Biological Features of Nanoparticles: Protein Corona Formation and Interaction with the Immune System
Abstract
:1. Introduction
2. The Protein Corona
2.1. General Features
2.2. Intrinsic NPs Properties That Affect the PC Formation
2.3. Extrinsic Factors which Affect the PC Formation
3. The Interaction of Nanoparticles with Components of Immune System
3.1. Opsonins
3.1.1. The Complement System
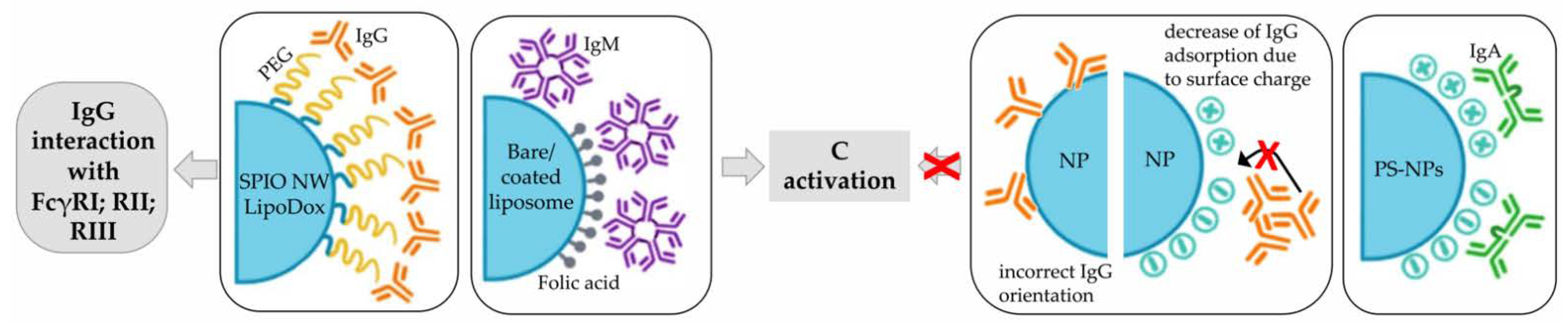
3.1.2. Immunoglobulins
3.1.3. Fibrinogen
3.2. Dysopsonins
3.2.1. HRG
3.2.2. Apolipoproteins
3.2.3. Albumin
3.2.4. SP-A and SP-D
3.3. Innate Immune System
3.3.1. Macrophages
3.3.2. Dendritic Cells
3.3.3. Neutrophils
3.4. Adaptive Immune System
3.4.1. B Lymphocytes
3.4.2. T Lymphocytes
4. Strategies to Evade Immune System Activation
5. Conclusions and Perspectives
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Viseu, A. Nanomedicine; Encyclopedia Britannica: Chicago, IL, USA, 2020. [Google Scholar]
- Gadekar, V.; Borade, Y.; Kannaujia, S.; Rajpoot, K.; Anup, N.; Tambe, V.; Kalia, K.; Tekade, R.K. Nanomedicines Accessible in the Market for Clinical Interventions. J. Control. Release 2021, 330, 372–397. [Google Scholar] [CrossRef]
- Doane, T.L.; Burda, C. The Unique Role of Nanoparticles in Nanomedicine: Imaging, Drug Delivery and Therapy. Chem. Soc. Rev. 2012, 41, 2885. [Google Scholar] [CrossRef]
- Mitchell, M.J.; Billingsley, M.M.; Haley, R.M.; Wechsler, M.E.; Peppas, N.A.; Langer, R. Engineering Precision Nanoparticles for Drug Delivery. Nat. Rev. Drug Discov. 2021, 20, 101–124. [Google Scholar] [CrossRef]
- DiMasi, J.A.; Grabowski, H.G.; Hansen, R.W. The Cost of Drug Development. N. Engl. J. Med. 2015, 372, 1972. [Google Scholar] [CrossRef]
- Metselaar, J.M.; Lammers, T. Challenges in Nanomedicine Clinical Translation. Drug Deliv. Transl. Res. 2020, 10, 721–725. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tenzer, S.; Docter, D.; Kuharev, J.; Musyanovych, A.; Fetz, V.; Hecht, R.; Schlenk, F.; Fischer, D.; Kiouptsi, K.; Reinhardt, C.; et al. Rapid Formation of Plasma Protein Corona Critically Affects Nanoparticle Pathophysiology. Nat. Nanotechnol. 2013, 8, 772–781. [Google Scholar] [CrossRef]
- Chen, F.; Wang, G.; Griffin, J.I.; Brenneman, B.; Banda, N.K.; Holers, V.M.; Backos, D.S.; Wu, L.; Moghimi, S.M.; Simberg, D. Complement Proteins Bind to Nanoparticle Protein Corona and Undergo Dynamic Exchange in Vivo. Nat. Nanotechnol. 2017, 12, 387–393. [Google Scholar] [CrossRef] [Green Version]
- Cai, R.; Ren, J.; Ji, Y.; Wang, Y.; Liu, Y.; Chen, Z.; Farhadi Sabet, Z.; Wu, X.; Lynch, I.; Chen, C. Corona of Thorns: The Surface Chemistry-Mediated Protein Corona Perturbs the Recognition and Immune Response of Macrophages. ACS Appl. Mater. Interfaces 2020, 12, 1997–2008. [Google Scholar] [CrossRef]
- Wang, H.; Ma, R.; Nienhaus, K.; Nienhaus, G.U. Formation of a Monolayer Protein Corona around Polystyrene Nanoparticles and Implications for Nanoparticle Agglomeration. Small 2019, 15, 1900974. [Google Scholar] [CrossRef]
- Fam, S.Y.; Chee, C.F.; Yong, C.Y.; Ho, K.L.; Mariatulqabtiah, A.R.; Tan, W.S. Stealth Coating of Nanoparticles in Drug-Delivery Systems. Nanomaterials 2020, 10, 787. [Google Scholar] [CrossRef]
- Pinals, R.L.; Chio, L.; Ledesma, F.; Landry, M.P. Engineering at the Nano-Bio Interface: Harnessing the Protein Corona towards Nanoparticle Design and Function. Analyst 2020, 145, 5090–5112. [Google Scholar] [CrossRef]
- Aoyama, M.; Hata, K.; Higashisaka, K.; Nagano, K.; Yoshioka, Y.; Tsutsumi, Y. Clusterin in the Protein Corona Plays a Key Role in the Stealth Effect of Nanoparticles against Phagocytes. Biochem. Biophys. Res. Commun. 2016, 480, 690–695. [Google Scholar] [CrossRef] [PubMed]
- Schöttler, S.; Becker, G.; Winzen, S.; Steinbach, T.; Mohr, K.; Landfester, K.; Mailänder, V.; Wurm, F.R. Protein Adsorption Is Required for Stealth Effect of Poly(Ethylene Glycol)- and Poly(Phosphoester)-Coated Nanocarriers. Nat. Nanotechnol. 2016, 11, 372–377. [Google Scholar] [CrossRef] [PubMed]
- Oh, J.Y.; Kim, H.S.; Palanikumar, L.; Go, E.M.; Jana, B.; Park, S.A.; Kim, H.Y.; Kim, K.; Seo, J.K.; Kwak, S.K.; et al. Cloaking Nanoparticles with Protein Corona Shield for Targeted Drug Delivery. Nat. Commun. 2018, 9, 4548. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, H.; Wang, Y.; Tang, Q.; Yin, D.; Tang, C.; He, E.; Zou, L.; Peng, Q. The Protein Corona and Its Effects on Nanoparticle-Based Drug Delivery Systems. Acta Biomater. 2021, 129, 57–72. [Google Scholar] [CrossRef]
- Walkey, C.D.; Chan, W.C.W. Understanding and Controlling the Interaction of Nanomaterials with Proteins in a Physiological Environment. Chem. Soc. Rev. 2012, 41, 2780–2799. [Google Scholar] [CrossRef]
- Kihara, S.; van der Heijden, N.J.; Seal, C.K.; Mata, J.P.; Whitten, A.E.; Köper, I.; McGillivray, D.J. Soft and Hard Interactions between Polystyrene Nanoplastics and Human Serum Albumin Protein Corona. Bioconjugate Chem. 2019, 30, 1067–1076. [Google Scholar] [CrossRef]
- Raoufi, M.; Hajipour, M.J.; Kamali Shahri, S.M.; Schoen, I.; Linn, U.; Mahmoudi, M. Probing Fibronectin Conformation on a Protein Corona Layer around Nanoparticles. Nanoscale 2018, 10, 1228–1233. [Google Scholar] [CrossRef]
- Vroman, L.; Adams, A.; Fischer, G.; Munoz, P. Interaction of High Molecular Weight Kininogen, Factor XII, and Fibrinogen in Plasma at Interfaces. Blood 1980, 55, 156–159. [Google Scholar] [CrossRef] [Green Version]
- Hu, Z.; Zhang, H.; Zhang, Y.; Wu, R.; Zou, H. Nanoparticle Size Matters in the Formation of Plasma Protein Coronas on Fe3O4 Nanoparticles. Colloids Surf. B Biointerfaces 2014, 121, 354–361. [Google Scholar] [CrossRef]
- Lundqvist, M.; Stigler, J.; Elia, G.; Lynch, I.; Cedervall, T.; Dawson, K.A. Nanoparticle Size and Surface Properties Determine the Protein Corona with Possible Implications for Biological Impacts. Proc. Natl. Acad. Sci. USA 2008, 105, 14265–14270. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Papini, E.; Tavano, R.; Mancin, F. Opsonins and Dysopsonins of Nanoparticles: Facts, Concepts, and Methodological Guidelines. Front. Immunol. 2020, 11, 567365. [Google Scholar] [CrossRef] [PubMed]
- Marichal, L.; Klein, G.; Armengaud, J.; Boulard, Y.; Chédin, S.; Labarre, J.; Pin, S.; Renault, J.-P.; Aude, J.-C. Protein Corona Composition of Silica Nanoparticles in Complex Media: Nanoparticle Size Does Not Matter. Nanomaterials 2020, 10, 240. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moustaoui, H.; Saber, J.; Djeddi, I.; Liu, Q.; Movia, D.; Prina-Mello, A.; Spadavecchia, J.; Lamy de la Chapelle, M.; Djaker, N. A Protein Corona Study by Scattering Correlation Spectroscopy: A Comparative Study between Spherical and Urchin-Shaped Gold Nanoparticles. Nanoscale 2019, 11, 3665–3673. [Google Scholar] [CrossRef]
- García-Álvarez, R.; Hadjidemetriou, M.; Sánchez-Iglesias, A.; Liz-Marzán, L.M.; Kostarelos, K. In Vivo Formation of Protein Corona on Gold Nanoparticles. The Effect of Their Size and Shape. Nanoscale 2018, 10, 1256–1264. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bewersdorff, T.; Glitscher, E.A.; Bergueiro, J.; Eravci, M.; Miceli, E.; Haase, A.; Calderón, M. The Influence of Shape and Charge on Protein Corona Composition in Common Gold Nanostructures. Mater. Sci. Eng. C 2020, 117, 111270. [Google Scholar] [CrossRef] [PubMed]
- Gessner, A.; Lieske, A.; Paulke, B.-R.; Müller, R.H. Functional Groups on Polystyrene Model Nanoparticles: Influence on Protein Adsorption: Influence of Functional Groups on Protein. J. Biomed. Mater. Res. 2003, 65A, 319–326. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.K.; Choi, E.-J.; Webster, T.J.; Kim, S.-H.; Khang, D. Effect of the Protein Corona on Nanoparticles for Modulating Cytotoxicity and Immunotoxicity. Int. J. Nanomed. 2015, 10, 97–113. [Google Scholar]
- Nierenberg, D.; Khaled, A.R.; Flores, O. Formation of a Protein Corona Influences the Biological Identity of Nanomaterials. Rep. Pract. Oncol. Radiother. 2018, 9, 300–308. [Google Scholar] [CrossRef]
- Bewersdorff, T.; Gruber, A.; Eravci, M.; Dumbani, M.; Klinger, D.; Haase, A. Amphiphilic Nanogels: Influence of Surface Hydrophobicity on Protein Corona, Biocompatibility and Cellular Uptake. IJN 2019, 14, 7861–7878. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lu, X.; Xu, P.; Ding, H.-M.; Yu, Y.-S.; Huo, D.; Ma, Y.-Q. Tailoring the Component of Protein Corona via Simple Chemistry. Nat. Commun. 2019, 10, 4520. [Google Scholar] [CrossRef] [PubMed]
- Prawatborisut, M.; Oberländer, J.; Jiang, S.; Graf, R.; Avlasevich, Y.; Morsbach, S.; Crespy, D.; Mailänder, V.; Landfester, K. Temperature-Responsive Nanoparticles Enable Specific Binding of Apolipoproteins from Human Plasma. Small 2022, 18, 2103138. [Google Scholar] [CrossRef] [PubMed]
- Piloni, A.; Wong, C.K.; Chen, F.; Lord, M.; Walther, A.; Stenzel, M.H. Surface Roughness Influences the Protein Corona Formation of Glycosylated Nanoparticles and Alter Their Cellular Uptake. Nanoscale 2019, 11, 23259–23267. [Google Scholar] [CrossRef] [PubMed]
- Clemments, A.M.; Botella, P.; Landry, C.C. Protein Adsorption From Biofluids on Silica Nanoparticles: Corona Analysis as a Function of Particle Diameter and Porosity. ACS Appl. Mater. Interfaces 2015, 7, 21682–21689. [Google Scholar] [CrossRef] [Green Version]
- Walkey, C.D.; Olsen, J.B.; Guo, H.; Emili, A.; Chan, W.C.W. Nanoparticle Size and Surface Chemistry Determine Serum Protein Adsorption and Macrophage Uptake. J. Am. Chem. Soc. 2012, 134, 2139–2147. [Google Scholar] [CrossRef]
- Givens, B.E.; Wilson, E.; Fiegel, J. The Effect of Salts in Aqueous Media on the Formation of the BSA Corona on SiO2 Nanoparticles. Colloids Surf. B Biointerfaces 2019, 179, 374–381. [Google Scholar] [CrossRef]
- Wang, W.; Huang, Z.; Li, Y.; Wang, W.; Shi, J.; Fu, F.; Huang, Y.; Pan, X.; Wu, C. Impact of Particle Size and PH on Protein Corona Formation of Solid Lipid Nanoparticles: A Proof-of-Concept Study. Acta Pharm. Sin. B 2021, 11, 1030–1046. [Google Scholar] [CrossRef] [PubMed]
- Tonigold, M.; Simon, J.; Estupiñán, D.; Kokkinopoulou, M.; Reinholz, J.; Kintzel, U.; Kaltbeitzel, A.; Renz, P.; Domogalla, M.P.; Steinbrink, K.; et al. Pre-Adsorption of Antibodies Enables Targeting of Nanocarriers despite a Biomolecular Corona. Nat. Nanotechnol. 2018, 13, 862–869. [Google Scholar] [CrossRef] [PubMed]
- Simon, J.; Müller, J.; Ghazaryan, A.; Morsbach, S.; Mailänder, V.; Landfester, K. Protein Denaturation Caused by Heat Inactivation Detrimentally Affects Biomolecular Corona Formation and Cellular Uptake. Nanoscale 2018, 10, 21096–21105. [Google Scholar] [CrossRef] [Green Version]
- Weiss, A.C.G.; Kelly, H.G.; Faria, M.; Besford, Q.A.; Wheatley, A.K.; Ang, C.-S.; Crampin, E.J.; Caruso, F.; Kent, S.J. Link between Low-Fouling and Stealth: A Whole Blood Biomolecular Corona and Cellular Association Analysis on Nanoengineered Particles. ACS Nano 2019, 13, 4980–4991. [Google Scholar] [CrossRef] [PubMed]
- Galbiati, V.; Cornaghi, L.; Gianazza, E.; Potenza, M.A.; Donetti, E.; Marinovich, M.; Corsini, E. In Vitro Assessment of Silver Nanoparticles Immunotoxicity. Food Chem. Toxicol. 2018, 112, 363–374. [Google Scholar] [CrossRef]
- Oberländer, J.; Champanhac, C.; da Costa Marques, R.; Landfester, K.; Mailänder, V. Temperature, Concentration, and Surface Modification Influence the Cellular Uptake and the Protein Corona of Polystyrene Nanoparticles. Acta Biomater. 2022, 148, 271–278. [Google Scholar] [CrossRef] [PubMed]
- Braun, N.J.; DeBrosse, M.C.; Hussain, S.M.; Comfort, K.K. Modification of the Protein Corona–Nanoparticle Complex by Physiological Factors. Mater. Sci. Eng. C 2016, 64, 34–42. [Google Scholar] [CrossRef] [Green Version]
- Palchetti, S.; Pozzi, D.; Capriotti, A.L.; Barbera, G.L.; Chiozzi, R.Z.; Digiacomo, L.; Peruzzi, G.; Caracciolo, G.; Laganà, A. Influence of Dynamic Flow Environment on Nanoparticle-Protein Corona: From Protein Patterns to Uptake in Cancer Cells. Colloids Surf. B Biointerfaces 2017, 153, 263–271. [Google Scholar] [CrossRef] [PubMed]
- Weiss, A.C.G.; Kempe, K.; Förster, S.; Caruso, F. Microfluidic Examination of the “Hard” Biomolecular Corona Formed on Engineered Particles in Different Biological Milieu. Biomacromolecules 2018, 19, 2580–2594. [Google Scholar] [CrossRef]
- Saha, K.; Rahimi, M.; Yazdani, M.; Kim, S.T.; Moyano, D.F.; Hou, S.; Das, R.; Mout, R.; Rezaee, F.; Mahmoudi, M.; et al. Regulation of Macrophage Recognition through the Interplay of Nanoparticle Surface Functionality and Protein Corona. ACS Nano 2016, 10, 4421–4430. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tavano, R.; Gabrielli, L.; Lubian, E.; Fedeli, C.; Visentin, S.; Polverino De Laureto, P.; Arrigoni, G.; Geffner-Smith, A.; Chen, F.; Simberg, D.; et al. C1q-Mediated Complement Activation and C3 Opsonization Trigger Recognition of Stealth Poly(2-Methyl-2-Oxazoline)-Coated Silica Nanoparticles by Human Phagocytes. ACS Nano 2018, 12, 5834–5847. [Google Scholar] [CrossRef] [PubMed]
- Lima, T.; Bernfur, K.; Vilanova, M.; Cedervall, T. Understanding the Lipid and Protein Corona Formation on Different Sized Polymeric Nanoparticles. Sci. Rep. 2020, 10, 1129. [Google Scholar] [CrossRef] [Green Version]
- Müller, L.K.; Simon, J.; Rosenauer, C.; Mailänder, V.; Morsbach, S.; Landfester, K. The Transferability from Animal Models to Humans: Challenges Regarding Aggregation and Protein Corona Formation of Nanoparticles. Biomacromolecules 2018, 19, 374–385. [Google Scholar] [CrossRef]
- González-García, L.E.; MacGregor, M.N.; Visalakshan, R.M.; Lazarian, A.; Cavallaro, A.A.; Morsbach, S.; Mierczynska-Vasilev, A.; Mailänder, V.; Landfester, K.; Vasilev, K. Nanoparticles Surface Chemistry Influence on Protein Corona Composition and Inflammatory Responses. Nanomaterials 2022, 12, 682. [Google Scholar] [CrossRef]
- Fedeli, C.; Segat, D.; Tavano, R.; Bubacco, L.; Franceschi, G.D.; de Laureto, P.; Lubian, E.; Selvestrel, F.; Mancin, F. The Functional Dissection of the Plasma Corona of SiO2-NPs Spots Histidine Rich Glycoprotein as a Major Player Able to Hamper Nanoparticles Capture by Macrophages. Nanoscale 2015, 7, 17710–17728. [Google Scholar] [CrossRef] [PubMed]
- Partikel, K.; Korte, R.; Mulac, D.; Humpf, H.-U.; Langer, K. Serum Type and Concentration Both Affect the Protein-Corona Composition of PLGA Nanoparticles. Beilstein J. Nanotechnol. 2019, 10, 1002–1015. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ernst, L.; Casals, E.; Italiani, P.; Boraschi, D.; Puntes, V. The Interactions between Nanoparticles and the Innate Immune System from a Nanotechnologist Perspective. Nanomaterials 2021, 11, 2991. [Google Scholar] [CrossRef] [PubMed]
- Botto, M.; Kirschfink, M.; Macor, P.; Pickering, M.C.; Würzner, R.; Tedesco, F. Complement in Human Diseases: Lessons from Complement Deficiencies. Mol. Immunol. 2009, 46, 2774–2783. [Google Scholar] [CrossRef] [PubMed]
- Macor, P.; Capolla, S.; Tedesco, F. Complement as a Biological Tool to Control Tumor Growth. Front. Immunol. 2018, 9, 2203. [Google Scholar] [CrossRef] [PubMed]
- Macor, P.; Tedesco, F. Complement as Effector System in Cancer Immunotherapy. Immunol. Lett. 2007, 111, 6–13. [Google Scholar] [CrossRef]
- Fornasier, M.; Biffi, S.; Bortot, B.; Macor, P.; Manhart, A.; Wurm, F.R.; Murgia, S. Cubosomes Stabilized by a Polyphosphoester-Analog of Pluronic F127 with Reduced Cytotoxicity. J. Colloid Interface Sci. 2020, 580, 286–297. [Google Scholar] [CrossRef]
- Ding, T.; Sun, J. Formation of Protein Corona on Nanoparticle Affects Different Complement Activation Pathways Mediated by C1q. Pharm. Res. 2020, 37, 10. [Google Scholar] [CrossRef]
- Merle, N.S.; Church, S.E.; Fremeaux-Bacchi, V.; Roumenina, L.T. Complement System Part I—Molecular Mechanisms of Activation and Regulation. Front. Immunol. 2015, 6, 262. [Google Scholar] [CrossRef] [Green Version]
- Klapper, Y.; Hamad, O.A.; Teramura, Y.; Leneweit, G.; Nienhaus, G.U.; Ricklin, D.; Lambris, J.D.; Ekdahl, K.N.; Nilsson, B. Mediation of a Non-Proteolytic Activation of Complement Component C3 by Phospholipid Vesicles. Biomaterials 2014, 35, 3688–3696. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vu, V.P.; Gifford, G.B.; Chen, F.; Benasutti, H.; Wang, G.; Groman, E.V.; Scheinman, R.; Saba, L.; Moghimi, S.M.; Simberg, D. Immunoglobulin Deposition on Biomolecule Corona Determines Complement Opsonization Efficiency of Preclinical and Clinical Nanoparticles. Nat. Nanotechnol. 2019, 14, 260–268. [Google Scholar] [CrossRef]
- Prozeller, D.; Rosenauer, C.; Morsbach, S.; Landfester, K. Immunoglobulins on the Surface of Differently Charged Polymer Nanoparticles. Biointerphases 2020, 15, 031009. [Google Scholar] [CrossRef]
- Wang, H.; Ding, T.; Guan, J.; Liu, X.; Wang, J.; Jin, P.; Hou, S.; Lu, W.; Qian, J.; Wang, W.; et al. Interrogation of Folic Acid-Functionalized Nanomedicines: The Regulatory Roles of Plasma Proteins Reexamined. ACS Nano 2020, 14, 14779–14789. [Google Scholar] [CrossRef]
- Ho, Y.T.; Azman, N.A.; Loh, F.W.Y.; Ong, G.K.T.; Engudar, G.; Kriz, S.A.; Kah, J.C.Y. Protein Corona Formed from Different Blood Plasma Proteins Affects the Colloidal Stability of Nanoparticles Differently. Bioconjugate Chem. 2018, 29, 3923–3934. [Google Scholar] [CrossRef]
- Fridman, W.H. Fc Receptors and Immunoglobulin Binding Factors. FASEB J. 1991, 5, 2684–2690. [Google Scholar] [CrossRef] [Green Version]
- Deng, Z.J.; Liang, M.; Monteiro, M.; Toth, I.; Minchin, R.F. Nanoparticle-Induced Unfolding of Fibrinogen Promotes Mac-1 Receptor Activation and Inflammation. Nat. Nanotechnol. 2011, 6, 39–44. [Google Scholar] [CrossRef] [PubMed]
- Levi, M.; van der Poll, T.; Büller, H.R. Bidirectional Relation Between Inflammation and Coagulation. Circulation 2004, 109, 2698–2704. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Amara, U.; Flierl, M.A.; Rittirsch, D.; Klos, A.; Chen, H.; Acker, B.; Brückner, U.B.; Nilsson, B.; Gebhard, F.; Lambris, J.D.; et al. Molecular Intercommunication between the Complement and Coagulation Systems. J. Immunol. 2010, 185, 5628–5636. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ortega, V.A.; Bahniuk, M.S.; Memon, S.; Unsworth, L.D.; Stafford, J.L.; Goss, G.G. Polymer-Coated Nanoparticle Protein Corona Formation Potentiates Phagocytosis of Bacteria by Innate Immune Cells and Inhibits Coagulation in Human Plasma. Biointerphases 2020, 15, 051003. [Google Scholar] [CrossRef] [PubMed]
- Bahniuk, M.S.; Alshememry, A.K.; Unsworth, L.D. Human Plasma Protein Adsorption to Elastin-like Polypeptide Nanoparticles. Biointerphases 2020, 15, 021007. [Google Scholar] [CrossRef] [PubMed]
- Vogt, C.; Pernemalm, M.; Kohonen, P.; Laurent, S.; Hultenby, K.; Vahter, M.; Lehtiö, J.; Toprak, M.S.; Fadeel, B. Proteomics Analysis Reveals Distinct Corona Composition on Magnetic Nanoparticles with Different Surface Coatings: Implications for Interactions with Primary Human Macrophages. PLoS ONE 2015, 10, e0129008. [Google Scholar] [CrossRef]
- Ekstrand-Hammarström, B.; Hong, J.; Davoodpour, P.; Sandholm, K.; Ekdahl, K.N.; Bucht, A.; Nilsson, B. TiO2 Nanoparticles Tested in a Novel Screening Whole Human Blood Model of Toxicity Trigger Adverse Activation of the Kallikrein System at Low Concentrations. Biomaterials 2015, 51, 58–68. [Google Scholar] [CrossRef] [PubMed]
- Cagliani, R.; Gatto, F.; Bardi, G. Protein Adsorption: A Feasible Method for Nanoparticle Functionalization? Materials 2019, 12, 1991. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, M.; Jin, X.; Liu, T.; Fan, F.; Gao, F.; Chai, S.; Yang, L. Nanoparticle Elasticity Affects Systemic Circulation Lifetime by Modulating Adsorption of Apolipoprotein A-I in Corona Formation. Nat. Commun. 2022, 13, 4137. [Google Scholar] [CrossRef]
- Thiele, L. Competitive Adsorption of Serum Proteins at Microparticles Affects Phagocytosis by Dendritic Cells. Biomaterials 2003, 24, 1409–1418. [Google Scholar] [CrossRef] [PubMed]
- Shen, L.; Tenzer, S.; Storck, W.; Hobernik, D.; Raker, V.K.; Fischer, K.; Decker, S.; Dzionek, A.; Krauthäuser, S.; Diken, M.; et al. Protein Corona–Mediated Targeting of Nanocarriers to B Cells Allows Redirection of Allergic Immune Responses. J. Allergy Clin. Immunol. 2018, 142, 1558–1570. [Google Scholar] [CrossRef] [Green Version]
- Hamilton, K.K.; Zhao, J.; Sims, P.J. Interaction between Apolipoproteins A-I and A-II and the Membrane Attack Complex of Complement. Affinity of the Apoproteins for Polymeric C9. J. Biol. Chem. 1993, 268, 3632–3638. [Google Scholar] [CrossRef]
- McDonald, J.F.; Nelsestuen, G.L. Potent Inhibition of Terminal Complement Assembly by Clusterin: Characterization of Its Impact on C9 Polymerization. Biochemistry 1997, 36, 7464–7473. [Google Scholar] [CrossRef]
- Peters, T., Jr. All about Albumin: Biochemistry, Genetics, and Medical Applications; Academic Press: San Diego, CA, USA, 1996; ISBN 978-0-12-552110-9. [Google Scholar]
- Bozzer, S.; Dal Bo, M.; Grimaldi, M.C.; Toffoli, G.; Macor, P. Nanocarriers as a Delivery Platform for Anticancer Treatment: Biological Limits and Perspectives in B-Cell Malignancies. Pharmaceutics 2022, 14, 1965. [Google Scholar] [CrossRef]
- Ogawara, K.; Furumoto, K.; Nagayama, S.; Minato, K.; Higaki, K.; Kai, T.; Kimura, T. Pre-Coating with Serum Albumin Reduces Receptor-Mediated Hepatic Disposition of Polystyrene Nanosphere: Implications for Rational Design of Nanoparticles. J. Control. Release 2004, 100, 451–455. [Google Scholar] [CrossRef] [PubMed]
- McKenzie, Z.; Kendall, M.; Mackay, R.-M.; Whitwell, H.; Elgy, C.; Ding, P.; Mahajan, S.; Morgan, C.; Griffiths, M.; Clark, H.; et al. Surfactant Protein A (SP-A) Inhibits Agglomeration and Macrophage Uptake of Toxic Amine Modified Nanoparticles. Nanotoxicology 2015, 9, 952–962. [Google Scholar] [CrossRef] [PubMed]
- Fedeli, C.; Segat, D.; Tavano, R.; De Franceschi, G.; de Laureto, P.P.; Lubian, E.; Selvestrel, F.; Mancin, F.; Papini, E. Variations of the Corona HDL:Albumin Ratio Determine Distinct Effects of Amorphous SiO 2 Nanoparticles on Monocytes and Macrophages in Serum. Nanomedicine 2014, 9, 2481–2497. [Google Scholar] [CrossRef] [PubMed]
- Ruge, C.A.; Kirch, J.; Cañadas, O.; Schneider, M.; Perez-Gil, J.; Schaefer, U.F.; Casals, C.; Lehr, C.-M. Uptake of Nanoparticles by Alveolar Macrophages Is Triggered by Surfactant Protein A. Nanomed. Nanotechnol. Biol. Med. 2011, 7, 690–693. [Google Scholar] [CrossRef]
- Ruge, C.A.; Hillaireau, H.; Grabowski, N.; Beck-Broichsitter, M.; Cañadas, O.; Tsapis, N.; Casals, C.; Nicolas, J.; Fattal, E. Pulmonary Surfactant Protein A-Mediated Enrichment of Surface-Decorated Polymeric Nanoparticles in Alveolar Macrophages. Mol. Pharm. 2016, 13, 4168–4178. [Google Scholar] [CrossRef]
- Pondman, K.M.; Paudyal, B.; Sim, R.B.; Kaur, A.; Kouser, L.; Tsolaki, A.G.; Jones, L.A.; Salvador-Morales, C.; Khan, H.A.; ten Haken, B.; et al. Pulmonary Surfactant Protein SP-D Opsonises Carbon Nanotubes and Augments Their Phagocytosis and Subsequent pro-Inflammatory Immune Response. Nanoscale 2017, 9, 1097–1109. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kendall, M.; Ding, P. Surfactant Protein D (SP-D) Alters Cellular Uptake of Particles and Nanoparticles. Nanotoxicology 2013, 7, 963–973. [Google Scholar] [CrossRef] [PubMed]
- Capolla, S.; Garrovo, G.; Zorzet, S.; Lorenzon, A.; Rampazzo, E.; Spretz, R.; Pozzato, G.; Nunez, L.; Tripodo, C.; Macor, P.; et al. Targeted Tumor Imaging of Anti-CD20-Polymeric Nanoparticles Developed for the Diagnosis of B-Cell Malignancies. Int. J. Nanomed. 2015, 10, 4099–4109. [Google Scholar] [CrossRef] [Green Version]
- Colombo, F.; Durigutto, P.; De Maso, L.; Biffi, S.; Belmonte, B.; Tripodo, C.; Oliva, R.; Bardini, P.; Marini, G.M.; Terreno, E.; et al. Targeting CD34+ Cells of the Inflamed Synovial Endothelium by Guided Nanoparticles for the Treatment of Rheumatoid Arthritis. J. Autoimmun. 2019, 103, 102288. [Google Scholar] [CrossRef] [PubMed]
- Baboci, L.; Capolla, S.; Di Cintio, F.; Colombo, F.; Mauro, P.; Dal Bo, M.; Argenziano, M.; Cavalli, R.; Toffoli, G.; Macor, P. The Dual Role of the Liver in Nanomedicine as an Actor in the Elimination of Nanostructures or a Therapeutic Target. J. Oncol. 2020, 2020, 4638192. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Macor, P.; Durigutto, P.; Argenziano, M.; Smith-Jackson, K.; Capolla, S.; Di Leonardo, V.; Marchbank, K.; Tolva, V.S.; Semeraro, F.; Ammollo, C.T.; et al. Plasminogen Activator-Coated Nanobubbles Targeting Cell-Bound Β2-Glycoprotein I as a Novel Thrombus-Specific Thrombolytic Strategy. Haematologica 2022. [Google Scholar] [CrossRef] [PubMed]
- Waegeneers, N.; Brasseur, A.; Van Doren, E.; Van der Heyden, S.; Serreyn, P.-J.; Pussemier, L.; Mast, J.; Schneider, Y.-J.; Ruttens, A.; Roels, S. Short-Term Biodistribution and Clearance of Intravenously Administered Silica Nanoparticles. Toxicol. Rep. 2018, 5, 632–638. [Google Scholar] [CrossRef]
- Freeman, S.A.; Grinstein, S. Phagocytosis: Receptors, Signal Integration, and the Cytoskeleton. Immunol. Rev. 2014, 262, 193–215. [Google Scholar] [CrossRef] [PubMed]
- Gustafson, H.H.; Holt-Casper, D.; Grainger, D.W.; Ghandehari, H. Nanoparticle Uptake: The Phagocyte Problem. Nano Today 2015, 10, 487–510. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Anderson, J.M.; Rodriguez, A.; Chang, D.T. Foreign Body Reaction to Biomaterials. Semin. Immunol. 2008, 20, 86–100. [Google Scholar] [CrossRef] [Green Version]
- Bozzer, S.; De Maso, L.; Grimaldi, M.C.; Capolla, S.; Dal Bo, M.; Toffoli, G.; Macor, P. Zebrafish: A Useful Animal Model for the Characterization of Drug-Loaded Polymeric NPs. Biomedicines 2022, 10, 2252. [Google Scholar] [CrossRef] [PubMed]
- Martinez, F.O. Regulators of Macrophage Activation. Eur. J. Immunol. 2011, 41, 1531–1534. [Google Scholar] [CrossRef] [PubMed]
- Taylor, P.R.; Martinez-Pomares, L.; Stacey, M.; Lin, H.-H.; Brown, G.D.; Gordon, S. Macrophage Receptors and Immune Recognition. Annu. Rev. Immunol. 2005, 23, 901–944. [Google Scholar] [CrossRef]
- Gallud, A.; Bondarenko, O.; Feliu, N.; Kupferschmidt, N.; Atluri, R.; Garcia-Bennett, A.; Fadeel, B. Macrophage Activation Status Determines the Internalization of Mesoporous Silica Particles of Different Sizes: Exploring the Role of Different Pattern Recognition Receptors. Biomaterials 2017, 121, 28–40. [Google Scholar] [CrossRef]
- Binnemars-Postma, K.A.; ten Hoopen, H.W.; Storm, G.; Prakash, J. Differential Uptake of Nanoparticles by Human M1 and M2 Polarized Macrophages: Protein Corona as a Critical Determinant. Nanomedicine 2016, 11, 2889–2902. [Google Scholar] [CrossRef]
- Lunov, O.; Syrovets, T.; Loos, C.; Beil, J.; Delacher, M.; Tron, K.; Nienhaus, G.U.; Musyanovych, A.; Mailänder, V.; Landfester, K.; et al. Differential Uptake of Functionalized Polystyrene Nanoparticles by Human Macrophages and a Monocytic Cell Line. ACS Nano 2011, 5, 1657–1669. [Google Scholar] [CrossRef]
- Shannahan, J.; Bai, W.; Brown, J. Implications of Scavenger Receptors in the Safe Development of Nanotherapeutics. Recept. Clin. Investig. 2015, 2, e811. [Google Scholar] [CrossRef]
- Lara, S.; Perez-Potti, A.; Herda, L.M.; Adumeau, L.; Dawson, K.A.; Yan, Y. Differential Recognition of Nanoparticle Protein Corona and Modified Low-Density Lipoprotein by Macrophage Receptor with Collagenous Structure. ACS Nano 2018, 12, 4930–4937. [Google Scholar] [CrossRef]
- Mortimer, G.M.; Butcher, N.J.; Musumeci, A.W.; Deng, Z.J.; Martin, D.J.; Minchin, R.F. Cryptic Epitopes of Albumin Determine Mononuclear Phagocyte System Clearance of Nanomaterials. ACS Nano 2014, 8, 3357–3366. [Google Scholar] [CrossRef]
- Park, J.; Park, S.J.; Park, J.Y.; Kim, S.; Kwon, S.; Jung, Y.; Khang, D. Unfolded Protein Corona Surrounding Nanotubes Influence the Innate and Adaptive Immune System. Adv. Sci. 2021, 8, 2004979. [Google Scholar] [CrossRef]
- Yan, Y.; Gause, K.T.; Kamphuis, M.M.J.; Ang, C.-S.; O’Brien-Simpson, N.M.; Lenzo, J.C.; Reynolds, E.C.; Nice, E.C.; Caruso, F. Differential Roles of the Protein Corona in the Cellular Uptake of Nanoporous Polymer Particles by Monocyte and Macrophage Cell Lines. ACS Nano 2013, 7, 10960–10970. [Google Scholar] [CrossRef] [PubMed]
- Obst, K.; Yealland, G.; Balzus, B.; Miceli, E.; Dimde, M.; Weise, C.; Eravci, M.; Bodmeier, R.; Haag, R.; Calderón, M.; et al. Protein Corona Formation on Colloidal Polymeric Nanoparticles and Polymeric Nanogels: Impact on Cellular Uptake, Toxicity, Immunogenicity, and Drug Release Properties. Biomacromolecules 2017, 18, 1762–1771. [Google Scholar] [CrossRef]
- Mo, J.; Xie, Q.; Wei, W.; Zhao, J. Revealing the Immune Perturbation of Black Phosphorus Nanomaterials to Macrophages by Understanding the Protein Corona. Nat. Commun. 2018, 9, 2480. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, H.; Lu, S.; Wang, S.; Liu, L.; Zhu, B.; Yu, S.; Yang, S.; Chang, J. Evolution of the Protein Corona Affects Macrophage Polarization. Int. J. Biol. Macromol. 2021, 191, 192–200. [Google Scholar] [CrossRef]
- Ueno, H.; Klechevsky, E.; Morita, R.; Aspord, C.; Cao, T.; Matsui, T.; Di Pucchio, T.; Connolly, J.; Fay, J.W.; Pascual, V.; et al. Dendritic Cell Subsets in Health and Disease. Immunol. Rev. 2007, 219, 118–142. [Google Scholar] [CrossRef]
- Bednarczyk, M.; Medina-Montano, C.; Fittler, F.J.; Stege, H.; Roskamp, M.; Kuske, M.; Langer, C.; Vahldieck, M.; Montermann, E.; Tubbe, I.; et al. Complement-Opsonized Nano-Carriers Are Bound by Dendritic Cells (DC) via Complement Receptor (CR)3, and by B Cell Subpopulations via CR-1/2, and Affect the Activation of DC and B-1 Cells. Int. J. Mol. Sci. 2021, 22, 2869. [Google Scholar] [CrossRef] [PubMed]
- Demento, S.L.; Eisenbarth, S.C.; Foellmer, H.G.; Platt, C.; Caplan, M.J.; Mark Saltzman, W.; Mellman, I.; Ledizet, M.; Fikrig, E.; Flavell, R.A.; et al. Inflammasome-Activating Nanoparticles as Modular Systems for Optimizing Vaccine Efficacy. Vaccine 2009, 27, 3013–3021. [Google Scholar] [CrossRef] [PubMed]
- Keshavan, S.; Calligari, P.; Stella, L.; Fusco, L.; Delogu, L.G.; Fadeel, B. Nano-Bio Interactions: A Neutrophil-Centric View. Cell Death Dis. 2019, 10, 569. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vlasova, I.I.; Mikhalchik, E.V.; Barinov, N.A.; Kostevich, V.A.; Smolina, N.V.; Klinov, D.V.; Sokolov, A.V. Adsorbed Plasma Proteins Modulate the Effects of Single-Walled Carbon Nanotubes on Neutrophils in Blood. Nanomed. Nanotechnol. Biol. Med. 2016, 12, 1615–1625. [Google Scholar] [CrossRef]
- Lu, N.; Sui, Y.; Tian, R.; Peng, Y.-Y. Adsorption of Plasma Proteins on Single-Walled Carbon Nanotubes Reduced Cytotoxicity and Modulated Neutrophil Activation. Chem. Res. Toxicol. 2018, 31, 1061–1068. [Google Scholar] [CrossRef] [PubMed]
- Jones, S.W.; Roberts, R.A.; Robbins, G.R.; Perry, J.L.; Kai, M.P.; Chen, K.; Bo, T.; Napier, M.E.; Ting, J.P.Y.; DeSimone, J.M.; et al. Nanoparticle Clearance Is Governed by Th1/Th2 Immunity and Strain Background. J. Clin. Investig. 2013, 123, 3061–3073. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, X.; Ishida, T.; Kiwada, H. Anti-PEG IgM Elicited by Injection of Liposomes Is Involved in the Enhanced Blood Clearance of a Subsequent Dose of PEGylated Liposomes. J. Control. Release 2007, 119, 236–244. [Google Scholar] [CrossRef] [PubMed]
- Grenier, P.; Viana, I.M.d.O.; Lima, E.M.; Bertrand, N. Anti-Polyethylene Glycol Antibodies Alter the Protein Corona Deposited on Nanoparticles and the Physiological Pathways Regulating Their Fate In Vivo. J. Control. Release 2018, 287, 121–131. [Google Scholar] [CrossRef]
- Bogart, L.K.; Pourroy, G.; Murphy, C.J.; Puntes, V.; Pellegrino, T.; Rosenblum, D.; Peer, D.; Lévy, R. Nanoparticles for Imaging, Sensing, and Therapeutic Intervention. ACS Nano 2014, 8, 3107–3122. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez, P.L.; Harada, T.; Christian, D.A.; Pantano, D.A.; Tsai, R.K.; Discher, D.E. Minimal “Self” Peptides That Inhibit Phagocytic Clearance and Enhance Delivery of Nanoparticles. Science 2013, 339, 971–975. [Google Scholar] [CrossRef] [Green Version]
- Panagi, Z.; Beletsi, A.; Evangelatos, G.; Livaniou, E.; Ithakissios, D.S.; Avgoustakis, K. Effect of Dose on the Biodistribution and Pharmacokinetics of PLGA and PLGA–MPEG Nanoparticles. Int. J. Pharm. 2001, 221, 143–152. [Google Scholar] [CrossRef] [PubMed]
- Pisani, A.; Bardi, G. Immunology of Biodegradable Nanoparticles: A Brief Overview on a Wide Growing Field. Explor. Immunol. 2021, 1, 48–60. [Google Scholar] [CrossRef]
- de la Rosa, V.R. Poly(2-Oxazoline)s as Materials for Biomedical Applications. J. Mater. Sci. Mater. Med. 2014, 25, 1211–1225. [Google Scholar] [CrossRef] [PubMed]
- Chapman, R.G.; Ostuni, E.; Takayama, S.; Holmlin, R.E.; Yan, L.; Whitesides, G.M. Surveying for Surfaces That Resist the Adsorption of Proteins. J. Am. Chem. Soc. 2000, 122, 8303–8304. [Google Scholar] [CrossRef]
- Najer, A.; Belessiotis-Richards, A.; Kim, H.; Saunders, C.; Fenaroli, F.; Adrianus, C.; Che, J.; Tonkin, R.L.; Høgset, H.; Lörcher, S.; et al. Block Length-Dependent Protein Fouling on Poly(2-oxazoline)-Based Polymersomes: Influence on Macrophage Association and Circulation Behavior. Small 2022, 18, 2201993. [Google Scholar] [CrossRef] [PubMed]
- Cao, Z.; Jiang, S. Super-Hydrophilic Zwitterionic Poly(Carboxybetaine) and Amphiphilic Non-Ionic Poly(Ethylene Glycol) for Stealth Nanoparticles. Nano Today 2012, 7, 404–413. [Google Scholar] [CrossRef]
- Jiang, S.; Cao, Z. Ultralow-Fouling, Functionalizable, and Hydrolyzable Zwitterionic Materials and Their Derivatives for Biological Applications. Adv. Mater. 2010, 22, 920–932. [Google Scholar] [CrossRef] [PubMed]
- Yuan, Y.-Y.; Mao, C.-Q.; Du, X.-J.; Du, J.-Z.; Wang, F.; Wang, J. Surface Charge Switchable Nanoparticles Based on Zwitterionic Polymer for Enhanced Drug Delivery to Tumor. Adv. Mater. 2012, 24, 5476–5480. [Google Scholar] [CrossRef] [PubMed]
- Simon, J.; Müller, L.K.; Kokkinopoulou, M.; Lieberwirth, I.; Morsbach, S.; Landfester, K.; Mailänder, V. Exploiting the Biomolecular Corona: Pre-Coating of Nanoparticles Enables Controlled Cellular Interactions. Nanoscale 2018, 10, 10731–10739. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aryal, S.; Hu, C.-M.J.; Fang, R.H.; Dehaini, D.; Carpenter, C.; Zhang, D.-E.; Zhang, L. Erythrocyte Membrane-Cloaked Polymeric Nanoparticles for Controlled Drug Loading and Release. Nanomedicine 2013, 8, 1271–1280. [Google Scholar] [CrossRef] [Green Version]
- Beh, C.Y.; Prajnamitra, R.P.; Chen, L.-L.; Hsieh, P.C.-H. Advances in Biomimetic Nanoparticles for Targeted Cancer Therapy and Diagnosis. Molecules 2021, 26, 5052. [Google Scholar] [CrossRef]
- Wang, Y.; Zhang, K.; Qin, X.; Li, T.; Qiu, J.; Yin, T.; Huang, J.; McGinty, S.; Pontrelli, G.; Ren, J.; et al. Biomimetic Nanotherapies: Red Blood Cell Based Core–Shell Structured Nanocomplexes for Atherosclerosis Management. Adv. Sci. 2019, 6, 1900172. [Google Scholar] [CrossRef] [PubMed]
- Hu, C.-M.J.; Zhang, L.; Aryal, S.; Cheung, C.; Fang, R.H.; Zhang, L. Erythrocyte Membrane-Camouflaged Polymeric Nanoparticles as a Biomimetic Delivery Platform. Proc. Natl. Acad. Sci. USA 2011, 108, 10980–10985. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cao, H.; Dan, Z.; He, X.; Zhang, Z.; Yu, H.; Yin, Q.; Li, Y. Liposomes Coated with Isolated Macrophage Membrane Can Target Lung Metastasis of Breast Cancer. ACS Nano 2016, 10, 7738–7748. [Google Scholar] [CrossRef] [PubMed]
- Gong, C.; Yu, X.; You, B.; Wu, Y.; Wang, R.; Han, L.; Wang, Y.; Gao, S.; Yuan, Y. Macrophage-Cancer Hybrid Membrane-Coated Nanoparticles for Targeting Lung Metastasis in Breast Cancer Therapy. J. Nanobiotechnol. 2020, 18, 92. [Google Scholar] [CrossRef]
- Mirshafiee, V.; Mahmoudi, M.; Lou, K.; Cheng, J.; Kraft, M.L. Protein Corona Significantly Reduces Active Targeting Yield. Chem. Commun. 2013, 49, 2557. [Google Scholar] [CrossRef]
- Hadjidemetriou, M.; Al-Ahmady, Z.; Kostarelos, K. Time-Evolution of In Vivo Protein Corona onto Blood-Circulating PEGylated Liposomal Doxorubicin (DOXIL) Nanoparticles. Nanoscale 2016, 8, 6948–6957. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chavez-Santoscoy, A.V.; Roychoudhury, R.; Pohl, N.L.B.; Wannemuehler, M.J.; Narasimhan, B.; Ramer-Tait, A.E. Tailoring the Immune Response by Targeting C-Type Lectin Receptors on Alveolar Macrophages Using “Pathogen-like” Amphiphilic Polyanhydride Nanoparticles. Biomaterials 2012, 33, 4762–4772. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Q.; Dehaini, D.; Zhang, Y.; Zhou, J.; Chen, X.; Zhang, L.; Fang, R.H.; Gao, W.; Zhang, L. Neutrophil Membrane-Coated Nanoparticles Inhibit Synovial Inflammation and Alleviate Joint Damage in Inflammatory Arthritis. Nat. Nanotechnol. 2018, 13, 1182–1190. [Google Scholar] [CrossRef] [PubMed]
- Corbo, C.; Molinaro, R.; Tabatabaei, M.; Farokhzad, O.C.; Mahmoudi, M. Personalized Protein Corona on Nanoparticles and Its Clinical Implications. Biomater. Sci. 2017, 5, 378–387. [Google Scholar] [CrossRef]
- Zheng, T.; Pierre-Pierre, N.; Yan, X.; Huo, Q.; Almodovar, A.J.O.; Valerio, F.; Rivera-Ramirez, I.; Griffith, E.; Decker, D.D.; Chen, S.; et al. Gold Nanoparticle-Enabled Blood Test for Early Stage Cancer Detection and Risk Assessment. ACS Appl. Mater. Interfaces 2015, 7, 6819–6827. [Google Scholar] [CrossRef] [PubMed]
- Hajipour, M.J.; Laurent, S.; Aghaie, A.; Rezaee, F.; Mahmoudi, M. Personalized Protein Coronas: A “Key” Factor at the Nanobiointerface. Biomater. Sci. 2014, 2, 1210. [Google Scholar] [CrossRef] [PubMed]
- Chantada-Vázquez, M.d.P.; García-Vence, M.; Vázquez-Estévez, S.; Bravo, S.B.; Núñez, C. Identification of a Profile of Neutrophil-Derived Granule Proteins in the Surface of Gold Nanoparticles after Their Interaction with Human Breast Cancer Sera. Nanomaterials 2020, 10, 1223. [Google Scholar] [CrossRef] [PubMed]
- Fogli, S.; Montis, C.; Paccosi, S.; Silvano, A.; Michelucci, E.; Berti, D.; Bosi, A.; Parenti, A.; Romagnoli, P. Inorganic Nanoparticles as Potential Regulators of Immune Response in Dendritic Cells. Nanomedicine 2017, 12, 1647–1660. [Google Scholar] [CrossRef]








| Name of the Protein | How Dysopsonins Affect Uptake of NPs in Macrophages | Type of NP Studied |
|---|---|---|
| HRG | Stealth properties | SiO2-NPs [51,52] |
| Apos | Modulation of C activation | PEG-NPs (Apo-J) [14] |
| PS-NPs [14] | ||
| SiO2-NPs (Apo-J) [74] | ||
| AgNPs (Apo-J) [74] | ||
| Decrease in clearance speed | Liposomes (Apo-AI) [75] | |
| HSA | Stealth properties | PS microparticles [76] |
| HA-NPs [77] | ||
| Decrease in clearance speed | PHBHHx NPs (BSA) [74] | |
| SP-A and SP-D | Hypothesis: inhibition of NPs’ agglomeration | PS-NPs (SP-A) [23] |
| CNTs (SP-D) [23] |
| Cell Type | Type of NP Studied | Cell’s Receptors Involved | Effect |
|---|---|---|---|
| Dendritic cells | C-opsonized FeO-DEX NPs [112] | CR3 | Increased NPs’ binding/uptake Lower MCHII and CD86 expression Lower responsiveness to stimuli |
| OVA-loaded LPS-modified (PLGA)-NPs [113] | TLRs and NLRs | Increased uptake Stimulation of CD8+ T cell responses | |
| Neutrophils | DEX-coated NPs BNF-starch NPs [112] | CR3 or SR-A | Increased clearance |
| IgG-coated PEG-SWCNTs [114,115,116] | IgG mediated Interaction | Neutrophil activation ROS and MPO release | |
| B cells | C-opsonized FeO-DEX NPs [112] | CR-1/2 | Decrease in CD86 Lower responsiveness to stimuli |
| T cells | Unfolded proteins-coated CNTs [106] | Not addressed | Increase in effector T helper cells Reduction of naïve T cells |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Panico, S.; Capolla, S.; Bozzer, S.; Toffoli, G.; Dal Bo, M.; Macor, P. Biological Features of Nanoparticles: Protein Corona Formation and Interaction with the Immune System. Pharmaceutics 2022, 14, 2605. https://doi.org/10.3390/pharmaceutics14122605
Panico S, Capolla S, Bozzer S, Toffoli G, Dal Bo M, Macor P. Biological Features of Nanoparticles: Protein Corona Formation and Interaction with the Immune System. Pharmaceutics. 2022; 14(12):2605. https://doi.org/10.3390/pharmaceutics14122605
Chicago/Turabian StylePanico, Sonia, Sara Capolla, Sara Bozzer, Giuseppe Toffoli, Michele Dal Bo, and Paolo Macor. 2022. "Biological Features of Nanoparticles: Protein Corona Formation and Interaction with the Immune System" Pharmaceutics 14, no. 12: 2605. https://doi.org/10.3390/pharmaceutics14122605
APA StylePanico, S., Capolla, S., Bozzer, S., Toffoli, G., Dal Bo, M., & Macor, P. (2022). Biological Features of Nanoparticles: Protein Corona Formation and Interaction with the Immune System. Pharmaceutics, 14(12), 2605. https://doi.org/10.3390/pharmaceutics14122605








