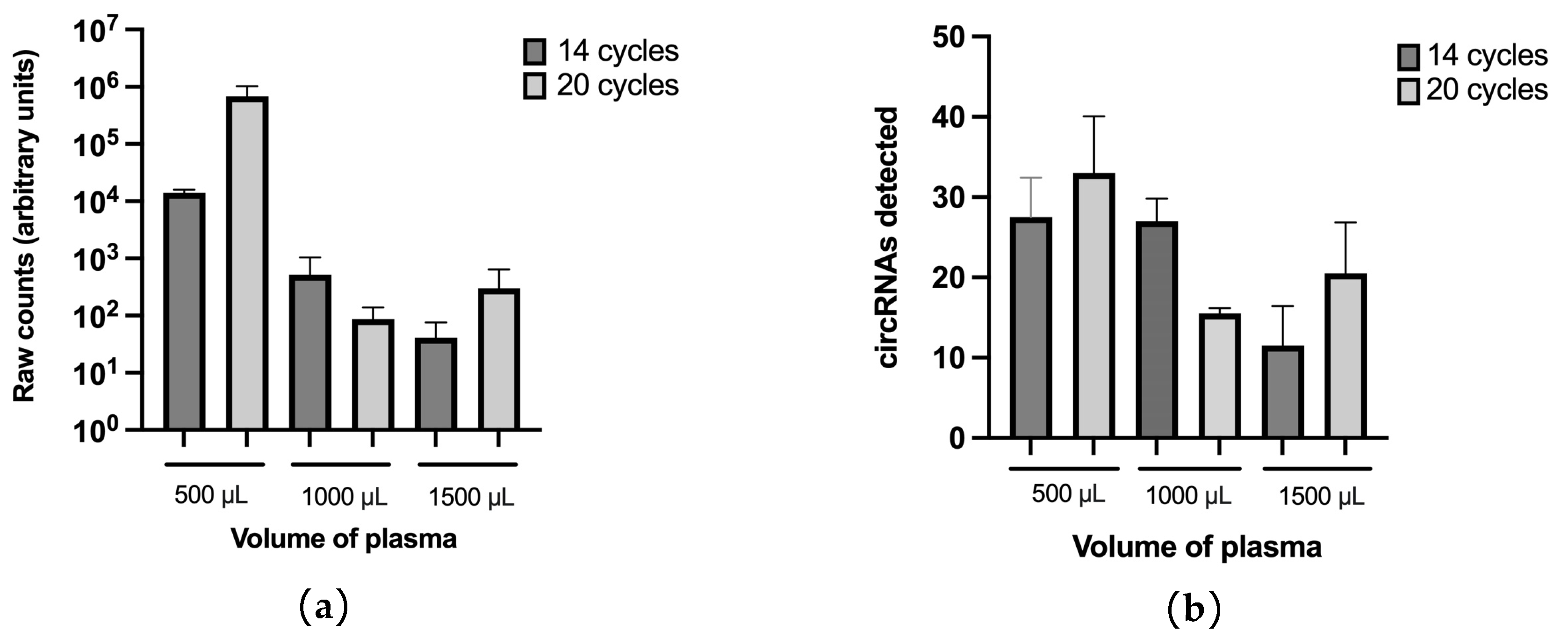
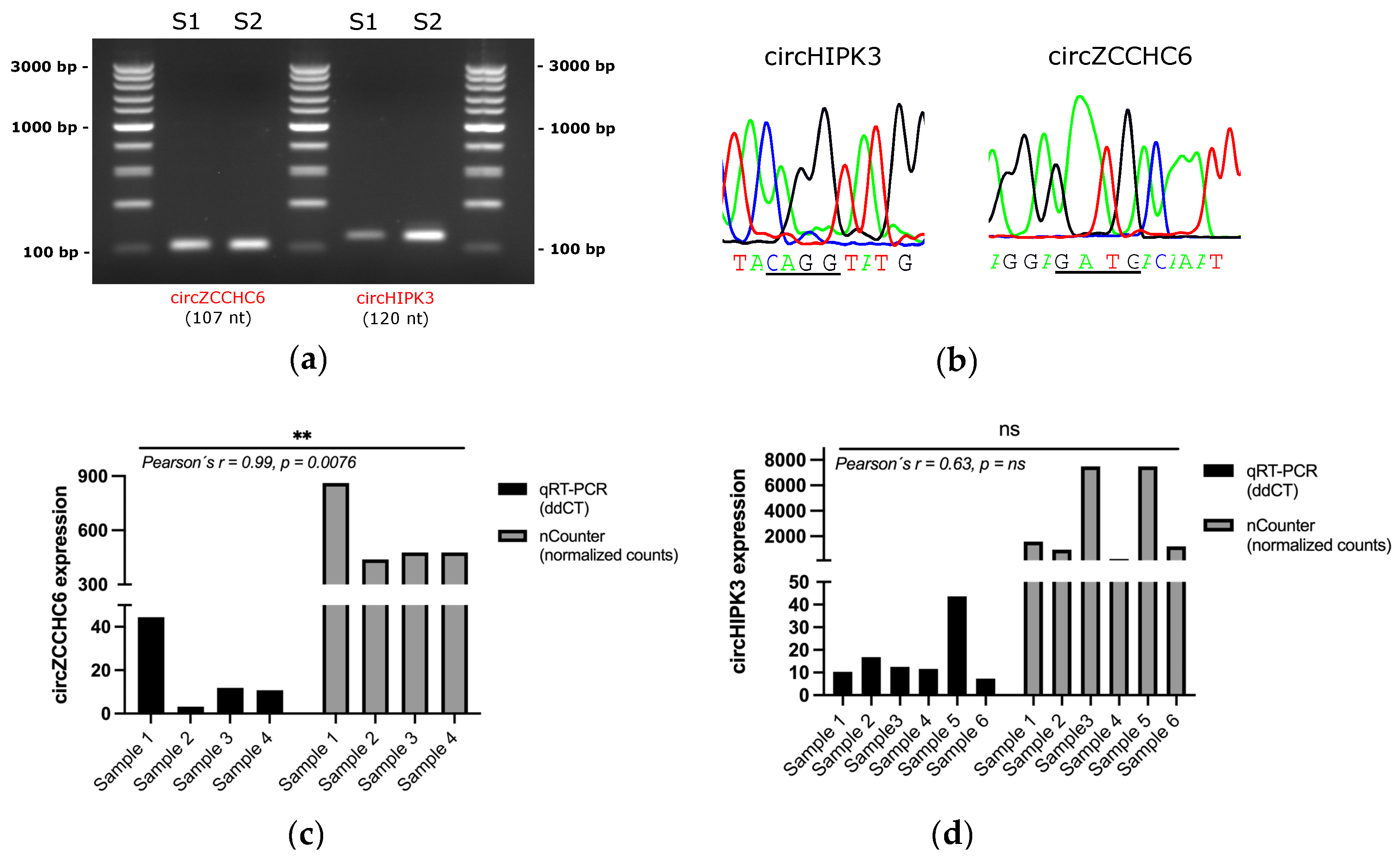
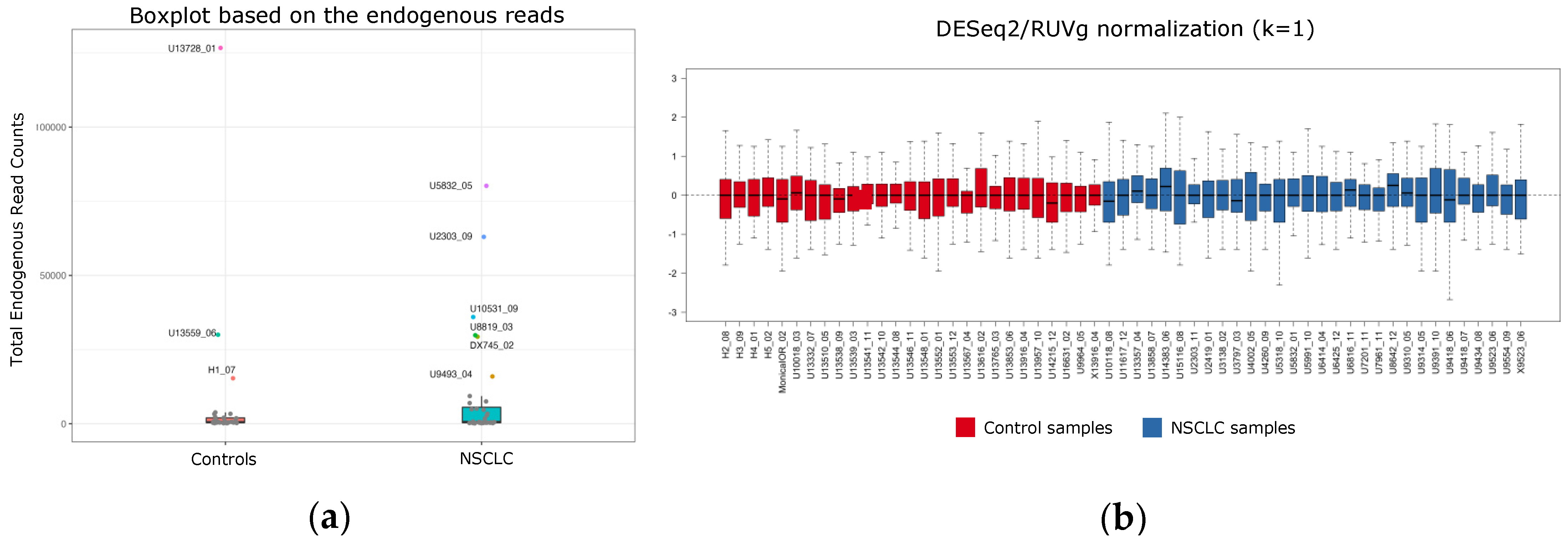
3.1. Enrichment of Plasma EVs and Workflow Development for nCounter CircRNA Analysis
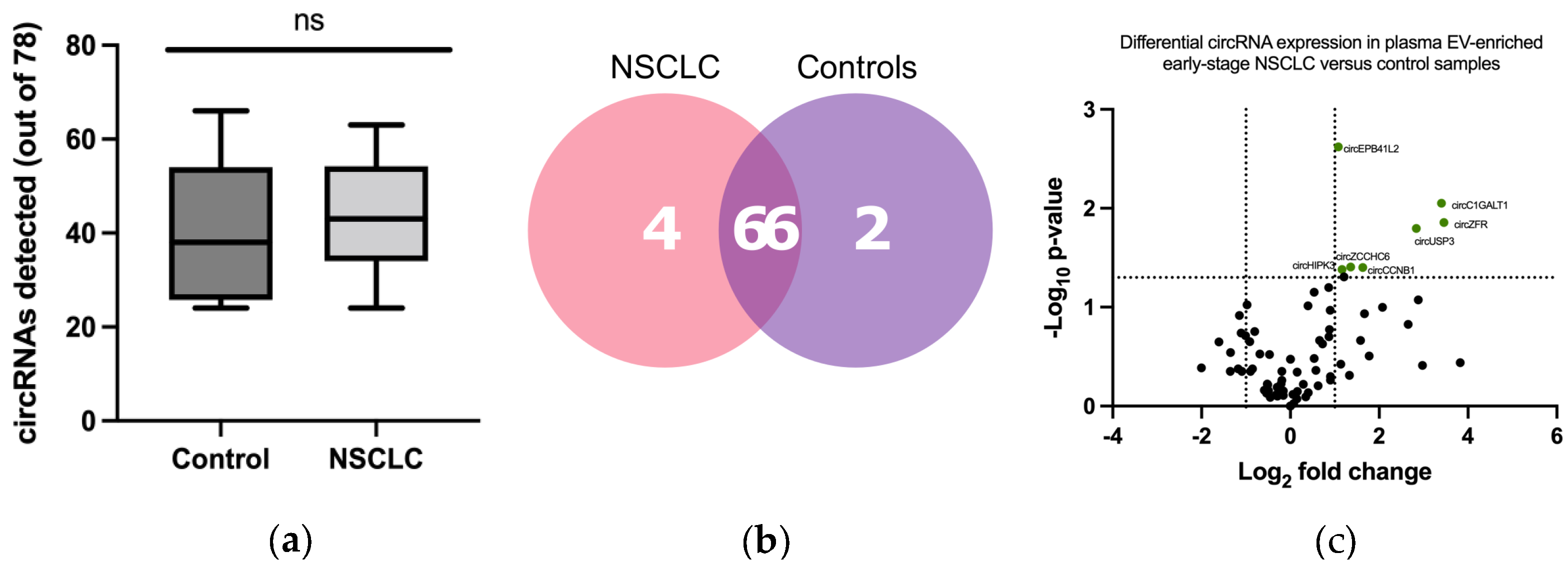
3.2. CircRNA Expression in Plasma EV Samples
3.3. Development of a CircRNA-Signature Associated with Early-Stage NSCLC
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Siegel, R.L.; Miller, K.D.; Fuchs, H.E.; Jemal, A. Cancer statistics, 2022. CA Cancer J. Clin. 2022, 72, 7–33. [Google Scholar] [CrossRef] [PubMed]
- European Union. ECIS—European Cancer Information System. Available online: https://ecis.jrc.ec.europa.eu (accessed on 30 June 2022).
- Crosby, D.; Bhatia, S.; Brindle Kevin, M.; Coussens Lisa, M.; Dive, C.; Emberton, M.; Esener, S.; Fitzgerald Rebecca, C.; Gambhir Sanjiv, S.; Kuhn, P.; et al. Early detection of cancer. Science 2022, 375, eaay9040. [Google Scholar] [CrossRef] [PubMed]
- Perakis, S.; Speicher, M.R. Emerging concepts in liquid biopsies. BMC Med. 2017, 15, 75. [Google Scholar] [CrossRef] [PubMed]
- Lone, S.N.; Nisar, S.; Masoodi, T.; Singh, M.; Rizwan, A.; Hashem, S.; El-Rifai, W.; Bedognetti, D.; Batra, S.K.; Haris, M.; et al. Liquid biopsy: A step closer to transform diagnosis, prognosis and future of cancer treatments. Mol. Cancer 2022, 21, 79. [Google Scholar] [CrossRef] [PubMed]
- Bracht, J.W.P.; Mayo-de-Las-Casas, C.; Berenguer, J.; Karachaliou, N.; Rosell, R. The Present and Future of Liquid Biopsies in Non-Small Cell Lung Cancer: Combining Four Biosources for Diagnosis, Prognosis, Prediction, and Disease Monitoring. Curr. Oncol. Rep. 2018, 20, 70. [Google Scholar] [CrossRef]
- Li, Y.; Zheng, Q.; Bao, C.; Li, S.; Guo, W.; Zhao, J.; Chen, D.; Gu, J.; He, X.; Huang, S. Circular RNA is enriched and stable in exosomes: A promising biomarker for cancer diagnosis. Cell Res. 2015, 25, 981–984. [Google Scholar] [CrossRef]
- Xiao, M.-S.; Wilusz, J.E. An improved method for circular RNA purification using RNase R that efficiently removes linear RNAs containing G-quadruplexes or structured 3′ ends. Nucleic Acids Res. 2019, 47, 8755–8769. [Google Scholar] [CrossRef]
- Hansen, T.B.; Jensen, T.I.; Clausen, B.H.; Bramsen, J.B.; Finsen, B.; Damgaard, C.K.; Kjems, J. Natural RNA circles function as efficient microRNA sponges. Nature 2013, 495, 384–388. [Google Scholar] [CrossRef]
- Memczak, S.; Jens, M.; Elefsinioti, A.; Torti, F.; Krueger, J.; Rybak, A.; Maier, L.; Mackowiak, S.D.; Gregersen, L.H.; Munschauer, M.; et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature 2013, 495, 333–338. [Google Scholar] [CrossRef]
- Zhou, R.; Wu, Y.; Wang, W.; Su, W.; Liu, Y.; Wang, Y.; Fan, C.; Li, X.; Li, G.; Li, Y.; et al. Circular RNAs (circRNAs) in cancer. Cancer Lett. 2018, 425, 134–142. [Google Scholar] [CrossRef]
- Pedraz-Valdunciel, C.; Rosell, R. Defining the landscape of circRNAs in non-small cell lung cancer and their potential as liquid biopsy biomarkers: A complete review including current methods. Extracell. Vesicles Circ. Nucleic Acids 2021, 2, 179–201. [Google Scholar] [CrossRef]
- Zhou, Q.; Ju, L.-L.; Ji, X.; Cao, Y.-L.; Shao, J.-G.; Chen, L. Plasma circRNAs as Biomarkers in Cancer. Cancer Manag. Res. 2021, 13, 7325–7337. [Google Scholar] [CrossRef] [PubMed]
- Kulkarni, M.M. Digital Multiplexed Gene Expression Analysis Using the NanoString nCounter System. Curr. Protoc. Mol. Biol. 2011, 94, 25B.10.1–25B.10.17. [Google Scholar] [CrossRef]
- Geiss, G.K.; Bumgarner, R.E.; Birditt, B.; Dahl, T.; Dowidar, N.; Dunaway, D.L.; Fell, H.P.; Ferree, S.; George, R.D.; Grogan, T.; et al. Direct multiplexed measurement of gene expression with color-coded probe pairs. Nat. Biotechnol. 2008, 26, 317–325. [Google Scholar] [CrossRef]
- Warren, S. Simultaneous, Multiplexed Detection of RNA and Protein on the NanoString® nCounter® Platform. In Gene Expression Analysis: Methods and Protocols; Raghavachari, N., Garcia-Reyero, N., Eds.; Springer: New York, NY, USA, 2018; pp. 105–120. [Google Scholar] [CrossRef]
- Giménez-Capitán, A.; Bracht, J.; García, J.J.; Jordana-Ariza, N.; García, B.; Garzón, M.; Mayo-de-las-Casas, C.; Viteri-Ramirez, S.; Martinez-Bueno, A.; Aguilar, A.; et al. Multiplex Detection of Clinically Relevant Mutations in Liquid Biopsies of Cancer Patients Using a Hybridization-Based Platform. Clin. Chem. 2021, 67, 554–563. [Google Scholar] [CrossRef]
- Porras, T.B.; Kaur, P.; Ring, A.; Schechter, N.; Lang, J.E. Challenges in using liquid biopsies for gene expression profiling. Oncotarget 2018, 9, 7036–7053. [Google Scholar] [CrossRef]
- Beck, T.N.; Boumber, Y.A.; Aggarwal, C.; Pei, J.; Thrash-Bingham, C.; Fittipaldi, P.; Vlasenkova, R.; Rao, C.; Borghaei, H.; Cristofanilli, M.; et al. Circulating tumor cell and cell-free RNA capture and expression analysis identify platelet-associated genes in metastatic lung cancer. BMC Cancer 2019, 19, 603. [Google Scholar] [CrossRef]
- Wu, T.-C.; Xu, K.; Martinek, J.; Young, R.R.; Banchereau, R.; George, J.; Turner, J.; Kim, K.I.; Zurawski, S.; Wang, X.; et al. IL1 Receptor Antagonist Controls Transcriptional Signature of Inflammation in Patients with Metastatic Breast Cancer. Cancer Res. 2018, 78, 5243–5258. [Google Scholar] [CrossRef]
- Kossenkov, A.V.; Qureshi, R.; Dawany, N.B.; Wickramasinghe, J.; Liu, Q.; Majumdar, R.S.; Chang, C.; Widura, S.; Kumar, T.; Horng, W.-H.; et al. A Gene Expression Classifier from Whole Blood Distinguishes Benign from Malignant Lung Nodules Detected by Low-Dose CT. Cancer Res. 2019, 79, 263–273. [Google Scholar] [CrossRef]
- Kamyabi, N.; Abbasgholizadeh, R.; Maitra, A.; Ardekani, A.; Biswal, S.L.; Grande-Allen, K.J. Isolation and mutational assessment of pancreatic cancer extracellular vesicles using a microfluidic platform. Biomed. Microdevices 2020, 22, 23. [Google Scholar] [CrossRef]
- Garcia-Contreras, M.; Shah, S.H.; Tamayo, A.; Robbins, P.D.; Golberg, R.B.; Mendez, A.J.; Ricordi, C. Plasma-derived exosome characterization reveals a distinct microRNA signature in long duration Type 1 diabetes. Sci. Rep. 2017, 7, 5998. [Google Scholar] [CrossRef] [PubMed]
- Vicentini, C.; Calore, F.; Nigita, G.; Fadda, P.; Simbolo, M.; Sperandio, N.; Luchini, C.; Lawlor, R.T.; Croce, C.M.; Corbo, V.; et al. Exosomal miRNA signatures of pancreatic lesions. BMC Gastroenterol. 2020, 20, 137. [Google Scholar] [CrossRef] [PubMed]
- Bracht, J.W.P.; Gimenez-Capitan, A.; Huang, C.-Y.; Potie, N.; Pedraz-Valdunciel, C.; Warren, S.; Rosell, R.; Molina-Vila, M.A. Analysis of extracellular vesicle mRNA derived from plasma using the nCounter platform. Sci. Rep. 2021, 11, 3712. [Google Scholar] [CrossRef]
- Hansen, E.B.; Fredsøe, J.; Okholm, T.L.H.; Ulhøi, B.P.; Klingenberg, S.; Jensen, J.B.; Kjems, J.; Bouchelouche, K.; Borre, M.; Damgaard, C.K.; et al. The transcriptional landscape and biomarker potential of circular RNAs in prostate cancer. Genome Med. 2022, 14, 8. [Google Scholar] [CrossRef]
- Berenguer, J.; Lagerweij, T.; Zhao, X.W.; Dusoswa, S.; van der Stoop, P.; Westerman, B.; Gooijer, M.C.d.; Zoetemelk, M.; Zomer, A.; Crommentuijn, M.H.W.; et al. Glycosylated extracellular vesicles released by glioblastoma cells are decorated by CCL18 allowing for cellular uptake via chemokine receptor CCR8. J. Extracell. Vesicles 2018, 7, 1446660. [Google Scholar] [CrossRef] [PubMed]
- Tian, Y.; Gong, M.; Hu, Y.; Liu, H.; Zhang, W.; Zhang, M.; Hu, X.; Aubert, D.; Zhu, S.; Wu, L.; et al. Quality and efficiency assessment of six extracellular vesicle isolation methods by nano-flow cytometry. J. Extracell. Vesicles 2020, 9, 1697028. [Google Scholar] [CrossRef] [PubMed]
- Aguado, C.; Giménez-Capitán, A.; Román, R.; Rodríguez, S.; Jordana-Ariza, N.; Aguilar, A.; Cabrera-Gálvez, C.; Rivas-Corredor, C.; Lianes, P.; Viteri, S.; et al. RNA-Based Multiplexing Assay for Routine Testing of Fusion and Splicing Variants in Cytological Samples of NSCLC Patients. Diagnostics 2020, 11, 15. [Google Scholar] [CrossRef]
- Pedraz-Valdunciel, C.; Giannoukakos, S.; Potie, N.; Giménez-Capitán, A.; Huang, C.-Y.; Hackenberg, M.; Fernandez-Hilario, A.; Bracht, J.; Filipska, M.; Aldeguer, E.; et al. Digital multiplexed analysis of circular RNAs in FFPE and fresh non-small cell lung cancer specimens. Mol. Oncol. 2022, 16, 2367–2383. [Google Scholar] [CrossRef]
- Margolis, L.; Sadovsky, Y. The biology of extracellular vesicles: The known unknowns. PLOS Biol. 2019, 17, e3000363. [Google Scholar] [CrossRef]
- Reclusa, P.; Sirera, R.; Araujo, A.; Giallombardo, M.; Valentino, A.; Sorber, L.; Bazo, I.G.; Pauwels, P.; Rolfo, C. Exosomes genetic cargo in lung cancer: A truly Pandora’s box. Transl. Lung Cancer Res. 2016, 5, 483–491. [Google Scholar] [CrossRef]
- Jeppesen, D.K.; Fenix, A.M.; Franklin, J.L.; Higginbotham, J.N.; Zhang, Q.; Zimmerman, L.J.; Liebler, D.C.; Ping, J.; Liu, Q.; Evans, R.; et al. Reassessment of Exosome Composition. Cell 2019, 177, 428–445.e418. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Zhang, Y.; Zhang, Y.; Zhang, S.; Qiu, L.; Zhuang, Z.; Wei, M.; Deng, X.; Wang, Z.; Han, J. The Key Role of Exosomes on the Pre-metastatic Niche Formation in Tumors. Front. Mol. Biosci. 2021, 8, 703640. [Google Scholar] [CrossRef] [PubMed]
- Costa-Silva, B.; Aiello, N.M.; Ocean, A.J.; Singh, S.; Zhang, H.; Thakur, B.K.; Becker, A.; Hoshino, A.; Mark, M.T.; Molina, H.; et al. Pancreatic cancer exosomes initiate pre-metastatic niche formation in the liver. Nat. Cell Biol. 2015, 17, 816–826. [Google Scholar] [CrossRef]
- Dahl, M.; Daugaard, I.; Andersen, M.S.; Hansen, T.B.; Grønbæk, K.; Kjems, J.; Kristensen, L.S. Enzyme-free digital counting of endogenous circular RNA molecules in B-cell malignancies. Lab. Investig. 2018, 98, 1657–1669. [Google Scholar] [CrossRef]
- Zhang, J.; Zhang, X.; Li, C.; Yue, L.; Ding, N.; Riordan, T.; Yang, L.; Li, Y.; Jen, C.; Lin, S.; et al. Circular RNA profiling provides insights into their subcellular distribution and molecular characteristics in HepG2 cells. RNA Biol. 2019, 16, 220–232. [Google Scholar] [CrossRef] [PubMed]
- Das Mahapatra, K.; Pasquali, L.; Søndergaard, J.N.; Lapins, J.; Nemeth, I.B.; Baltás, E.; Kemény, L.; Homey, B.; Moldovan, L.-I.; Kjems, J.; et al. A comprehensive analysis of coding and non-coding transcriptomic changes in cutaneous squamous cell carcinoma. Sci. Rep. 2020, 10, 3637. [Google Scholar] [CrossRef]
- Moldovan, L.-I.; Hansen, T.B.; Venø, M.T.; Okholm, T.L.H.; Andersen, T.L.; Hager, H.; Iversen, L.; Kjems, J.; Johansen, C.; Kristensen, L.S. High-throughput RNA sequencing from paired lesional- and non-lesional skin reveals major alterations in the psoriasis circRNAome. BMC Med. Genom. 2019, 12, 174. [Google Scholar] [CrossRef]
- Ahmadov, U.; Bendikas, M.M.; Ebbesen, K.K.; Sehested, A.M.; Kjems, J.; Broholm, H.; Kristensen, L.S. Distinct circular RNA expression profiles in pediatric ependymomas. Brain Pathol. 2021, 31, 387–392. [Google Scholar] [CrossRef]
- Pedraz-Valdunciel, C.; Huang, C.; Ito, M.; Bracht, J.; Giménez-Capitán, A.; Aldeguer, E.; Filipska, M.; Xu, W.; Molina-Vila, M.A.; Rosell, R. P65.04 Tracking circRNAs in Lung Adenocarcinoma Samples as Promising Biomarkers for Cancer Detection using the NanoString nCounter®. Thorac. Oncol. 2021, 16, S555–S556. [Google Scholar] [CrossRef]
- Helwa, I.; Cai, J.; Drewry, M.D.; Zimmerman, A.; Dinkins, M.B.; Khaled, M.L.; Seremwe, M.; Dismuke, W.M.; Bieberich, E.; Stamer, W.D.; et al. A Comparative Study of Serum Exosome Isolation Using Differential Ultracentrifugation and Three Commercial Reagents. PLoS ONE 2017, 12, e0170628. [Google Scholar] [CrossRef]
- Melo, S.A.; Luecke, L.B.; Kahlert, C.; Fernandez, A.F.; Gammon, S.T.; Kaye, J.; LeBleu, V.S.; Mittendorf, E.A.; Weitz, J.; Rahbari, N.; et al. Glypican-1 identifies cancer exosomes and detects early pancreatic cancer. Nature 2015, 523, 177–182. [Google Scholar] [CrossRef] [PubMed]
- Fasihi, A.; Soltani, B.M.; Ranjbaran, Z.S.; Bahonar, S.; Norouzi, R.; Nasiri, S. Hsa-miR-942 fingerprint in colorectal cancer through Wnt signaling pathway. Gene 2019, 712, 143958. [Google Scholar] [CrossRef] [PubMed]
- Ge, C.; Wu, S.; Wang, W.; Liu, Z.; Zhang, J.; Wang, Z.; Li, R.; Zhang, Z.; Li, Z.; Dong, S.; et al. miR-942 promotes cancer stem cell-like traits in esophageal squamous cell carcinoma through activation of Wnt/β-catenin signalling pathway. Oncotarget 2015, 6, 10964–10977. [Google Scholar] [CrossRef] [PubMed]
- Lu, H.; Han, X.; Ren, J.; Ren, K.; Li, Z.; Sun, Z. Circular RNA HIPK3 induces cell proliferation and inhibits apoptosis in non-small cell lung cancer through sponging miR-149. Cancer Biol. Ther. 2020, 21, 113–121. [Google Scholar] [CrossRef]
- Chen, X.; Mao, R.; Su, W.; Yang, X.; Geng, Q.; Guo, C.; Wang, Z.; Wang, J.; Kresty, L.A.; Beer, D.G.; et al. Circular RNA circHIPK3 modulates autophagy via MIR124-3p-STAT3-PRKAA/AMPKα signaling in STK11 mutant lung cancer. Autophagy 2020, 16, 659–671. [Google Scholar] [CrossRef]
- Yu, H.; Chen, Y.; Jiang, P. Circular RNA HIPK3 exerts oncogenic properties through suppression of miR-124 in lung cancer. Biochem. Biophys. Res. Commun. 2018, 506, 455–462. [Google Scholar] [CrossRef]
- Guo, Y.; Xue, W.; Sun, S.; Chen, X.; Li, H.; Yan, C. Circular RNA circZCCHC6 contributes to tumorigenesis by regulating LPCAT1 via miR-433-3p in non-small cell lung cancer. Clin. Exp. Med. 2022. [Google Scholar] [CrossRef]
- Wu, R.-R.; Zhong, Q.; Liu, H.-F.; Liu, S.-B. Role of miR-579-3p in the development of squamous cell lung carcinoma and the regulatory mechanisms. Eur. Rev. Med. Pharmacol. Sci. 2019, 23, 9464–9470. [Google Scholar]
- Wei, S.; Zhang, Z.-y.; Fu, S.-l.; Xie, J.-g.; Liu, X.-s.; Xu, Y.-j.; Zhao, J.-p.; Xiong, W.-n. Erratum: Hsa-miR-623 suppresses tumor progression in human lung adenocarcinoma. Cell Death Dis. 2017, 8, e2829. [Google Scholar] [CrossRef]
- Sun, B.; Hua, J.; Cui, H.; Liu, H.; Zhang, K.; Zhou, H. MicroRNA-1197 downregulation inhibits proliferation and migration in human non- small cell lung cancer cells by upregulating HOXC11. Biomed. Pharmacother. 2019, 117, 109041. [Google Scholar] [CrossRef]
- Li, C.-g.; Pu, M.-f.; Li, C.-z.; Gao, M.; Liu, M.-x.; Yu, C.-z.; Yan, H.; Peng, C.; Zhao, Y.; Li, Y.; et al. MicroRNA-1304 suppresses human non-small cell lung cancer cell growth in vitro by targeting heme oxygenase-1. Acta Pharmacol. Sin. 2017, 38, 110–119. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Yang, H.; Xu, Z.; Li, D.; Zhou, M.; Xiao, K.; Shi, Z.; Zhu, L.; Yang, L.; Zhou, R. microRNA-548l is involved in the migration and invasion of non-small cell lung cancer by targeting the AKT1 signaling pathway. J. Cancer Res. Clin. Oncol. 2015, 141, 431–441. [Google Scholar] [CrossRef] [PubMed]
- Liao, Y.; Cao, L.; Wang, F.; Pang, R. miR-605-5p promotes invasion and proliferation by targeting TNFAIP3 in non–small-cell lung cancer. J. Cell. Biochem. 2020, 121, 779–787. [Google Scholar] [CrossRef]
- Wang, C.; Li, S.; Xu, J.; Niu, W. microRNA-935 is reduced in non-small cell lung cancer tissue, is linked to poor outcome, and acts on signal transduction mediator E2F7 and the AKT pathway. Br. J. Biomed. Sci. 2019, 76, 17–23. [Google Scholar] [CrossRef] [PubMed]
- Badillo, S.; Banfai, B.; Birzele, F.; Davydov, I.I.; Hutchinson, L.; Kam-Thong, T.; Siebourg-Polster, J.; Steiert, B.; Zhang, J.D. An Introduction to Machine Learning. Clin. Pharmacol. Ther. 2020, 107, 871–885. [Google Scholar] [CrossRef]
- Hang, D.; Zhou, J.; Qin, N.; Zhou, W.; Ma, H.; Jin, G.; Hu, Z.; Dai, J.; Shen, H. A novel plasma circular RNA circFARSA is a potential biomarker for non-small cell lung cancer. Cancer Med. 2018, 7, 2783–2791. [Google Scholar] [CrossRef] [PubMed]
- Chen, T.; Liu, Y.; Li, C.; Xu, C.; Ding, C.; Chen, J.; Zhao, J. Tumor-derived exosomal circFARSA mediates M2 macrophage polarization via the PTEN/PI3K/AKT pathway to promote non-small cell lung cancer metastasis. Cancer Treat. Res. Commun. 2021, 28, 100412. [Google Scholar] [CrossRef] [PubMed]


| Clinicopathological Characteristics | NSCLC Patients (n = 36) | Non-Cancer Controls (n = 30) |
|---|---|---|
| Gender—no. (%) | ||
| Male | 18 (50.0) | 13 (43.3) |
| Female | 18 (50.0) | 17 (56.7) |
| Age—yr. | ||
| Median | 71.5 | 38 |
| Range | 32–91 | 23–57 |
| Histological type | ||
| Adenocarcinoma | 27 (75.0) | - |
| Squamous carcinoma | 4 (11.1) | - |
| Not information | 5 (13.9) | - |
| Smoking status—no. (%) | ||
| Former- or current smoker | 20 (55.5) | 11 (36.6) |
| Never smoker | 13 (36.2) | 17 (56.7) |
| Not information | 3 (8.3) | 2 (6.7) |
| Tumor stage—no. (%) | ||
| I | 19 (52.8) | - |
| II | 2 (5.5) | - |
| IIIA | 15 (41.7) | - |
| CircRNA | Primers and Probes | Sequence |
|---|---|---|
| circHIPK3 | Forward | 5′CGGCCAGTCATGTATCAAAGAC 3′ |
| Reverse | 5′AAAGGCACTTGACTGAGTTTGATAAA 3′ | |
| Probe | FAM 5′AATCTCGGTACTACAGGTATG 3′ MGB | |
| circZCCHC6 | Forward | 5′AGATGTTGTCGAATTTGTGGAAAA 3′ |
| Reverse | 5′TCTTCTACCATTGATAAAAGCCTTCAT 3′ | |
| Probe | FAM 5′GAGGAGAAATGACAAATT 3′ MGB |
| Model | ETC | RF | KNN |
|---|---|---|---|
| No. concordant samples | 49 | 44 | 30 |
| No. discordant samples | 8 | 13 | 27 |
| AUC ROC | 0.86 | 0.83 | 0.54 |
| Accuracy | 86% | 77% | 53% |
| Sensitivity | 90% (CI = 73.47–97.89%) | 83% (CI = 65.28–94.36%) | 50% CI = 31.30–68.70%) |
| Specificity | 81% (CI = 61.92–93.70%) | 70% (CI = 49.82–86.25%) | 56% CI = 41.83–68.49%) |
| PPV | 84% (CI = 70.81–92.32%) | 76% (CI = 63.10–85.10%) | 56% (CI = 41.83–68.49%) |
| NPV | 88% (CI = 71.18–95.61%) | 79% (CI = 62.20–89.77%) | 50% (CI = 37.95–62.02%) |
| Cohen’s κ | 0.72 (CI = 0.458–0.976) | 0.54 (CI = 0.281–0.798) | 0.06 (CI =−0.202–0.313) |
| Statistic | DF | Value | p-Value |
|---|---|---|---|
| Chi-Square | 1 | 32.245 | <0.0001 |
| Likelihood Ratio Chi-Square | 1 | 41.232 | <0.0001 |
| Parameter | DF | Estimate | Standard Error | Wald Chi-Square | p-Value |
|---|---|---|---|---|---|
| Age | 1 | 0.356 | 1.301 | 0.075 | 0.7840 |
| Cancer status | 1 | 3.427 | 1.178 | 8.462 | 0.0036 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pedraz-Valdunciel, C.; Giannoukakos, S.; Giménez-Capitán, A.; Fortunato, D.; Filipska, M.; Bertran-Alamillo, J.; Bracht, J.W.P.; Drozdowskyj, A.; Valarezo, J.; Zarovni, N.; et al. Multiplex Analysis of CircRNAs from Plasma Extracellular Vesicle-Enriched Samples for the Detection of Early-Stage Non-Small Cell Lung Cancer. Pharmaceutics 2022, 14, 2034. https://doi.org/10.3390/pharmaceutics14102034
Pedraz-Valdunciel C, Giannoukakos S, Giménez-Capitán A, Fortunato D, Filipska M, Bertran-Alamillo J, Bracht JWP, Drozdowskyj A, Valarezo J, Zarovni N, et al. Multiplex Analysis of CircRNAs from Plasma Extracellular Vesicle-Enriched Samples for the Detection of Early-Stage Non-Small Cell Lung Cancer. Pharmaceutics. 2022; 14(10):2034. https://doi.org/10.3390/pharmaceutics14102034
Chicago/Turabian StylePedraz-Valdunciel, Carlos, Stavros Giannoukakos, Ana Giménez-Capitán, Diogo Fortunato, Martyna Filipska, Jordi Bertran-Alamillo, Jillian W. P. Bracht, Ana Drozdowskyj, Joselyn Valarezo, Natasa Zarovni, and et al. 2022. "Multiplex Analysis of CircRNAs from Plasma Extracellular Vesicle-Enriched Samples for the Detection of Early-Stage Non-Small Cell Lung Cancer" Pharmaceutics 14, no. 10: 2034. https://doi.org/10.3390/pharmaceutics14102034
APA StylePedraz-Valdunciel, C., Giannoukakos, S., Giménez-Capitán, A., Fortunato, D., Filipska, M., Bertran-Alamillo, J., Bracht, J. W. P., Drozdowskyj, A., Valarezo, J., Zarovni, N., Fernández-Hilario, A., Hackenberg, M., Aguilar-Hernández, A., Molina-Vila, M. Á., & Rosell, R. (2022). Multiplex Analysis of CircRNAs from Plasma Extracellular Vesicle-Enriched Samples for the Detection of Early-Stage Non-Small Cell Lung Cancer. Pharmaceutics, 14(10), 2034. https://doi.org/10.3390/pharmaceutics14102034











