1. Introduction
Bacteria are essential for human survival, but they can also cause severe and potentially fatal diseases [
1]. One of the greatest achievements in medical history was the discovery of antibiotics and their clinical introduction to treat infections [
2]. Nowadays, however, infections that were once treatable are no longer responsive to many antibiotics, due to the increase in antimicrobial resistance (AMR). Indeed, roughly 700,000 people die every year due to drug-resistant infections, with predictions of up to 10 million deaths per year by 2050 [
3]. The rise in antimicrobial resistant bacteria, especially multidrug-resistant (MDR) bacteria, together with the failure to develop new antibiotics has motivated researcher aimed at the development of novel therapeutic strategies, including those based on new antimicrobial agents [
4,
5,
6,
7], such as antimicrobial peptides (AMPs) [
8,
9]. A number of AMPs are currently in use or under study in clinical trials as a viable option to overcome resistance, either in combination with or as an alternative to existing antibiotics [
10]. AMPs are produced by most living organisms as natural defensive molecules that protect against pathogenic microorganisms [
11]. Despite their small size (2–9 kDa), AMPs are versatile, due to their structural and physicochemical properties. Moreover, the sequence of AMPs facilitates their amphipathic nature [
12,
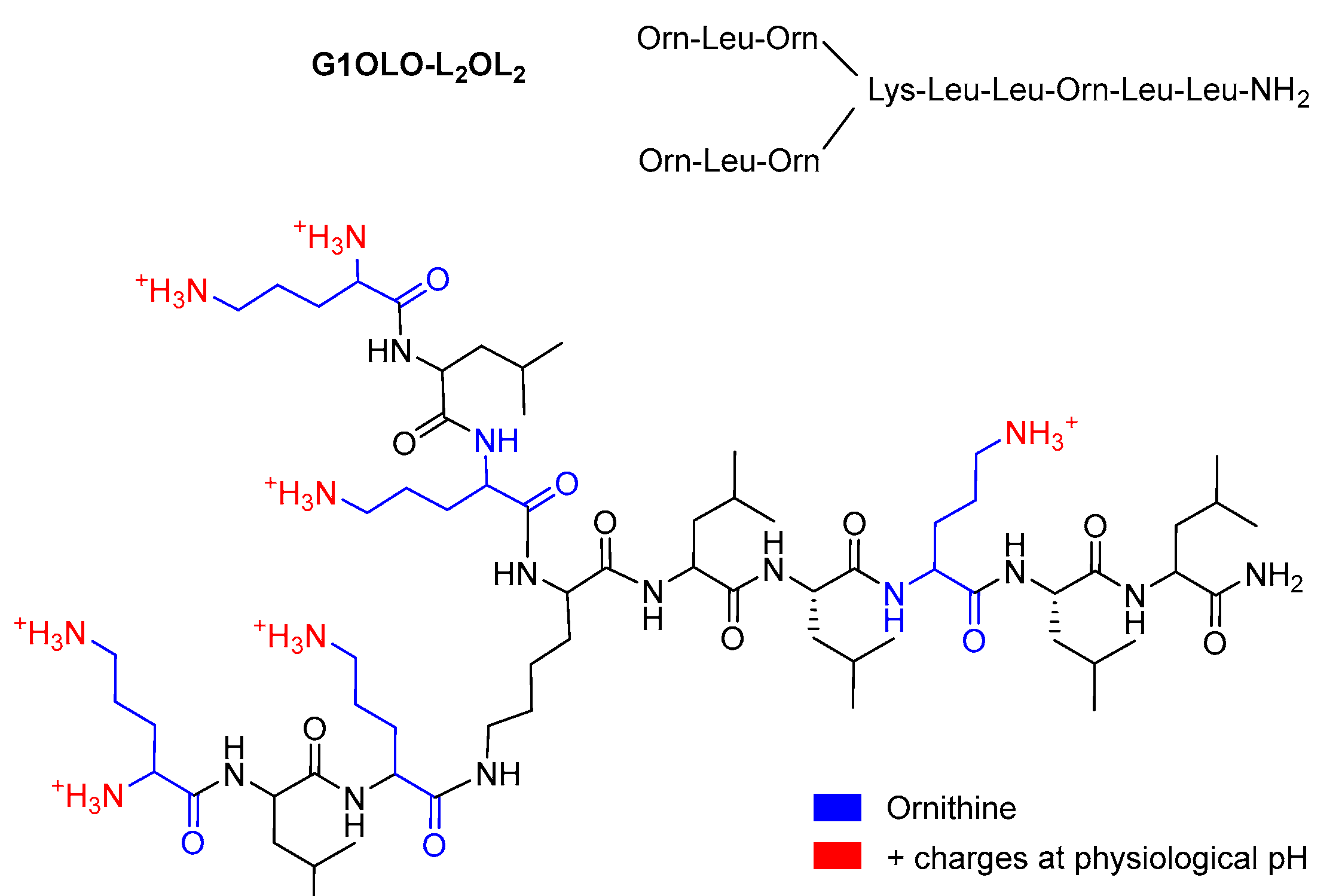
13]. Nearly all AMPs have a positive net charge that allows them to interact selectively with bacterial membranes or other negatively charged structures. In order to obtain new antibacterial molecules, we have investigated branched peptides which has led us to the synthesis of what we call super-cationic peptide dendrimers (SCPDs) that appear to be broad-spectrum antibacterial compounds acting on Gram-positive and Gram-negative bacteria. Nevertheless, time–kill kinetics and growth curves, revealed considerable differences in their action, showing higher activity against Gram-negative bacteria. Among a long series of molecules G1OLO-L
2OL
2 displayed excellent microbiological results. The most prominent characteristic of SCPDs is their number of positive charges.
The bacterial membrane mediates interactions with both other organisms and the environment and is a key factor in the development of drug resistance [
14]. The physical and chemical properties of biological membranes are directly linked to their functions [
15]. In this work, we investigated the membrane interactions of the branched AMP rich in Ornitine (OLO)
2KLLOLL-NH
2 (G1OLO-L
2OL
2), from the recently described super-cationic peptide dendrimers (SCPDS) family [
16]. To do so, we monitored the steady-state fluorescence of the polarity-sensitive probe Laurdan, and the fluorescence anisotropy of liposome-incorporated fluorescent molecules as a function of temperature. Membrane-peptide interactions were followed using atomic force microscopy (AFM).
2. Materials and Methods
2.1. Materials
Lipids 1-palmitoyl-2-oleoyl-
sn-glycero-3-phosphoethanolamine (POPE) and 1-palmitoyl-2-oleoyl-
sn-glycero-3-[phospho-rac-(1-glycerol)] (sodium salt) (POPG) were purchased from Avanti Polar Lipids (Alabaster, AL, USA) and dissolved in a chloroform:methanol (3:1,
v/
v) solution to a final concentration of 1 mg/mL. G1OLO-L
2OL
2 was synthesized as previously described [
16].
Figure 1 shows the chemical structure of the G1OLO-L
2OL
2 peptide. The buffer used throughout the experiments was 10 mM Tris-HCl (pH 7.40) supplemented with 150 mM NaCl, prepared in ultrapure water (Milli-Q reverse osmosis system, 18.2 mΩ·cm resistivity). 1,6-Diphenyl-1,3,5-hexatriene (DPH), 1-(4-trimethylammoniumphenyl)-6-phenyl1,3,5-hexatriene p-toluenesulfonate (TMA-DPH), 6-dodecanoyl-2-dimethylaminonaphthalene (Laurdan), and 1-anilinonaphthalene-8-sulfonic acid (ANS) were purchased from Thermo Fisher Scientific, Inc. (Waltham, MA, USA). G1OLO-L
2OL
2 lyophilized peptide was fully dissolved in ultrapure water supplemented with 5% acetic acid to a final concentration of 5 mM. Volumes from this stock solution were used for all the experiments. Concentration in experiments was 0.005 µM but for AFM of SLB (0.0005 µM).
2.2. Bacterial Strains
Two imipenem-resistant Pseudomonas aeruginosa isolates (846VH, 536SJD) and, as a quality control, one collection strain of P. aeruginosa (ATCC 27853) were studied. For the AFM bacterial studies, Escherichia coli strain ATCC 25922 was also used. All isolates were stored as tryptic soy broth (TSB)-glycerol (15%) stocks at −80 °C and subcultured for use in the experiments.
2.3. Liposome Preparation
Liposomes were prepared as follows: the corresponding volume of each phospholipid was prepared in a conical glass tube as described above. The solvent was then evaporated under a stream of oxygen-free N2 during constant rotation of the tube. The tube was kept under a vacuum overnight and protected from light. The dry lipid film was then resuspended in buffer to a final concentration of 500 µM. Multilamellar vesicles (MLVs) were formed after several freeze–thaw cycles below and above the transition temperature of the lipids (22 °C). The MLVs were then extruded through an Avanti® Mini-extruder (Avanti Polar Lipids, Inc.), using polycarbonate membranes with a pore size of 100 nm.
The mean particle size and polydispersity of the liposomes were measured by dynamic light scattering, using a Nanosizer Nano S (Malvern Instruments, Malvern, UK). Electrophoretic mobility, indicating the effective surface electrical charge (potential), was determined using a Zetasizer Nano ZS90 (Malvern Instruments, UK). Each sample was measured in triplicate.
2.4. Electrostatic Surface Membrane Potential (∆Ψ) Measurements
The interaction of liposomes with macromolecules, including peptides, will depend on the balance of electrostatic repulsion vs. attraction forces and thus on the surface charge of the structures involved. In this study, the surface charge of the liposomes in the presence or absence of G1OLO-L
2OL
2, was determined using ANS, a negatively charged fluorescent probe with a low fluorescence yield in polar environments. Liposomes (500 µM) in the presence or absence of G1OLO-L
2OL
2 (incubated under the same conditions as in the DPH experiments) were titrated with 5 mM ANS in methanol and fluorescence was monitored at λ
ex and λ
em of 380 and 480 nm, respectively, using a lipid:probe ratio of 300/1 (mol/mol). The concentration of bound ANS ([
ANS]
B) vs. free ANS ([
ANS]
free) was adjusted using a modified Langmuir isotherm (Equation (1)):
where
Cmax is the maximum concentration of ANS bound to the liposomes,
k is the binding constant, and
b is a parameter value related to the cooperativity of the process.
[
ANS]
B values can be calculated as shown in Equation (2):
where
Fb and
F0 are the fluorescence intensities, and
Ab and
A0 are the emission coefficients of ANS in the presence or absence of lipid, respectively. Emission coefficient
Ab can be evaluated as the slope of a high-lipid-concentration sample (2 mM) titrated with a diluted ANS solution (0.1–1 µM), and
A0 from the same slope in the absence of lipid.
The change in the electrostatic surface potential (ΔΨ) of the liposomes due to incorporation of the peptide can be calculated as shown in Equation (3):
where
kpeptide and
kliposomes are the ANS binding constants for liposomes in the presence or absence of the peptide, respectively,
R is the universal gas constant,
T is the absolute temperature, and
F is Faraday’s constant.
All fluorescent determinations were carried out using an SLM-Aminco 8100 spectrofluorimeter equipped with a jacketed cuvette holder. The temperature was controlled (±0.1 °C) using a circulating water bath (Haake, Thermo Scientific, Waltham, MA, USA). The excitation and emission slits were 8/8 and 4/4, respectively.
2.5. Fluorescence Measurements
DPH tends to embed in the phospholipid bilayer, whereas TMA-DPH tends to anchor at its aqueous interface [
17]. This difference was used to investigate the liposome phase behavior of the hydrocarbon domain of the bilayer. The liposomes were incubated with 0.005 µM of G1OLO-L
2OL
2 at 37 °C overnight. DPH or TMA-DPH was then added to the sample to a final lipid-to-probe ratio of 300/1 (mol:mol), followed by incubation for 30 min at 37 °C to allow the probe to interact with the liposomes. The anisotropy (
r) of the samples over a temperature range of 3–45 °C was recorded at excitation and emission wavelengths of 381 nm and 526 nm, respectively. Vertically and horizontally polarized emission intensities were corrected for background scattering by subtracting the corresponding polarized intensities of a blank containing the unlabeled suspension (liposomes in buffer without the probe). The
r values were calculated as shown in Equation (4):
where
Iij is the fluorescence intensity when the excitation (
i) and emission (
j) polarizers are fixed in the vertical (
V) or horizontal (
H) position, and
G is the instrument sensitivity ratio of the detection system for vertically and horizontally polarized light.
The values of
r as a function of temperature were adjusted using a modified Boltzmann equation (Equation (5)):
where
r1 and
r2 are the maximum and minimum values of
r,
Tm is the
Lβ-to-
Lα phase transition temperature of the sample, and
b is a parameter that provides information on the cooperativity of the transition process.
Laurdan is a polarity-sensitive probe with an affinity for the glycerol backbone of the bilayer; its lauric acid tail anchors to the phospholipid acyl chain region [
18]. In this study, Laurdan was used to monitor the bilayer fluidity related to a fluorescence shift, by taking advantage of its dipolar relaxation characteristics. Laurdan excitation was measured over a range of 320–420 nm, using emission wavelengths of 440 nm and 490 nm. The lipid concentration in the liposome suspension was adjusted to 250 μM, with Laurdan added to obtain a lipid:probe ratio of 300:1. The generalized polarization (
GPex) for the emission spectra was calculated as shown in Equation (6)
where
I440 and
I490 are the fluorescence intensities at emission wavelengths of 440 nm (gel phase,
Lβ) and 490 nm (liquid crystalline phase
Lα), respectively.
GPex values depend on the excitation wavelength (λ
ex). In lipid mixtures and at constant temperature, positive slope values of
GPex vs. λ
ex indicate the coexistence of domains of different composition, and negative slope values a thermal transition towards a more fluid phase.
2.6. Atomic Force Microscopy Imaging
2.6.1. Bacteria
The in vivo effects of G1OLO-L2OL2 onto the surfaces of Pseudomonas strain 27,853 and E. coli strain ATCC 25,922 after 4 h of exposure to the peptide at concentrations of 0.02 μM (2 × MIC) and 0.04 μM (4 × MIC) and 0.01 μM (2 × MIC) and 0.02 μM (4 × MIC), respectively, were assessed using AFM. Both strains were grown on Muller-Hinton broth cation-adjusted (MHBCA) medium to a concentration of 106 colony-forming units (CFU)/mL. After incubation of the bacteria in fresh medium at 37 °C for 24 h to obtain cultures in the exponential growth phase, G1OLO-L2OL2 was added to the cultures for 4 h at an incubation temperature of 37 °C. The cells were then harvested by centrifugation, resuspended in 2% glutaraldehyde in 0.2 M PBS overnight, and washed three times with distilled water to remove cell debris. The pellets were then suspended in 1.5 mL of distilled water. A 10 μL drop of the suspension was placed on a Thermanox® coverslip and glued to a mica disc for AFM imaging.
The samples were imaged in air using an atomic force microscope XE-70 (Park Systems, Suwon, Korea). Images were obtained in non-contact mode using pyramidal-shaped silicon cantilevers with a spring constant of ±40 N/m and a resonance frequency of ±300 kHz; the upper sides were coated with aluminum to enhance the reflectivity of the laser beam. AFM images were acquired with a scan size of 5 μm2 at a scan rate of 0.3–0.6 Hz. Data acquired during surface scanning were converted into images of topography and amplitude and analyzed using XEP and XEI software (Park Systems, Korea). The topography images were then used to observe the shape, structure, and surface of the planktonic bacteria. In addition, they were used to determine the average surface nano-roughness (Ra) of the treated and untreated planktonic bacteria, with (Ra) calculated as the average distance from the roughness profile to the center plane of the profile.
2.6.2. Liposomes
AFM was carried out on a commercial multimode atomic force microscope controlled by Nanoscope V electronics (Bruker AXS Corp., Madison, WI, USA). Freshly cleaved mica discs (1 cm2) mounted on round Teflon discs were glued to steel discs. Liposome suspensions were incubated overnight on the mica discs at 37 °C. To prevent sample evaporation, the steel discs containing the mica discs and the sample were enclosed in a small Petri dish placed inside a larger Petri dish with a small amount of water at the bottom as a reservoir. The large Petri dish was then sealed with Teflon ribbon and placed inside an oven (Termaks AS, Bergen, Norway) for 20 min with a temperature control of ±0.2 °C. Non-adsorbed liposomes were removed by gently rinsing the samples with buffer, covering the mica surface with 60 µL of buffer. The samples were then directly mounted on the AFM scanner (“E” scanner, 10 µm) and allowed to stabilize. Images were acquired in liquid using MSNL-10 sharpened silicon nitride tips (Bruker AXS Probes, Camarillo, CA, USA) with a mean spring constant of 30 pN nm−1, in contact mode at a 0° scan angle, and with a scan rate of 1.5 Hz. To minimize the applied force on the sample, the set point was continuously adjusted during imaging. Peptide was injected to a final concentration of 5 nM. All images were processed using NanoScope analysis software (Bruker AXS Corp., Santa Barbara, CA, USA).
2.7. Synergy Study
Checkerboard testing was used to assess the susceptibility of the planktonic, imipenem-resistant bacteria (
P. aeruginosa strains 846VH and 536SJD) to G1OLO-L
2OL
2, added in combination with imipenem. Bacteria at a concentration of 10
6 CFU/mL were added together with G1OLO-L
2OL
2 in imipenem-containing MHB (pH of 7.3 ± 0.2) to the wells of a 96-well round-bottom microtiter plate. Concentrations assayed were between 0.5 µg/mL to 16 µg/mL (Imipenem) and between 0.125 µg/mL to 64 µg/mL of the peptide. All experiments were performed in triplicate. The interactions of the bacteria with the peptide were quantitatively evaluated by calculating the fractional inhibitory concentration index (FICi) according to the following formula: FICi = ([MIC drug X in combination)/(MIC of drug X alone])+([MIC of drug Y in combination)/(MIC of drug Y alone]). An FICi < 0.5 was considered to indicate a synergistic interaction, an FICi > 4 an antagonistic interaction, and an FICi ≥ 0.5 and ≤ 4 an indifferent interaction [
19].
4. Discussion
In a previous work [
16] we described a new AMP family, grouped under the name super-cationic peptide dendrimers (SCPDs). Although all members exert antibacterial activity, some of them were shown to be selective for Gram-negative species but with virtually no cytotoxicity in HepG2 and HEK293 human cells. These results suggested that SCPD peptides could serve as a valuable class of AMPs. Among the SCPDs, G1OLO-L
2OL
2 was one of the most promising candidates for the development of an antibacterial agent against Gram-negative bacteria. The most prominent difference between Gram-negative and Gram-positive bacteria is the presence in the former of an outer membrane that acts as a permeability barrier—although the mechanism that ultimately kills bacteria may act on the internal (cytoplasmic) membrane. Our final aim was to decipher the mechanisms by which G1OLO-L
2OL
2 is able to kill Gram-negative MDR and, particularly in this work we aimed to gain insights on its interaction with model membranes and living cells. Using biophysical approaches, we examined the interaction of G1OLO-L
2OL
2 with model membranes mimicking the inner membrane of
E. coli (POPE:POPG 3:1, mol/mol) [
19]. When G1OLO-L
2OL
2 is incubated with liposomes in suspension, it preferentially interacts with the latter’s phospholipid head groups, as can be seen by the shift towards a higher melting transition temperature detected with the TMA-DPH probe. Something that does not occur with the DPH probe that resides in the core of the bilayer. This indicated an increase in the rigidity of the headgroup region of the liposome due to its interaction with the peptide. In their interactions with liposomes, peptides can either be adsorbed onto the membrane or be partially absorbed into the lipid-water interface, close to the upper portion of the fatty acyl chains, where TMA-DPH tends to localize. However, it cannot be excluded that this increase in the Tm, might be attributed to a charge screening selective effect due to the interaction of the peptide with the negatively charged POPG. The fluorescence results showed that the peptides were not found in the hydrophobic core region of the liposome, although their ability to permeate the membrane by forming pores cannot be excluded. In spite of the structural differences, a similar behavior was observed for a series of three nine residue peptides that contained unnatural amino acids in the primary sequence [
20]. Nevertheless, the addition of G1OLO-L
2OL
2 to reconstituted black lipid bilayers did not generate noticeable electrophysiological phenomena, thus suggesting that the peptide was unable to generate true transmembrane channels (data not shown).
In the AFM experiments, G1OLO-L
2OL
2 was injected in situ inside the AFM liquid cell at the same concentration used with liposomes in solution. In the absence of injected peptide, the AFM images revealed two lipid domains in the SLB surfaces, in concordance with our previous study [
21]. Since the Laurdan fluorescence provided no evidence of the existence of domains of different lipid composition on the liposomes, these lipid domains may have been: (i) domains induced by changes in temperature of the same lipid composition, in which lipids in the taller domains were in a more rigid phase than those in the shorter domains or (ii) domains of different lipid composition in a different lipid phase, in which the presence of the mica surface decreased the lateral diffusion of the lipids, thus promoting the formation of segregated lipid domains differing in their lipid composition. The work performed in this study and in previously published work [
22] indicated that the taller lipid domains were POPG enriched, and the more extended domains POPE enriched.
When added to the SLBs, G1OLO-L
2OL
2 interacted with their surfaces, without formation of pores, in agreement with the synergy studies. According to these observations, the peptide, at the concentrations studied, was able to adsorb onto or be partially absorb into the surface of the SLBs. However, a dose-dependent effect of the peptide was also observed, as higher concentrations (
Figure 8) induced the erosion and solubilization of the SLBs. A similar effect was observed in the fluorescence experiments (data not shown), in which higher G1OLO-L
2OL
2 concentrations induced the erratic behavior of the liposomes, most likely attributable to their destabilization.
In agreement with these observations, determinations of the zeta potential of the liposomes were consistent with the incorporation of G1OLO-L2OL2 into the vesicles. However, to confirm that the peptide was in close contact with the lipid membrane of the liposome and not located in the hydration layer (where the zeta potential is actually measured), the surface potential was determined in an ANS fluorescence assay, which also showed that the peptide was present on the liposome surface.
Finally, the effects of G1OLO-L
2OL
2 on living bacteria were evaluated by AFM. The peptide had a more destructive effect on
P. aeruginosa than on
E. coli. While at 2 × MIC
P. aeruginosa was destroyed, while
E. coli retained its shape and cell integrity to a certain degree. However, it should be noted that AFM reveals only the topography of the bacterial surface, not bacterial viability. It is therefore possible that the bactericidal effect was similar in
P. aeruginosa and
E. coli, but the destruction of the lipid outer membrane differed. Studies of the differences in the membrane lipid composition in the two species in model membranes could help to explain the differences in the observed behaviors. In fact, although membrane permeabilization is the main mechanism of action of AMPs against pathogens, additional mechanisms have been described in detail. This includes membrane destabilization, inhibition of macromolecular synthesis and intracellular translocation and inhibition of the biosynthesis of nucleic acids and proteins [
13]. The bacterial cytoplasm possesses a high osmotic potential that is maintained by the function of bacterial envelopes. The alteration of the membrane and/or the cell wall may determine a water influx and generate hydrostatic pressures incompatible with bacterial growth and even with survival. This is known as osmotic stress and has in bacteria some characteristics clearly different from those in the eukaryotic cells [
23]. Here, the presence of low concentrations of G1OLO-L
2OL
2 at the outer membrane surface could induce osmotic stress and thereby facilitate a destabilization of cell integrity at higher peptide concentrations. In spite of this, the clear effect of G1OLO-L
2OL
2 on bacterial membranes, and its action on other targets cannot be ruled out and should be further investigated.