Nosustrophine: An Epinutraceutical Bioproduct with Effects on DNA Methylation, Histone Acetylation and Sirtuin Expression in Alzheimer’s Disease
Abstract
:1. Introduction
2. Materials and Methods
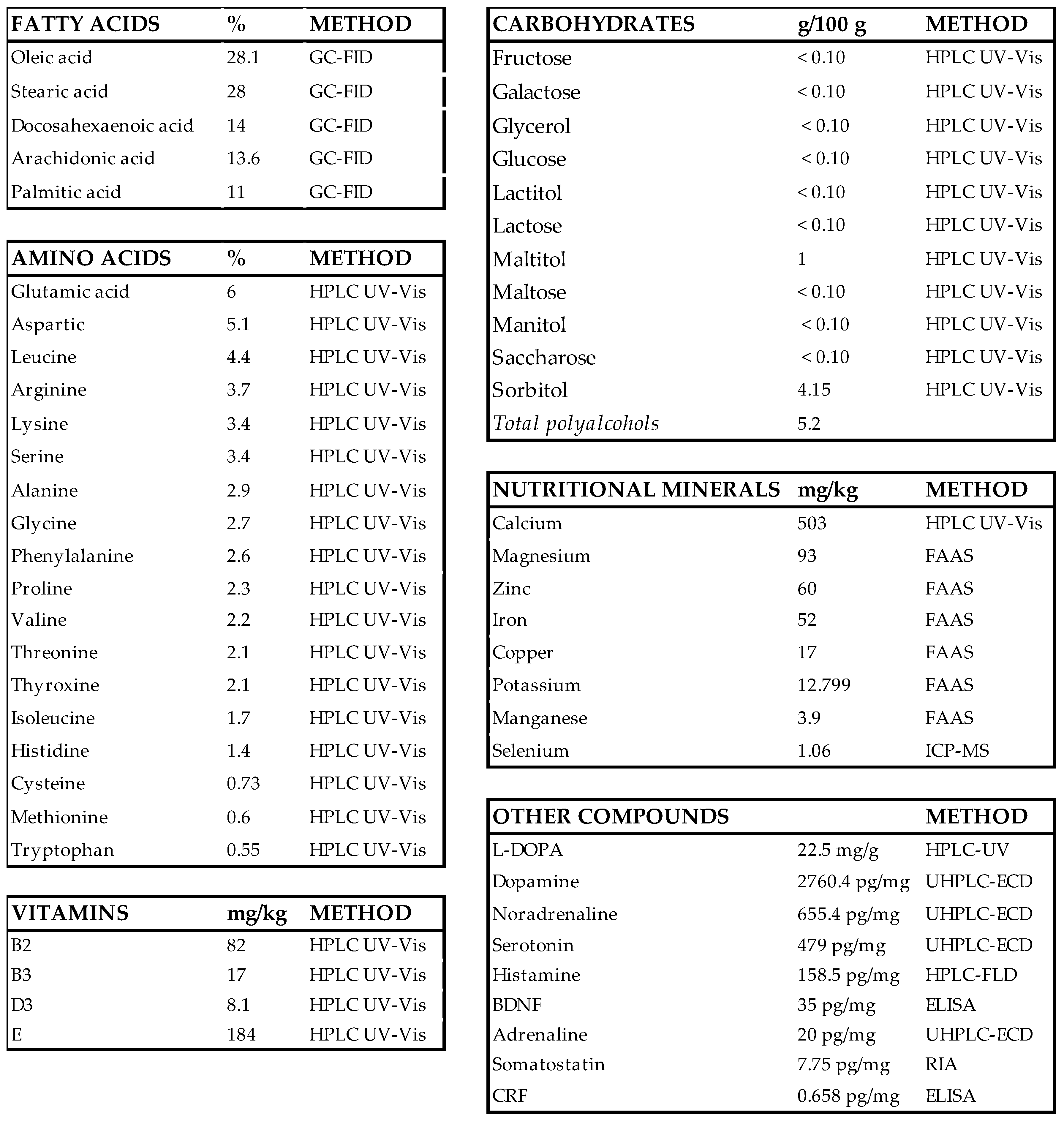
2.1. Nutritional Analysis of Nosustrophine
2.2. Catecholamine, Serotonin and L-DOPA Analyses
2.3. Neurotrophic Factor Analysis
2.4. Proteomic Analysis
2.4.1. Sample Preparation
2.4.2. Liquid Chromatography-Tandem Mass Spectrometry (LC-MS/MS) Analysis
2.4.3. LC-MS/MS Data Analysis
2.5. Animals and Genotyping
2.6. Preparation of Nosustrophine and Treatments In Vivo
2.7. Immunofluorescence
2.8. Western Blotting and Dot Blot Characterization
2.9. Quantification of Global DNA Methylation (5mC)
2.10. Quantitative Real Time RT-PCR (qPCR)
2.11. Nuclear Protein Extraction
2.12. Quantification of HDAC and Sirtuin Activity
2.13. Statistical Analysis
3. Results
3.1. Screen for the Composition of Nosustrophine
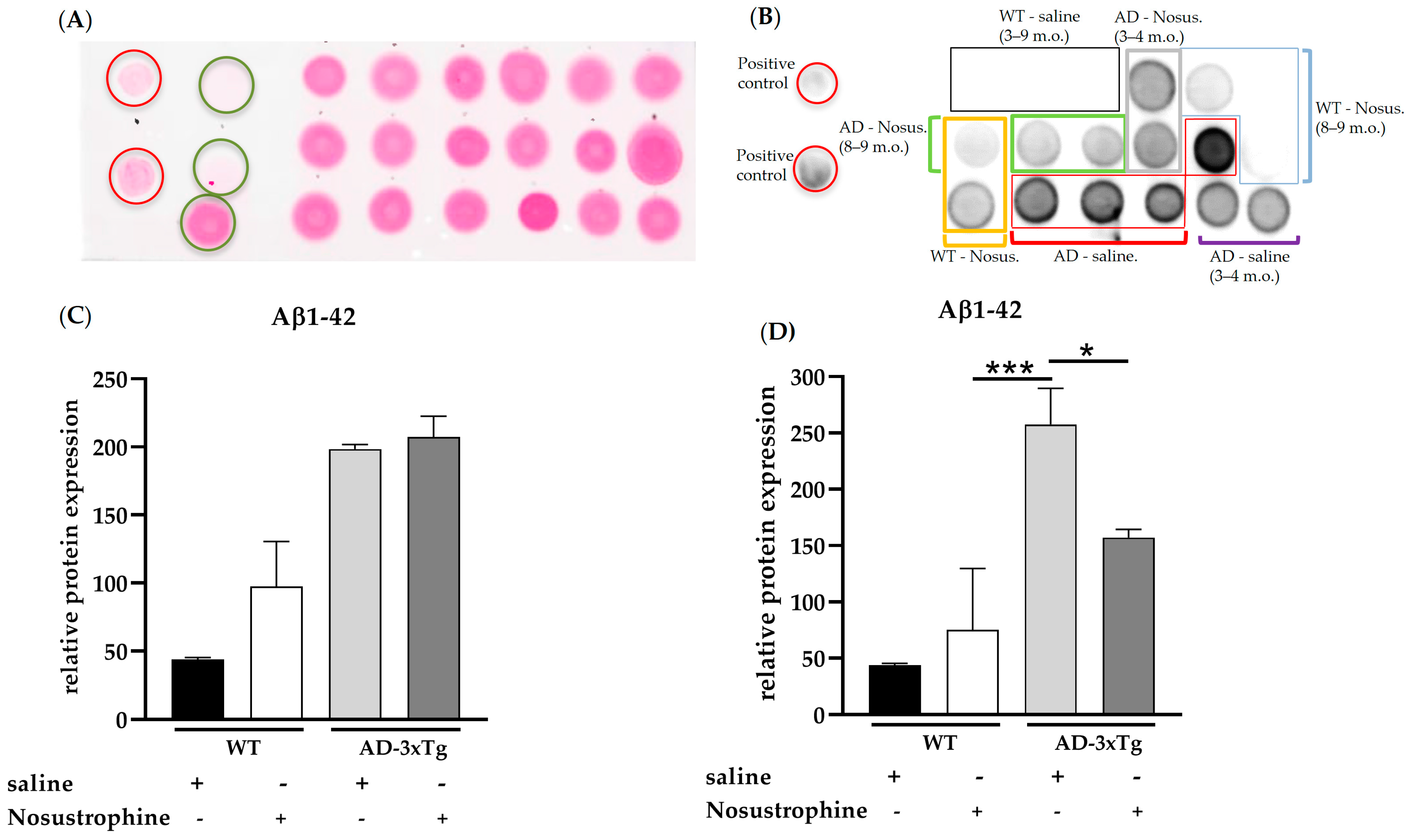
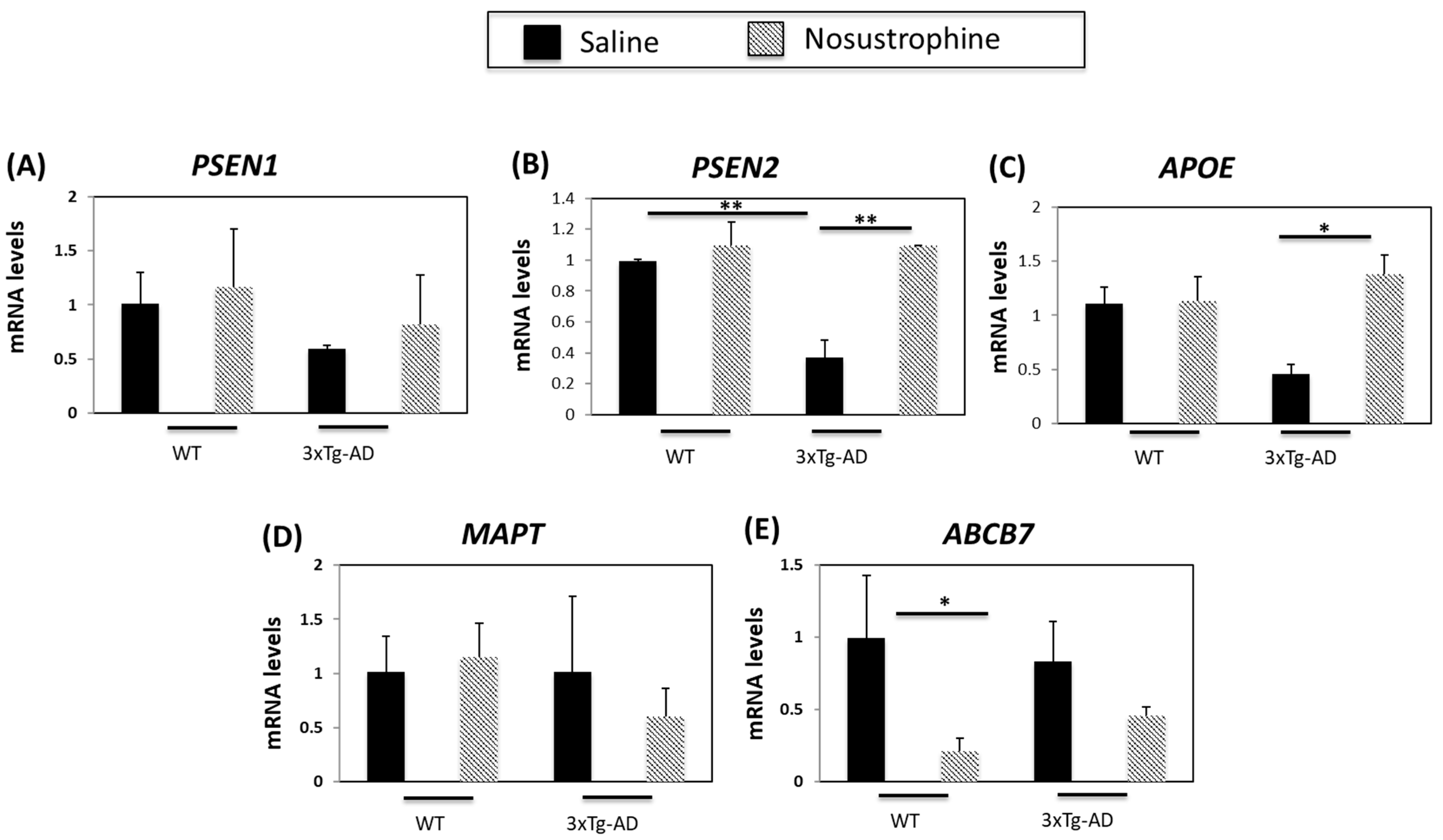
3.2. Nosustrophine Regulates AD-Related Gene Expression In Vivo
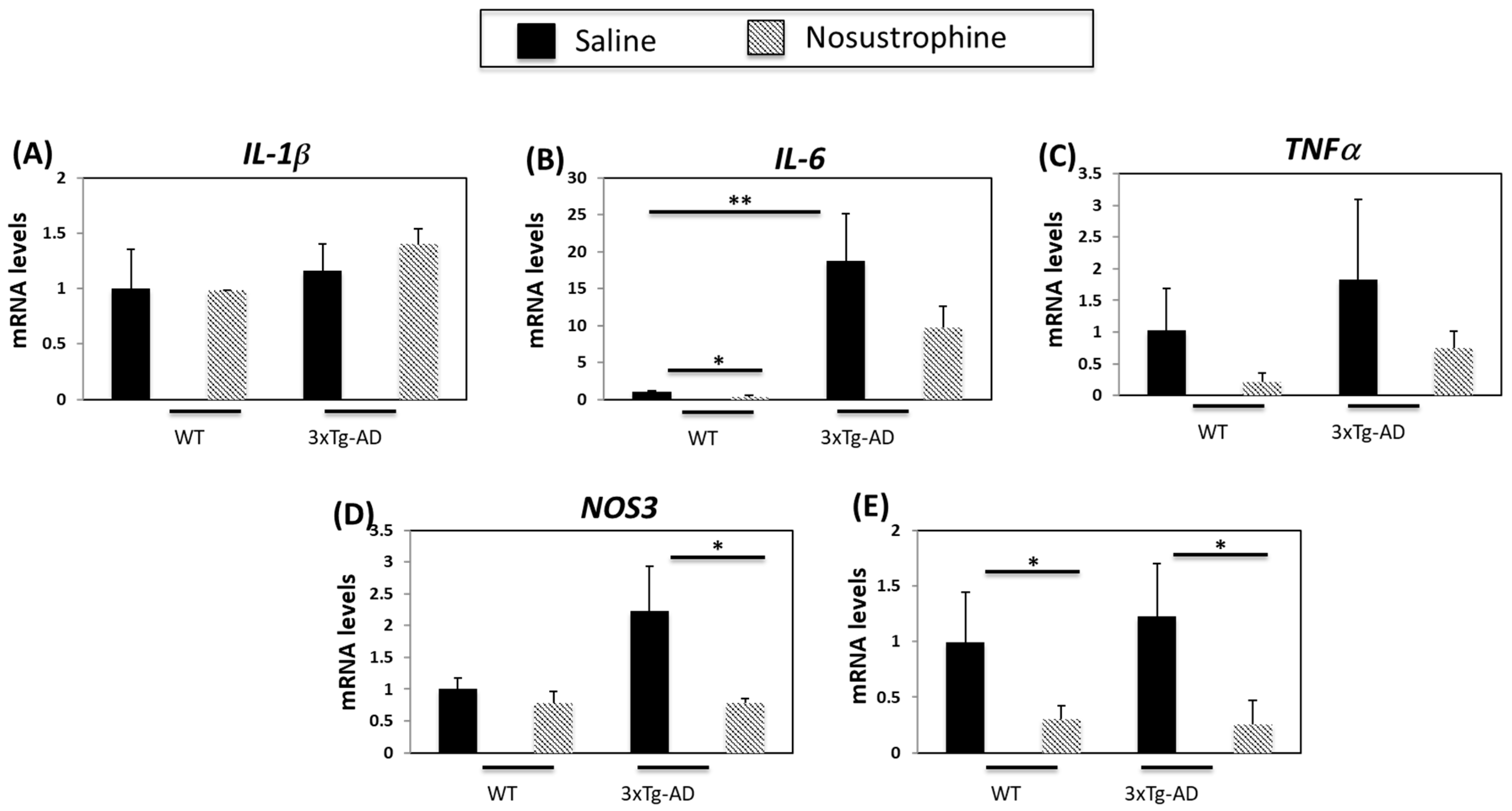
3.3. Nosustrophine Regulates Inflammation-Related Gene Expression in Transgenic Mice with AD
3.4. Nosustrophine Regulates DNA Methylation in Trigenic AD Mice
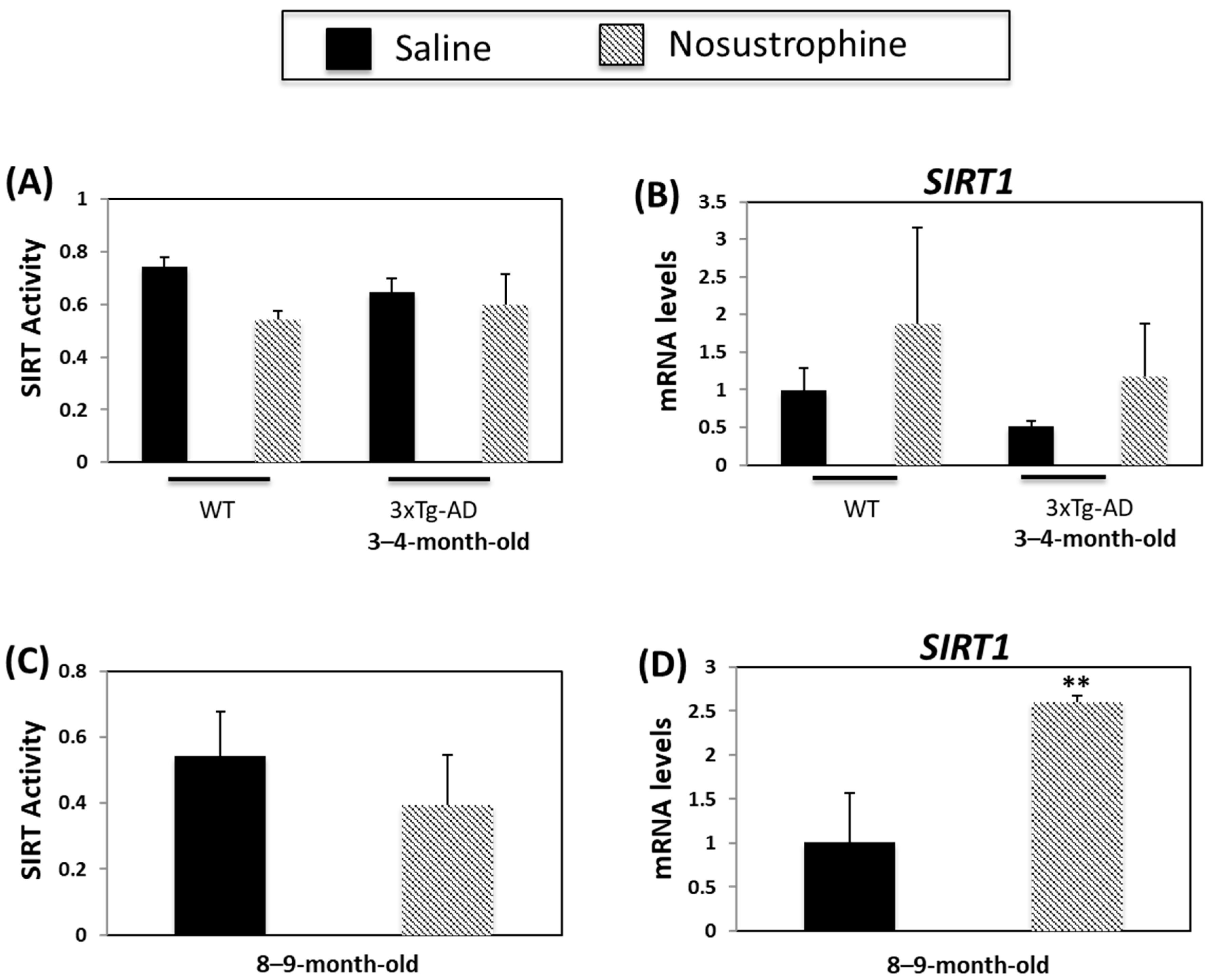
3.5. Nosustrophine Regulates SIRT Activity
3.6. Nosustrophine Regulates HDAC Activity
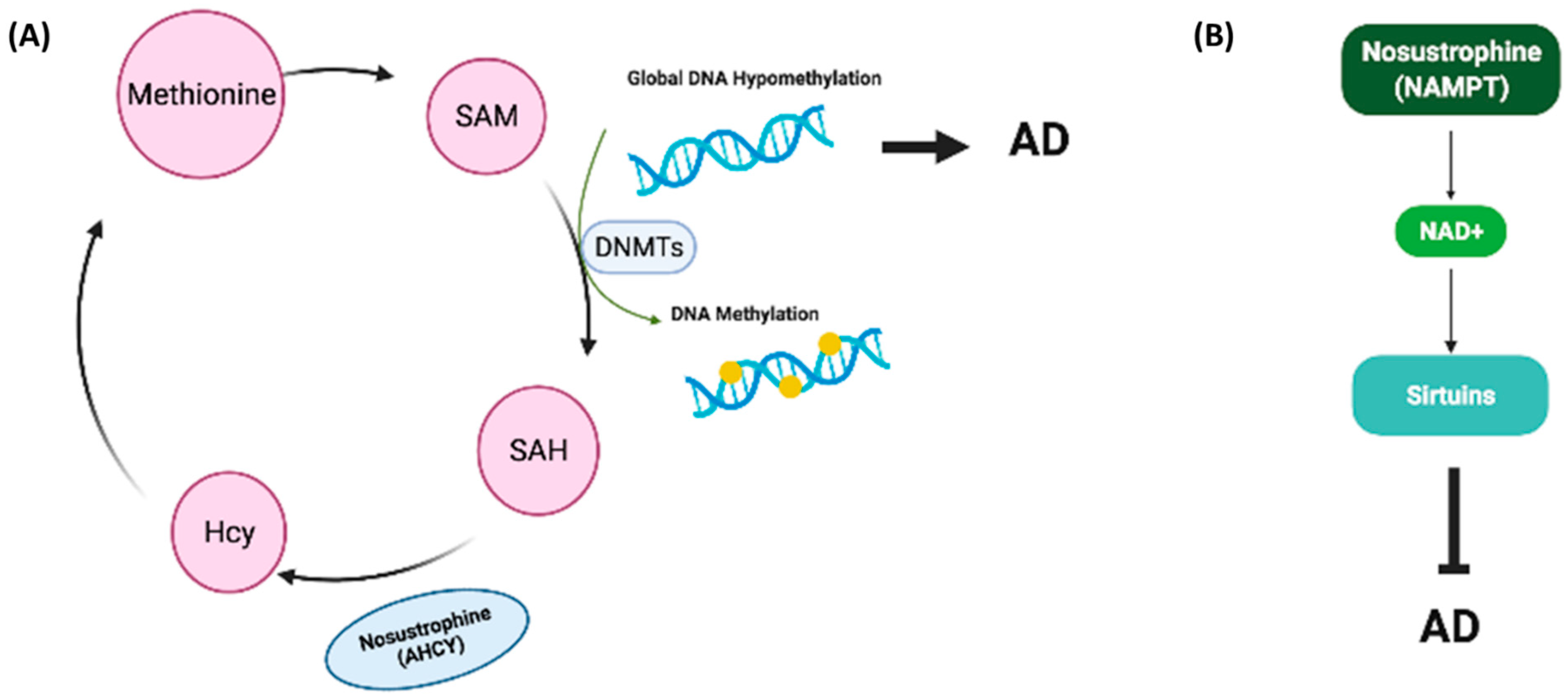
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Conflicts of Interest
References
- Rosende-Roca, M.; Abdelnour, C.; Esteban, E.; Tartari, J.P.; Alarcon, E.; Martínez-Atienza, J.; González-Pérez, A.; Sáez, M.E.; Lafuente, A.; Buendía, M.; et al. The role of sex and gender in the selection of Alzheimer patients for clinical trial pre-screening. Alzheimers Res. Ther. 2021, 13, 95. [Google Scholar] [CrossRef] [PubMed]
- GBD 2019 Dementia Forecasting Collaborators. Estimation of the global prevalence of dementia in 2019 and forecasted prevalence in 2050: An analysis for the Global Burden of Disease Study 2019. Lancet Public Health 2022, 7, e105–e125. [Google Scholar] [CrossRef]
- Cacabelos, R. Pharmacogenomics of Alzheimer’s and Parkinson’s diseases. Neurosci. Lett. 2020, 726, 133807. [Google Scholar] [CrossRef] [PubMed]
- Cacabelos, R.; Fernandez-Novoa, L.; Lombardi, V.; Kubota, Y.; Takeda, M. Molecular genetics of Alzheimer’s disease and ageing. Methods Find Exp. Clin. Pharm. 2005, 27, 1673. [Google Scholar]
- Morales, M.; Margolis, E.B. Ventral tegmental area: Cellular heterogeneity, connectivity and behaviour. Nat. Rev. Neurosci. 2017, 18, 73–85. [Google Scholar] [CrossRef]
- Broussard, J.I.; Yang, K.; Levine, A.T.; Tsetsenis, T.; Jenson, D.; Cao, F.; Garcia, I.; Arenkiel, B.R.; Zhou, F.M.; De Biasi, M.; et al. Regulates Aversive Contextual Learning and Associated In Vivo Synaptic Plasticity in the Hippocampus. Cell Rep. 2016, 14, 1930–1939. [Google Scholar] [CrossRef] [Green Version]
- Russo, S.J.; Nestler, E.J. The brain reward circuitry in mood disorders. Nat. Rev. Neurosci. 2013, 14, 609–625. [Google Scholar] [CrossRef] [Green Version]
- Nobili, A.; Latagliata, E.C.; Viscomi, M.T.; Cavallucci, V.; Cutuli, D.; Giacovazzo, G.; Krashia, P.; Rizzo, F.R.; Marino, R.; Federici, M.; et al. Dopamine neuronal loss contributes to memory and reward dysfunction in a model of Alzheimer’s disease. Nat. Commun. 2017, 8, 14727. [Google Scholar] [CrossRef] [Green Version]
- Bozzali, M.; Serra, L.; Cercignani, M. Quantitative MRI to understand Alzheimer’s disease pathophysiology. Curr. Opin. Neurol. 2016, 29, 437–444. [Google Scholar] [CrossRef]
- Cacabelos, R. Pharmacogenomics of Cognitive Dysfunction and Neuropsychiatric Disorders in Dementia. Int. J. Mol. Sci. 2020, 21, 3059. [Google Scholar] [CrossRef]
- Niki, K.; Castillo, S.; Hoie, E.; O’Brien, K.K. Alzheimer’s Disease: Current treatments and potential new agents. US Pharm. 2019, 44, 20–23. [Google Scholar]
- Sadhukhan, P.; Saha, S.; Dutta, S.; Mahalanobish, S.; Sil, P.C. Nutraceuticals: An emerging therapeutic approach against the pathogenesis of Alzheimer’s disease. Pharm. Res. 2018, 129, 100–114. [Google Scholar] [CrossRef] [PubMed]
- Farias, G.A.; Guzmán-Martínez, L.; Delgado, C.; Maccioni, R.B. Nutraceuticals: A novel concept in prevention and treatment of Alzheimer’s disease and related disorders. J. Alzheimers Dis. 2014, 42, 357–367. [Google Scholar] [CrossRef] [PubMed]
- Carrera, I.; Martínez, O.; Cacabelos, R. Neuroprotection with natural antioxidants and nutraceuticals in the context of brain cell degeneration: The epigenetic connection. Curr. Top. Med. Chem. 2019, 19, 2999–3011. [Google Scholar] [CrossRef] [PubMed]
- Carlos-Reyes, Á.; López-González, J.S.; Meneses-Flores, M.; Gallardo-Rincón, D.; Ruíz-García, E.; Marchat, L.A.; Astudillo-De La Vega, H.; Hernández de la Cruz, O.N.; López-Camarillo, C. compounds as epigenetic modulating agents in cancer. Front. Genet. 2019, 10, 79. [Google Scholar] [CrossRef] [Green Version]
- Urdinguio, R.G.; Sanxhez-Mut, J.V.; Esteller, M. Epigenetic mechanisms in neurological diseases: Genes, syndromes, and therapies. Lancet Neurol. 2009, 8, 1056–1072. [Google Scholar] [CrossRef]
- Sweatt, J.D. The emerging field of neuroepigenetics. Neuron 2013, 80, 624–632. [Google Scholar] [CrossRef] [Green Version]
- Cacabelos, R.; Tellado, I.; Cacabelos, P.R. The epigenetic machinery in the life cycle and pharmacoepigenetics. In Pharmacoepigenetics; Academic Press: Cambridge, MA, USA, 2019; Chapter 1; pp. 1–100. [Google Scholar]
- Delgado-Morales, R.; Esteller, M. Opening up the DNA methylome of dementia. Mol. Psychiatry 2017, 22, 485–496. [Google Scholar] [CrossRef]
- Wang, J.; Yu, J.T.; Tan, M.S.; Jiang, T.; Tan, L. Epigenetic mechanisms in Alzheimer’s disease: Implications for pathogenesis and therapy. Ageing Res. Rev. 2013, 12, 1024–1041. [Google Scholar] [CrossRef]
- Cacabelos, R.; Carril, J.C.; Cacabelos, N.; Kazantsev, A.G.; Vostrov, A.V.; Corzo, L.; Cacabelos, P.; Goldgaber, D. Sirtuins in Alzheimer’s Disease: SIRT2-Related GenoPhenotypes and Implications for PharmacoEpiGenetics. Int. J. Mol. Sci. 2019, 20, 1249. [Google Scholar] [CrossRef] [Green Version]
- Wu, C.Y.; Hu, H.Y.; Chow, L.H.; Chou, Y.J.; Huang, N.; Wang, P.N.; Li, C.P. The Effects of Anti-Dementia and Nootropic Treatments on the Mortality of Patients with Dementia: A Population-Based Cohort Study in Taiwan. PLoS ONE 2015, 10, e0130993. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Colucci, L.; Bosco, M.; Ziello, A.R.; Rea, R.; Amenta, F.; Fasanaro, A.M. Effectiveness of nootropic drugs with cholinergic activity in treatment of cognitive deficit: A review. J. Exp. Pharm. 2012, 4, 163–172. [Google Scholar] [CrossRef] [PubMed]
- Akhondzadeh, S.; Sabet, M.S.; Harirchian, M.H.; Togha, M.; Cheraghmakani, H.; Razeghi, S.; Hejazi, S.S.; Yousefi, M.H.; Alimardani, R.; Jamshidi, A.; et al. A 22-week, multicenter, randomized, double-blind controlled trial of Crocus sativus in the treatment of mild-to-moderate Alzheimer’s disease. Psychopharmacology 2010, 207, 637–643. [Google Scholar] [CrossRef] [PubMed]
- Schaeffer, E.L.; Forlenza, O.V.; Gattaz, W.F. Phospholipase A2 activation as a therapeutic approach for cognitive enhancement in early-stage Alzheimer disease. Psychopharmacology 2009, 202, 37–51. [Google Scholar] [CrossRef]
- Yurko-Mauro, K. Cognitive and cardiovascular benefits of docosahexaenoic acid in aging and cognitive decline. Curr. Alzheimer Res. 2010, 7, 190–196. [Google Scholar] [CrossRef]
- Fiani, B.; Covarrubias, C.; Wong, A.; Doan, T.; Reardon, T.; Nikolaidis, D.; Sarno, E. Cerebrolysin for stroke, neurodegeneration, and traumatic brain injury: Review of the literature and outcomes. Neurol. Sci. 2021, 42, 1345–1353. [Google Scholar] [CrossRef]
- Gevaert, B.; D’Hondt, M.; Bracke, N.; Yao, H.; Wynendaele, E.; Vissers, J.P.; De Cecco, M.; Claereboudt, J.; De Spiegeleer, B. Peptide profiling of internet-obrained cerebrolysin using high performance liquid chromatography-electrospray ionization ion trap and ultra high performance liquid chromatography-ion mobility-quadrupole time of flight mass spectrometry. Drug Test Anal. 2015, 7, 835–842. [Google Scholar] [CrossRef]
- Mitra, S.; Behbahani, H.; Eriksdotter, M. Innovative Therapy for Alzheimer’s Disease-With Focus on Biodelivery of NGF. Front. Neurosci. 2019, 13, 38. [Google Scholar] [CrossRef] [Green Version]
- Gonzalez, S.; McHugh, T.L.M.; Yang, T.; Suriani, W.; Massa, S.M.; Longo, F.M.; Simmons, D.A. Small molecule modulation of TrkB and TrkC neurotrophin receptors prevents cholinergic neuron atrophy in an Alzheimer’s disease mouse model at an advanced pathological stage. Neurobiol. Dis. 2022, 162, 105563. [Google Scholar] [CrossRef]
- Hou, Y.; Aboukhatwa, M.A.; Lei, D.-L.; Manaye, K.; Khan, I.; Luo, Y. Anti-depressant natural flavonols modulate BDNF and beta amyloid in neurons and hippocampus of double TgAD mice. Neuropharmacology 2010, 58, 911–932. [Google Scholar] [CrossRef] [Green Version]
- Kazim, S.F.; Iqbal, K. Neurotrophic factor small-molecule mimetics mediated neuroregeneration and synaptic repair: Emerging therapeutic modality for Alzheimer’s Disease. Mol. Neurodeg. 2016, 11, 50. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Burbano, C.; Cuadrado, C.; Murquiz, M.; Cubero, J.I. Variation of favism-inducing factors (vicine, convicine and L-DOPA) during pod development in Vicia faba L. Plant Foods Hum. Nutr. 1995, 47, 265–275. [Google Scholar] [CrossRef] [PubMed]
- Flurkey, K.; Currer, J.M.; Harrison, D.E. Mouse models in aging research. In The Mouse in Biomedical Research, 2nd ed.; Academic Press: Cambridge, MA, USA, 2007; pp. 637–672. [Google Scholar]
- Zhou, Z.D.; Chan, C.H.; Ma, Q.H.; Xu, X.H.; Xiao, Z.C.; Tan, E.K. The roles of amyloid precursor protein (APP) in neurogenesis, implications to pathogenesis and therapy of Alzheimer disease (AD). Cell Adh. Migr. 2011, 5, 280–292. [Google Scholar] [CrossRef] [Green Version]
- Mahajan, U.V.; Varma, V.R.; Griswold, M.E.; Blackshear, C.T.; An, Y.; Oommen, A.M.; Varma, S.; Troncoso, J.C.; Pletnikova, O.; O’Brien, R.; et al. Dysregulation of multiple metabolic networks related to brain transmethylation and polyamine pathways in Alzheimer disease: A targeted metabolomic and transcriptomic study. PLoS Med. 2020, 17, e1003012. [Google Scholar]
- Yamazaki, Y.; Zhao, N.; Caufield, T.R.; Liu, C.-C.; Bu, G. Apolipoprotein E and Alzheimer disease: Pathobiology and targeting strategies. Nat. Rev. Neurol. 2019, 15, 501–518. [Google Scholar] [CrossRef] [PubMed]
- Sadik, G.; Kaji, H.; Takeda, K.; Yamagata, F.; Kameoka, Y.; Hashimoto, K.; Miyanaga, K.; Shinoda, T. In vitro processing of amyloid precursor protein by cathepsin D. Int. J. Biochem. Cell Biol. 1999, 31, 1327–1337. [Google Scholar] [CrossRef]
- Ferreira-Vieira, T.H.; Guimaraes, I.M.; Silva, F.R.; Ribeiro, F.M. Alzheimer’s disease: Targeting the Cholinergic System. Curr. Neuropharmacol. 2016, 14, 101–115. [Google Scholar] [CrossRef] [Green Version]
- Helwig, M.; Hoshino, A.; Berridge, C.; Lee, S.N.; Lorenzen, N.; Otzen, D.E.; Eriksen, J.L.; Lindberg, I. The neuroendocrine protein 7B2 suppresses the aggregation of neurodegenerative disease-related proteins. J. Biol. Chem. 2013, 288, 1114–1124. [Google Scholar] [CrossRef] [Green Version]
- Xing, S.; Hu, Y.; Huang, X.; Shen, D.; Chen, C. Nicotinamide phosphoribosyltransferase-related signaling pathway in early Alzheimer’s disease mouse models. Mol. Med. Rep. 2019, 20, 5163–5171. [Google Scholar] [CrossRef] [Green Version]
- De Strooper, B.; Iwatsubo, T.; Wolfe, M.S. Presenilins and γ-secretase: Structure, function, and role in Alzheimer Disease. Cold Spring Harb. Perspect. Med. 2012, 2, a006304. [Google Scholar] [CrossRef]
- Carrera, I.; Fernández-Novoa, L.; Teijido, O.; Sampedro, C.; Seoane, S.; Lakshmana, M.; Cacabelos, R. Comparative Characterization Profile of Transgenic Mouse Models of Alzheimer ‘s Disease. J. Genom. Med. Pharm. 2017, 2, 331–337. [Google Scholar]
- Martínez-Iglesias, O.; Naidoo, V.; Carrera, I.; Cacabelos, R. Epigenetic studies in the male APP/BIN1/COPS5 triple-transgenic mouse model of Alzheimer’s disease. Int. J. Mol. Sci 2022, 23, 2446. [Google Scholar] [CrossRef] [PubMed]
- Pan, X.; Kaminga, A.C.; Wen, S.W.; Wu, X.; Achaempong, K.; Liu, A. Dopamine and Dopamine Receptors in Alzheimer’s Disease: A Systematic Review and Network Meta-Analysis. Front. Aging Neurosci. 2019, 11, 175. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Caligiore, D.; Silvetti, M.; D’Amelio, M.; Puglisi-Allegra, S.; Baldassarre, G. Computational Modeling of Catecholamines Dysfunction in Alzheimer’s Disease at Pre-Plaque Stage. J. Alzheimers Dis. 2020, 77, 275–290. [Google Scholar] [CrossRef]
- Ambrée, O.; Richter, H.; Sachser, N.; Lewejohann, L.; Dere, E.; de Souza Silva, M.A.; Herring, A.; Keyvani, K.; Paulus, W.; Schäbitz, W.R. Levodopa ameliorates learning and memory deficits in a murine model of Alzheimer’s disease. Neurobiol. Aging 2009, 30, 1192–1204. [Google Scholar] [CrossRef]
- Lista, S.; O’Bryant, S.E.; Blennow, K.; Dubois, B.; Hugon, J.; Zetterberg, H.; Hampel, H. Biomarkers in sporadic and familial Alzheimer’s disease. J. Alzheimers Dis. 2015, 47, 291–317. [Google Scholar] [CrossRef]
- Cooper-Knock, J.; Kirby, J.; Ferraiuolo, L.; Heath, P.R.; Rattray, M.; Shaw, P.J. Gene expression profiling in human neurodegenerative disease. Nat. Rev. Neurol. 2012, 8, 518–530. [Google Scholar] [CrossRef]
- Duff, K.; Eckman, C.; Zehr, C.; Yu, X.; Prada, C.M.; Perez-Tur, J.; Hutton, M.; Buee, L.; Harigaya, Y.; Yager, D.; et al. Increased amyloid-beta42(43) in brains of mice expressing mutant presenilin 1. Nature 1996, 383, 710–713. [Google Scholar] [CrossRef]
- Liu, C.C.; Kanekiyo, T.; Xu, H.; Bu, G. Apolipoprotein E and Alzheimer’s disease: Risk, mechanisms, and therapy. Nat. Rev. Neurol. 2013, 9, 106–118. [Google Scholar] [CrossRef] [Green Version]
- Zhou, F.; Wang, D. The association between the MAPT polymorphisms and Alzheimer’s disease risk: A meta-analysis. Oncotarget 2017, 8, 43506–43520. [Google Scholar] [CrossRef] [Green Version]
- Pereira, C.; Martins, F.; Wiltfang, J.; da Cruz e Silva, O.; Rebelo, S. ABC transporters are key players in Alzheimer’s disease. J. Alzheimers Dis. 2018, 61, 463–485. [Google Scholar] [CrossRef] [PubMed]
- Akiyama, H.; Barger, S.; Barnum, S.; Bradt, B.; Bauer, J.; Cole, G.M.; Cooper, N.R.; Eikelenboom, P.; Emmerling, M.; Fiebich, B.L.; et al. Inflammation and Alzheimer’s disease. Neurobiol. Aging 2000, 21, 383–421. [Google Scholar] [CrossRef]
- El Idrissi, F.; Gressier, B.; Devos, D.; Belarbi, K. A computational exploration of the molecular network associated to neuroinflammation in Alzheimer’s Disease. Front. Pharm. 2021, 12, 630003. [Google Scholar] [CrossRef] [PubMed]
- O’Banion, M.K. COX-2 and Alzheimer’s disease: Potential roles in inflammation and neurodegeneration. Exp. Opin. Investig. Drugs 1999, 8, 1521–1536. [Google Scholar] [CrossRef]
- Martínez-Iglesias, O.; Carrera, I.; Carril, J.C.; Férnandez-Novoa, L.; Cacabelos, N.; Cacabelos, R. DNA methylation in neurodegenerative and cerebrovascular disorders. Int. J. Mol. Sci. 2020, 21, 2220. [Google Scholar] [CrossRef] [Green Version]
- Martínez-Iglesias, O.; Naidoo, V.; Cacabelos, N.; Cacabelos, R. Epigenetic Biomarkers as diagnostic tools for Neurodegenerative Disorders. Int. J. Mol. Sci. 2022, 23, 13. [Google Scholar] [CrossRef]
- Lalla, R.; Donmez, G. The role of sirtuins in Alzheimer’s disease. Front. Aging Neurosci. 2013, 5, 16. [Google Scholar] [CrossRef] [Green Version]
- Kurukulasooeiya, K.F.; Yasamamdana, S.W. Sirtuins as Potential Therapeutic Targets for Mitigating Neuroinflammation Associated with Alzheimer’s Disease. Front. Cell Neurosci. 2021, 15, 746631. [Google Scholar]
- Xu, K.; Dai, X.-L.; Huang, H.-C.; Jiang, Z.-F. Targeting HDACs: A promising therapy for Alzheimer’s Disease. Oxidative Med. Cell. Longev. 2011, 2011, 143269. [Google Scholar] [CrossRef] [Green Version]
- Cacabelos, R. What have we learnt from past failures in Alzheimer’s disease drug discovery? Expert Opin. Drug Discov. 2022, 6, 1–15. [Google Scholar] [CrossRef]
- Wu, Y.-Y.; Hsu, J.-L.; Wang, H.-C.; Wu, S.-J.; Hong, C.-J.; Cheng, I. Alterations of the neuroinflammatory markers IL-6 and TRAIL in Alzheimer’s Disease. Dement. Geriatr. Cogn. Dis. Extra 2015, 5, 424–434. [Google Scholar] [CrossRef]
- Park, J.C.; Han, S.H.; Mook-Jung, I. Peripheral inflammatory biomarkers in Alzheimer’s disease: A brief review. BMB Rep. 2020, 53, 10–19. [Google Scholar] [CrossRef] [Green Version]
- Mrak, R.E.; Griffin, W.S.T. Interleukin-1, neuroinflammation, and Alzheimer’s disease. Neurobiol. Aging 2001, 22, 903–908. [Google Scholar] [CrossRef]
- López-González, I.; Schlüter, A.; Aso, E.; Garcia-Esparcia, P.; Ansoleaga, B.; LLorens, F.; Carmona, M.; Moreno, J.; Fuso, A.; Portero-Otin, M.; et al. Neuroinflammatory singnals in Alzheimer’s Disease and APP/PS1 Transgenic Mice: Correlations with plaques, tangles, and oligomeric species. J. Meuropath. Exp. Neurol. 2015, 74, 319–344. [Google Scholar] [CrossRef] [Green Version]
- De la Monte, S.; Chiche, J.-D.; von dem Buscsche, A.; Sanyal, S.; Lahousse, S.; Janssens, S.; Bloch, D. Nitric oxide synthase-3 overexpression causes apoptosis and impairs neuronal mitochondrial function: Relevance to alxheimer’s type degeneration. Lab. Investig. 2003, 83, 287–298. [Google Scholar] [CrossRef]
- Nogawa, S.; Takao, M.; Suzuki, S.; Tanaka, K.; Koto, A.; Fukuuchi, Y.; Hayakawa, I. COX-2 expression in brains of patients with familiar Alzheimer’s disease. Int. Congr. Ser. 2003, 1252, 363–372. [Google Scholar] [CrossRef]
- Hoozemans, J.J.; O’Banion, M.K. The role of COX-1 and COX-2 in Alzheimer’s Disease Patholgy and the therapeutic potentials of non-steroidal anti-inflammatory drugs. Curr. Drugs Targ CNS Neurol. Dis. 2005, 4, 307–315. [Google Scholar] [CrossRef]
- Aisen, P.S. Evaluation of selective COX-2 inhibitors for the treatment of Alzheimer’s disease. J. Pain Symptom Manag. 2002, 23, S35–S40. [Google Scholar] [CrossRef]
- Woodling, N.S.; Colas, D.; Wang, Q.; Minhas, P.; Panchal, M. Cyclooxygenase inhibition targets neurons to prevent early behavioural decline in Alzheimer’s disease model mice. Brain 2016, 139, 2063–2081. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ahmed, S.; Jing, Y.; Mockett, B.G.; Zhang, H.; Abraham, W.C.; Liu, P. Partial Endothelial Nitric Oxide Synthase Deficiency Exacerbates Cognitive Deficit and Amyloid Pathology in the APPswe/PS1ΔE9 Mouse Model of Alzheimer’s Disease. Int. J. Mol. Sci. 2022, 23, 7316. [Google Scholar] [CrossRef]
- Martínez-Iglesias, O.; Cacabelos, R.O. Epigenetic treatment of neurodegenerative disorders. Histone Modif. Ther. 2019, 311, 335. [Google Scholar]
- Chouliaras, L.; Mastroeni, D.; Delvaux, E.; Grover, A.; Kenis, G.; Hof, P.R.; Steinbusch, H.W.; Coleman, P.D.; Rutten, B.P.; van den Hove, D.L. Consistent decrease in global DNA methylation and hydroxymethylation in the hippocampus of Alzheimer’s disease patients. Neurobiol. Aging 2013, 34, 2091–2099. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bayraktar, G.; Kreutz, M.R. Neuronal DNA Methyltransferases: Epigenetic Mediators between Synaptic Activity and Gene Expression? Neuroscientist 2017, 24, 171–185. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cui, D.; Xu, X. DNA methyltransferases, DNA methylation and age-associated cognitive function. Int. J. Mol. Sci. 2018, 19, 1315. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Davinelli, S.; Calabrese, V.; Zella, D.; Scarpagnini, G. Epigenetic nutraceutical diets in Alzheimer’s disease. J. Nutr. Health Aging 2014, 18, 800–805. [Google Scholar] [CrossRef] [PubMed]
- Vizán, P.; Di Croce, L.; Aranda, S. Functional and pathological roles of AHCY. Front. Cell Dev. Biol. 2021, 9, 654344. [Google Scholar] [CrossRef]
- Ponnulari, V.K.; Estéve, P.-O.; Ruse, C.I.; Pradhan, S. S-adenosylhomocysteine hydrolase partcipitas in DNA methylation inheritance. J. Mol. Biol. 2018, 430, 2051–2065. [Google Scholar] [CrossRef]
- Ghazi, T.; Arumugam, T.; Foolchand, A.; Chuturgoon, A. The impact of natural dietary compounds on food-borne mycotoxins on DNA methylation and cancer. Cells 2020, 9, 2004. [Google Scholar] [CrossRef]
- Zhou, J.; Yang, L.; Zhong, T.; Mueller, M.; Men, Y.; Zhang, N.; Xie, J.; Giang, K.; Chung, H.; Sun, X.; et al. H19IncRNA alters DNA methylation genome wide by regulating S-adenosylhomocysteine hydroase. Nat. Commun. 2015, 6, 10221. [Google Scholar] [CrossRef] [Green Version]
- Pukhalskaia, A.E.; Diatlova, A.S.; Linkova, B.S.; Kvetnoy, I.M. Sirtuins: Role in the regulation of oxidative stress and the pathogenesis of neurodegenerative disorders. Neurosci. Behav. Physiol. 2022, 52, 164–174. [Google Scholar] [CrossRef]
- Grabowska, W.; Sikora, E.; Bielak-Zmijewska, A. Sirtuins, a promising traget in slowing down the ageing process. Biogerontology 2017, 18, 447–476. [Google Scholar] [CrossRef] [Green Version]
- Donmez, G. The effects of SIRT1 on Alzheimer’s Disease models. Int. J. Alzheimers Dis. 2012, 2012, 509529. [Google Scholar]
- Dai, H.; Sinclair, D.A.; Ellis, J.L.; Steegborn, C. Sirtuin activators and inhibitors: Promises, achievements and challenges. Pharm. Ther. 2018, 188, 140–154. [Google Scholar] [CrossRef]
- Herrschaft, H.; Nacu, A.; Likhachev, S.; Sholomov, I.; Hoerr, R.; Xhlaefke, S. Ginkgo biloba extract EGb 761® in dementia with neuropsychiatric features: A randomised, placebo-controlled trial to confirm the efficacy and safety of a daily dose of 240 mg. J. Psychiatr. Res. 2012, 46, 716–723. [Google Scholar] [CrossRef]
- Kandiah, N.; Ong, P.A.; Yuda, T.; Ng, L.L.; Mamun, K.; Merchant, R.A.; Chen, C.; Dominguez, J.; Marasigan, S.; Ampil, E.; et al. Treatment of dementia and mild cognitive impairment with or without cerebrovascular disease: Expert consensus on the use of Ginkgo biloba extract, EGb 761®. CNS Neurosci. Ther. 2019, 25, 288–298. [Google Scholar] [CrossRef] [Green Version]
- Longpré, F.; Garneau, P.; Christen, Y.; Ramassamy, C. Protection by EGb 761 against beta-amyloid-induced neurotoxicity: Involvement of NF-kappaB, SIRT1, and MAPKs pathways and inhibition of amyloid fibril formation. Free Radic. Biol. Med. 2006, 41, 1781–1794. [Google Scholar] [CrossRef]
- Lagouge, M.; Argmann, C.; Gerhart-Hines, Z.; Meziane, H.; Lerin, C.; Daussin, F.; Messadeq, N.; Milne, J.; Lambert, P.; Elliott, P.; et al. Resveratrol improves mitochondrial function and protects against metabolic disease by activating SIRT1 and PGC 1alpha. Cell 2006, 127, 1109–1122. [Google Scholar] [CrossRef]
- Gräff, J.; Tsai, L.H. Histone acetylation: Molecular mnemonics on the chromatin. Nat. Rev. Neurosci. 2013, 14, 97–111. [Google Scholar] [CrossRef]
- Manjula, R.; Anuja, K.; Alcain, F.J. SIRT1 and SIRT2 Activity Control in Neurodegenerative Diseases. Front. Pharmacol. 2021, 11, 585821. [Google Scholar] [CrossRef]
- Kim, D.; Nguyen, M.D.; Dobbin, M.M.; Fischer, A.; Sananbenesi, F.; Rodgers, J.T.; Delalle, I.; Baur, J.A.; Sui, G.; Armour, S.M.; et al. SIRT1 deacetylase protects against neurodegeneration in models for Alzheimer’s disease and amyotrophic lateral sclerosis. EMBO J. 2007, 26, 3169–3179. [Google Scholar] [CrossRef] [Green Version]
- Brunet, A.; Sweeney, L.B.; Sturgill, J.F.; Chua, K.F.; Greer, P.L.; Lin, Y.; Tran, H.; Ross, S.E.; Mostoslavsky, R.; Cohen, H.Y.; et al. Stress-dependent regulation of FOXO transcription factors by the SIRT1 deacetylase. Science 2004, 303, 2011–2015. [Google Scholar] [CrossRef] [Green Version]
- Menssen, A.; Hydbring, P.; Kapelle, K.; Vervoorts, J.; Diebold, J.; Lüscher, B.; Larsson, L.G.; Hermeking, H. The c-MYC oncoprotein, the NAMPT enzyme, the SIRT-inhibitor DBC1, and the SIRT1 deacetylase form a positive feedbach loop. Proc. Natl. Acad. Sci. USA 2012, 109, 187–196. [Google Scholar] [CrossRef] [Green Version]
- Stein, L.R.; Imai, S.I. Specific ablation of Nampt in adult neural stem cells recapitulates their functional defects during aging. EMBO J. 2014, 33, 1321–1340. [Google Scholar] [CrossRef] [Green Version]
- Imai, S.-C.; Guarente, L. It takes two to tango: NAD+ and sirtuins in aging/longevity control. NPJ Aging Mech. Dis. 2016, 2, 16017. [Google Scholar] [CrossRef] [Green Version]
- Ghosh, D.; Levault, K.R.; Brewer, G.J. Relative importance of redox buffers GSH and NAD(P)H in age-related neurodegeneration and Alzheimer’s disease-like mouse neurons. Aging Cell 2014, 13, 631–640. [Google Scholar] [CrossRef]
- Imai, S.-I.; Guarante, L. NAD+ and sirtuins in aging and disease. Trends Cell Biol. 2014, 24, 464–471. [Google Scholar] [CrossRef]
- Burdge, G.; Lillycrop, K. Fatty acids and epigenetics. Curr. Opin. Clin. Nutr. Metab. Care 2014, 17, 156–161. [Google Scholar] [CrossRef]
- Mohajeri, M.; Troesch, B.; Weber, P. Inadequate supply of vitamins and DHA in the elderly: Implications for brain aging and Alzheimer-type dementia. Nutrition 2015, 31, 261–275. [Google Scholar] [CrossRef] [Green Version]
- Cossío, F.P.; Esteller, M.; Berdasco, M. Towards a more precise therapy in cancer: Exploring epigenetic complexity. Curr. Opin. Chem. Biol. 2020, 57, 41–49. [Google Scholar] [CrossRef]
- Furtado, C.L.; Luciano, M.C.; Santos, R.; Furtado, G.; Moraes, M.O.; Pessoa, C. Epidrugs: Targeting epigenetic marks in cancer treatment. Epigenetics 2019, 14, 1164–1176. [Google Scholar] [CrossRef]
- de los Santos, M.; Martinez-Iglesias, O.; Aranda, A. Anti-estrogenic actions of histone deacetylase inhibitors in MCF-7 breast cancer cells. Endocr. Relat. Cancer 2007, 14, 1021–1028. [Google Scholar] [CrossRef] [Green Version]
- Mota, M.; Porrini, V.; Parrella, E.; Benarese, M.; Bellucci, A.; Rhein, S.; Schwaninger, M.; Pizzi, M. Neuroprotective epi-drugs quench the inflammatory response and microglial/macrophage activation in a mouse model of permanent brain ischemia. J. Neuroinflamm. 2020, 17, 161. [Google Scholar] [CrossRef]
- Dragicevic, N.; Smith, A.; Lin, X.; Yuan, F.; Copes, N.; Delic, V.; Tan, J.; Cao, C.; Shytle, R.D.; Bradshaw, P.C. Green tea epigallocatechin-3-gallate (EGCG) and other flavonoids reduce Alzheimer’s amyloid-induced mitochondrial dysfunction. J. Alzheimers Dis. 2011, 26, 507–521. [Google Scholar] [CrossRef]
- Zhang, X.; Wu, M.; Lu, F.; Luo, N.; He, Z.-P.; Yang, H. Involvement of alpha7 nAChR signaling cascade in epigallocatechin gallate suppression of beta-amyloid-induced apoptotic cortical neuronal insults. Mol. Neurobiol. 2014, 49, 66–77. [Google Scholar] [CrossRef]
- Sood, P.K.; Nahar, U.; Nehru, B. Curcumin attenuates aluminium induced oxidative stress and mitochondrial dysfunction in rat brain. Neurotox. Res. 2011, 20, 351–361. [Google Scholar] [CrossRef]
- Palomera-ávalos, V.; Griñán-Ferré, C.; Izquierdo, V.; Camins, A.; Sanfeliu, C.; Pallás, M. Metabolic Stress Induces Cognitive Disturbances and Inflammation in Aged Mice: Protective Role of Resveratrol. Rejuvenation Res. 2017, 10, 202–217. [Google Scholar] [CrossRef]
- Roboz, G.J. Epigenetic targeting and personalized approaches for AML. Hematology 2014, 44, 51. [Google Scholar] [CrossRef]
- Martínez-Iglesias, O.; Carrera, I.; Naidoo, V. Cacabelos AntiGan: An epinutraceutical bioproduct with antitumor properties in cultured cell lines. Life 2022, 12, 97. [Google Scholar] [CrossRef]
- Peedicayil, J. Epigenetic drugs for multiple sclerosis. Curr. Neuropharmacol. 2016, 14, 3–9. [Google Scholar] [CrossRef] [Green Version]
- Singh, P.; Konar, A.; Kumar, A.; Srivas, S.; Thakur, M.K. Hippocampal chromatin-modifying enzimes are pivotal for scopolamine-induced synaptic plasticity gene expression changes and memory impairment. J. Neurochem. 2015, 134, 642–651. [Google Scholar] [CrossRef]
- Atlante, A.; Amadoro, G.; Bobba, A.; Latina, V. Functional foods: An approach to modulate molecular mechanisms of Alzheimer’s disease. Cells 2020, 9, 2347. [Google Scholar] [CrossRef] [PubMed]
- May, B.H.; Feng, M.; Hyde, A.J.; Hügel, H.; Chang, S.Y.; Dong, L.; Guo, X.; Zhang, A.L.; Lu, C.; Xue, C.C. Comparisons between traditional medicines and pharmacotherapies for Alzheimer disease: A systematic review and meta-analysis of cognitive outcomes. Int. J. Geriatr. Psychiatry 2018, 33, 449–458. [Google Scholar] [CrossRef] [PubMed]
- Fan, F.; Liu, H.; Shi, X.; Ai, Y.; Liu, Q.; Cheng, Y. The efficacy and safety of Alzheimer’s Disease Therapies: An updated umbrella review. J. Alzheimer Dis. 2022, 85, 1195–1204. [Google Scholar] [CrossRef]
- Reinhoud, N.J.; Brouwer, H.-J.; Heerwaarden, L.M.; Korte-Bows, G.A.H. Analysis of Glutamate, GABA, Noradrenaline, Dopamine, Serotonin and Metabolites using microbore UHPLC with Electrochemical Detection. ACS Chem. Neurosci. 2013, 15, 888–894. [Google Scholar] [CrossRef] [Green Version]
- Yamatodani, A.; Fukuda, H.; Wada, H.; Iwaewda, T.; Watanabe, T. High-performance liquid chromatographic determination of plasma and brain histamine without previous purification of biological samples: Cation-exchange chromatography coupled with post-column derivatization fluorometry. J. Chromatogr. 1985, 344, 115–123. [Google Scholar] [CrossRef]
- Allison, J.; Kaliszewska, A.; Uceda, S.; Reiriz, M.; Arias, N. Targeting DNA methylation in the adult brain through diet. Nutrients 2021, 13, 3979. [Google Scholar] [CrossRef]
- Littlejohns, T.J.; Henley, W.E.; Lang, I.A.; Annweiler, C.; Beauchet, O.; Chaves, P.H.; Fried, L.; Kestenbaum, B.R.; Kuller, L.H.; Langa, K.M.; et al. Vitamin D and the risk of dementia and Alzheimer’s disease. Neurology 2014, 83, 920–928. [Google Scholar] [CrossRef]
- Joven, J.; Micol, V.; Segura-Carretero, A.; Alonso-Villaverde, C.; Menéndez, C. Biocative Food Complements Platform. Polyphenols and the modulation of gene expression pathways: Can we eat our way out of the danger of chronic disease? Crit. Rev. Food Sci. Nutr. 2014, 54, 985–1001. [Google Scholar] [CrossRef]
- Bollati, V.; Favero, C.; Albetti, B.; Tarantini, L.; Moroni, A.; Byun, H.M.; Motta, V.; Conti, D.M.; Tirelli, A.S.; Vigna, L.; et al. Nutrients intake is associated with DNA methylation of candidate inflammatory genes in a population of obese subjects. Nutrients 2014, 6, 4625–4639. [Google Scholar] [CrossRef] [Green Version]
- Cacabelos, R. Have there been improvements in Alzheimer’s disease drug discovery over the past 5 years? Expert Opin. Drug Discov. 2018, 13, 523–538. [Google Scholar] [CrossRef]
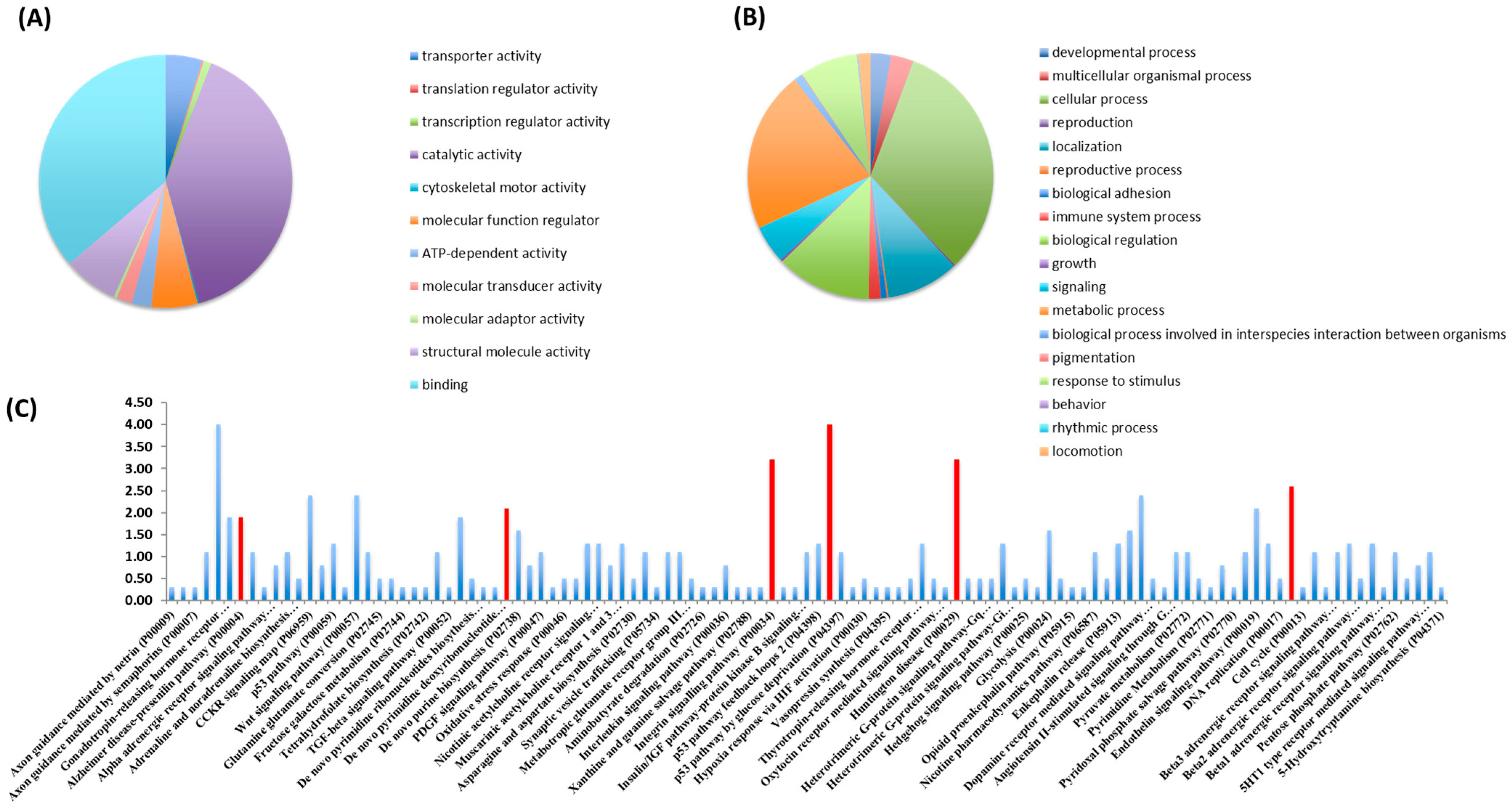








| Protein | Functions | Ref. |
|---|---|---|
| Aβ A4 | Cell surface receptor. Regulates neurite growth, neuronal adhesion and axonogenesis. Promotes synaptogenesis and contributes to cell mobility and regulation of transcription. Couples to apoptosis-inducing pathways. | [35] |
| Adenosylhomocysteinase | Competitively inhibits S-adenosyl-L-methionine-dependent methyl transferase reactions. Regulates and maintains DNA methylation by controlling intracellular adenosylhomocysteine concentrations. | [36] |
| Apolipoprotein E | Lipid-transporting glycoprotein. Involved in the production, conversion and clearance of plasma lipoproteins. Three isoforms confer risk (ε2 < ε3 < ε4) for developing AD. Regulates clearance of misfolded proteins, synaptic function, inflammation and neurogenesis. | [37] |
| Cathepsin D | Lysosomal protease. Mediates protein degradation and turnover. Regulates autophagy and neuroinflammation, and implicated in AD pathogenesis. | [38] |
| Choline O-acetyltransferase | Enzyme that catalyzes the synthesis of acetylcholine in cholinergic neurons. Loss of cholinergic neurons in the nucleus basalis of Meynert is a feature of AD. | [39] |
| Neuroendocrine protein 7B2 | Blocks aggregation of certain secreted proteins. Inhibits α-synuclein aggregation and suppresses Aβ1-40, Aβ1-42 fibrillation in the AD brain. | [40] |
| Nicotinamide phosphoribosyltransferase | Rate-limiting enzyme in the NAD+ pathway. Regulates NAD+ synthesis, thereby contributing to energy metabolism. Reduced NAD+ levels in the brain are prevalent with increasing age, and contribute to AD pathophysiology. NAD+ levels and NAMPT expression are low in the hippocampus and cortex in mice with early stage AD than in wild-type animals. | [41] |
| Presenilin-2 | Catalytic subunit of the γ-secretase complex that processes APP. One of three genes in which mutations cause early onset AD. Negatively regulates apoptosis. | [42] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Martínez-Iglesias, O.; Naidoo, V.; Carrera, I.; Corzo, L.; Cacabelos, R. Nosustrophine: An Epinutraceutical Bioproduct with Effects on DNA Methylation, Histone Acetylation and Sirtuin Expression in Alzheimer’s Disease. Pharmaceutics 2022, 14, 2447. https://doi.org/10.3390/pharmaceutics14112447
Martínez-Iglesias O, Naidoo V, Carrera I, Corzo L, Cacabelos R. Nosustrophine: An Epinutraceutical Bioproduct with Effects on DNA Methylation, Histone Acetylation and Sirtuin Expression in Alzheimer’s Disease. Pharmaceutics. 2022; 14(11):2447. https://doi.org/10.3390/pharmaceutics14112447
Chicago/Turabian StyleMartínez-Iglesias, Olaia, Vinogran Naidoo, Iván Carrera, Lola Corzo, and Ramón Cacabelos. 2022. "Nosustrophine: An Epinutraceutical Bioproduct with Effects on DNA Methylation, Histone Acetylation and Sirtuin Expression in Alzheimer’s Disease" Pharmaceutics 14, no. 11: 2447. https://doi.org/10.3390/pharmaceutics14112447
APA StyleMartínez-Iglesias, O., Naidoo, V., Carrera, I., Corzo, L., & Cacabelos, R. (2022). Nosustrophine: An Epinutraceutical Bioproduct with Effects on DNA Methylation, Histone Acetylation and Sirtuin Expression in Alzheimer’s Disease. Pharmaceutics, 14(11), 2447. https://doi.org/10.3390/pharmaceutics14112447








