Chlorotoxin and Lung Cancer: A Targeting Perspective for Drug Delivery
Abstract
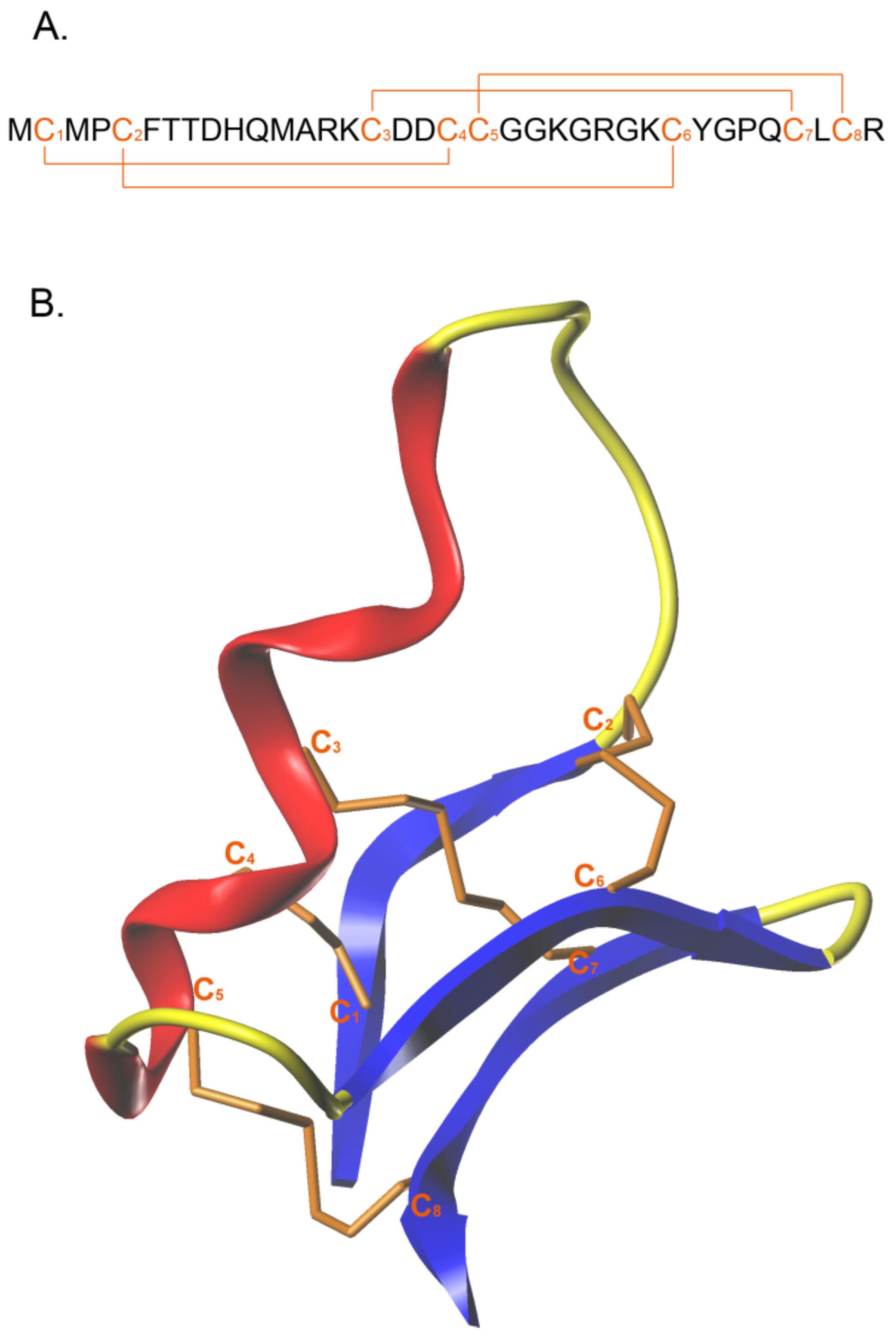
1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Cell Culture
2.3. Cell Viability
2.4. Quantitative Phase Imaging Microscopy
2.5. Statstical Analysis
3. Discussion
3.1. Chlorotoxin Selectively Binds to Cancer Cells—Receptor Targeting
3.2. Chlorotoxin and Lung Cancer: Promising but Limited Research
3.3. Identification and Comparison of CTX’s Targeting Capacity
3.4. CTX Activity on Lung Cancer Cells
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Mattheolabakis, G.; Rigas, B.; Constantinides, P.P. Nanodelivery strategies in cancer chemotherapy: Biological rationale and pharmaceutical perspectives. Nanomedicine 2012, 7, 1577–1590. [Google Scholar] [CrossRef] [PubMed]
- Torchilin, V.P. Recent advances with liposomes as pharmaceutical carriers. Nat. Rev. Drug Discov. 2005, 4, 145–160. [Google Scholar] [CrossRef] [PubMed]
- Suk, J.S.; Xu, Q.; Kim, N.; Hanes, J.; Ensign, L.M. PEGylation as a strategy for improving nanoparticle-based drug and gene delivery. Adv. Drug. Deliv. Rev. 2016, 99, 28–51. [Google Scholar] [CrossRef] [PubMed]
- Karakoti, A.S.; Das, S.; Thevuthasan, S.; Seal, S. PEGylated inorganic nanoparticles. Angew. Chem. Int. Ed. Engl. 2011, 50, 1980–1994. [Google Scholar] [CrossRef]
- Thakur, S.; Kesharwani, P.; Tekade, R.K.; Jain, N.K. Impact of pegylation on biopharmaceutical properties of dendrimers. Polymer 2015, 59, 67–92. [Google Scholar] [CrossRef]
- Harris, J.M.; Chess, R.B. Effect of pegylation on pharmaceuticals. Nat. Rev. Drug Discov. 2003, 2, 214–221. [Google Scholar] [CrossRef]
- Bazak, R.; Houri, M.; El Achy, S.; Kamel, S.; Refaat, T. Cancer active targeting by nanoparticles: A comprehensive review of literature. J. Cancer Res. Clin. Oncol. 2015, 141, 769–784. [Google Scholar] [CrossRef]
- Irache, J.M.; Salman, H.H.; Gamazo, C.; Espuelas, S. Mannose-targeted systems for the delivery of therapeutics. Expert Opin. Drug. Deliv. 2008, 5, 703–724. [Google Scholar] [CrossRef]
- Mukherjee, B.; Satapathy, B.S.; Mondal, L.; Dey, N.S.; Maji, R. Potentials and challenges of active targeting at the tumor cells by engineered polymeric nanoparticles. Curr. Pharm. Biotechnol. 2013, 14, 1250–1263. [Google Scholar] [CrossRef]
- Madaan, K.; Kumar, S.; Poonia, N.; Lather, V.; Pandita, D. Dendrimers in drug delivery and targeting: Drug-dendrimer interactions and toxicity issues. J. Pharm. Bioallied Sci. 2014, 6, 139–150. [Google Scholar] [CrossRef]
- Montaseri, H.; Kruger, C.A.; Abrahamse, H. Inorganic Nanoparticles Applied for Active Targeted Photodynamic Therapy of Breast Cancer. Pharmaceutics 2021, 13, 296. [Google Scholar] [CrossRef] [PubMed]
- Gaspar, N.; Zambito, G.; Lowik, C.; Mezzanotte, L. Active Nano-targeting of Macrophages. Curr. Pharm. Des. 2019, 25, 1951–1961. [Google Scholar] [CrossRef] [PubMed]
- Caminschi, I.; Maraskovsky, E.; Heath, W.R. Targeting Dendritic Cells in vivo for Cancer Therapy. Front. Immunol. 2012, 3, 13. [Google Scholar] [CrossRef] [PubMed]
- Simone, E.; Ding, B.S.; Muzykantov, V. Targeted delivery of therapeutics to endothelium. Cell Tissue Res. 2009, 335, 283–300. [Google Scholar] [CrossRef]
- Kan, S.; Hariyadi, D.M.; Grainge, C.; Knight, D.A.; Bartlett, N.W.; Liang, M. Airway epithelial-targeted nanoparticles for asthma therapy. Am. J. Physiol. Lung Cell Mol. Physiol. 2020, 318, L500–L509. [Google Scholar] [CrossRef]
- Lau, J.L.; Dunn, M.K. Therapeutic peptides: Historical perspectives, current development trends, and future directions. Bioorg. Med. Chem. 2018, 26, 2700–2707. [Google Scholar] [CrossRef]
- London, N.; Raveh, B.; Schueler-Furman, O. Modeling peptide-protein interactions. Methods Mol. Biol. 2012, 857, 375–398. [Google Scholar] [CrossRef]
- Scodeller, P.; Asciutto, E.K. Targeting Tumors Using Peptides. Molecules 2020, 25, 808. [Google Scholar] [CrossRef]
- Liu, M.; Fang, X.; Yang, Y.; Wang, C. Peptide-Enabled Targeted Delivery Systems for Therapeutic Applications. Front. Bioeng. Biotechnol. 2021, 9, 701504. [Google Scholar] [CrossRef]
- Calandro, P.; Iovenitti, G.; Zamperini, C.; Candita, F.; Dreassi, E.; Chiariello, M.; Angelucci, A.; Schenone, S.; Botta, M.; Mancini, A. Plasmin-Binding Tripeptide-Decorated Liposomes Loading Pyrazolo[3,4-d]pyrimidines for Targeting Hepatocellular Carcinoma. ACS Med. Chem. Lett. 2018, 9, 646–651. [Google Scholar] [CrossRef]
- Bordon, K.C.F.; Cologna, C.T.; Fornari-Baldo, E.C.; Pinheiro-Junior, E.L.; Cerni, F.A.; Amorim, F.G.; Anjolette, F.A.P.; Cordeiro, F.A.; Wiezel, G.A.; Cardoso, I.A.; et al. From Animal Poisons and Venoms to Medicines: Achievements, Challenges and Perspectives in Drug Discovery. Front. Pharmacol. 2020, 11, 1132. [Google Scholar] [CrossRef] [PubMed]
- Herzig, V.; Cristofori-Armstrong, B.; Israel, M.R.; Nixon, S.A.; Vetter, I.; King, G.F. Animal toxins—Nature’s evolutionary-refined toolkit for basic research and drug discovery. Biochem. Pharmacol. 2020, 181, 114096. [Google Scholar] [CrossRef] [PubMed]
- Siegel, R.L.; Miller, K.D.; Fuchs, H.E.; Jemal, A. Cancer Statistics, 2021. CA Cancer. J. Clin. 2021, 71, 7–33. [Google Scholar] [CrossRef]
- Gregory, A.J.; Voit-Ostricki, L.; Lovas, S.; Watts, C.R. Effects of Selective Substitution of Cysteine Residues on the Conformational Properties of Chlorotoxin Explored by Molecular Dynamics Simulations. Int. J. Mol. Sci. 2019, 20, 1261. [Google Scholar] [CrossRef] [PubMed]
- Dardevet, L.; Rani, D.; Aziz, T.A.; Bazin, I.; Sabatier, J.M.; Fadl, M.; Brambilla, E.; De Waard, M. Chlorotoxin: A helpful natural scorpion peptide to diagnose glioma and fight tumor invasion. Toxins 2015, 7, 1079–1101. [Google Scholar] [CrossRef]
- Cohen, G.; Burks, S.R.; Frank, J.A. Chlorotoxin-A Multimodal Imaging Platform for Targeting Glioma Tumors. Toxins 2018, 10, 496. [Google Scholar] [CrossRef]
- Soroceanu, L.; Gillespie, Y.; Khazaeli, M.B.; Sontheimer, H. Use of Chlorotoxin for Targeting of Primary Brain Tumors. Cancer Res. 1998, 58, 4871–4879. [Google Scholar]
- Veiseh, M.; Gabikian, P.; Bahrami, S.B.; Veiseh, O.; Zhang, M.; Hackman, R.C.; Ravanpay, A.C.; Stroud, M.R.; Kusuma, Y.; Hansen, S.J.; et al. Tumor paint: A chlorotoxin:Cy5.5 bioconjugate for intraoperative visualization of cancer foci. Cancer Res. 2007, 67, 6882–6888. [Google Scholar] [CrossRef]
- McFerrin, M.B.; Sontheimer, H. A role for ion channels in glioma cell invasion. Neuron Glia. Biol. 2006, 2, 39–49. [Google Scholar] [CrossRef]
- Webb, A.H.; Gao, B.T.; Goldsmith, Z.K.; Irvine, A.S.; Saleh, N.; Lee, R.P.; Lendermon, J.B.; Bheemreddy, R.; Zhang, Q.; Brennan, R.C.; et al. Inhibition of MMP-2 and MMP-9 decreases cellular migration, and angiogenesis in in vitro models of retinoblastoma. BMC Cancer 2017, 17, 434. [Google Scholar] [CrossRef]
- Merchant, N.; Nagaraju, G.P.; Rajitha, B.; Lammata, S.; Jella, K.K.; Buchwald, Z.S.; Lakka, S.S.; Ali, A.N. Matrix metalloproteinases: Their functional role in lung cancer. Carcinogenesis 2017, 38, 766–780. [Google Scholar] [CrossRef] [PubMed]
- Hofmann, U.B.; Westphal, J.R.; Waas, E.T.; Zendman, A.J.; Cornelissen, I.M.; Ruiter, D.J.; van Muijen, G.N. Matrix metalloproteinases in human melanoma cell lines and xenografts: Increased expression of activated matrix metalloproteinase-2 (MMP-2) correlates with melanoma progression. Br. J. Cancer 1999, 81, 774–782. [Google Scholar] [CrossRef] [PubMed]
- Ren, F.; Tang, R.; Zhang, X.; Madushi, W.M.; Luo, D.; Dang, Y.; Li, Z.; Wei, K.; Chen, G. Overexpression of MMP Family Members Functions as Prognostic Biomarker for Breast Cancer Patients: A Systematic Review and Meta-Analysis. PLoS ONE 2015, 10, e0135544. [Google Scholar] [CrossRef]
- Baum, O.; Hlushchuk, R.; Forster, A.; Greiner, R.; Clezardin, P.; Zhao, Y.; Djonov, V.; Gruber, G. Increased invasive potential and up-regulation of MMP-2 in MDA-MB-231 breast cancer cells expressing the beta3 integrin subunit. Int. J. Oncol. 2007, 30, 325–332. [Google Scholar] [PubMed]
- Kenny, H.A.; Lengyel, E. MMP-2 functions as an early response protein in ovarian cancer metastasis. Cell Cycle 2009, 8, 683–688. [Google Scholar] [CrossRef] [PubMed]
- Perigny, M.; Bairati, I.; Harvey, I.; Beauchemin, M.; Harel, F.; Plante, M.; Tetu, B. Role of immunohistochemical overexpression of matrix metalloproteinases MMP-2 and MMP-11 in the prognosis of death by ovarian cancer. Am. J. Clin. Pathol. 2008, 129, 226–231. [Google Scholar] [CrossRef] [PubMed]
- Ellenrieder, V.; Alber, B.; Lacher, U.; Hendler, S.F.; Menke, A.; Boeck, W.; Wagner, M.; Wilda, M.; Friess, H.; Buchler, M.; et al. Role of MT-MMPs and MMP-2 in pancreatic cancer progression. Int. J. Cancer 2000, 85, 14–20. [Google Scholar] [CrossRef]
- Juuti, A.; Lundin, J.; Nordling, S.; Louhimo, J.; Haglund, C. Epithelial MMP-2 expression correlates with worse prognosis in pancreatic cancer. Oncology 2006, 71, 61–68. [Google Scholar] [CrossRef]
- Xie, T.; Dong, B.; Yan, Y.; Hu, G.; Xu, Y. Association between MMP-2 expression and prostate cancer: A meta-analysis. Biomed. Rep. 2016, 4, 241–245. [Google Scholar] [CrossRef]
- Ross, J.S.; Kaur, P.; Sheehan, C.E.; Fisher, H.A.; Kaufman, R.A., Jr.; Kallakury, B.V. Prognostic significance of matrix metalloproteinase 2 and tissue inhibitor of metalloproteinase 2 expression in prostate cancer. Mod. Pathol. 2003, 16, 198–205. [Google Scholar] [CrossRef]
- Han, L.; Sheng, B.; Zeng, Q.; Yao, W.; Jiang, Q. Correlation between MMP2 expression in lung cancer tissues and clinical parameters: A retrospective clinical analysis. BMC Pulm. Med. 2020, 20, 283. [Google Scholar] [CrossRef]
- Kesavan, K.; Ratliff, J.; Johnson, E.W.; Dahlberg, W.; Asara, J.M.; Misra, P.; Frangioni, J.V.; Jacoby, D.B. Annexin A2 is a molecular target for TM601, a peptide with tumor-targeting and anti-angiogenic effects. J. Biol. Chem. 2010, 285, 4366–4374. [Google Scholar] [CrossRef]
- McGonigle, S.; Majumder, U.; Kolber-Simonds, D.; Wu, J.; Hart, A.; Noland, T.; TenDyke, K.; Custar, D.; Li, D.; Du, H.; et al. Neuropilin-1 drives tumor-specific uptake of chlorotoxin. Cell Commun. Signal 2019, 17, 67. [Google Scholar] [CrossRef]
- Lyons, S.A.; O’Neal, J.; Sontheimer, H. Chlorotoxin, a scorpion-derived peptide, specifically binds to gliomas and tumors of neuroectodermal origin. Glia 2002, 39, 162–173. [Google Scholar] [CrossRef]
- Gribbin, T.E.; Senzer, N.; Raizer, J.J.; Shen, S.; Nabors, L.B.; Wiranowska, M.; Fiveash, J.B. A phase I evaluation of intravenous (IV) 131I-chlorotoxin delivery to solid peripheral and intracranial tumors. J. Clin. Oncol. 2009, 27, e14507. [Google Scholar] [CrossRef]
- Wiranowska, M.; Colina, L.O.; Johnson, J.O. Clathrin-mediated entry and cellular localization of chlorotoxin in human glioma. Cancer Cell Int. 2011, 11, 27. [Google Scholar] [CrossRef] [PubMed]
- Graf, N.; Mokhtari, T.E.; Papayannopoulos, I.A.; Lippard, S.J. Platinum(IV)-chlorotoxin (CTX) conjugates for targeting cancer cells. J. Inorg. Biochem. 2012, 110, 58–63. [Google Scholar] [CrossRef] [PubMed]
- Gandhi, L.; Rodriguez-Abreu, D.; Gadgeel, S.; Esteban, E.; Felip, E.; De Angelis, F.; Domine, M.; Clingan, P.; Hochmair, M.J.; Powell, S.F.; et al. Pembrolizumab plus Chemotherapy in Metastatic Non-Small-Cell Lung Cancer. N. Engl. J. Med. 2018, 378, 2078–2092. [Google Scholar] [CrossRef]
- Dilruba, S.; Kalayda, G.V. Platinum-based drugs: Past, present and future. Cancer Chemother. Pharmacol. 2016, 77, 1103–1124. [Google Scholar] [CrossRef]
- Mattheolabakis, G.; Ling, D.; Ahmad, G.; Amiji, M. Enhanced Anti-Tumor Efficacy of Lipid-Modified Platinum Derivatives in Combination with Survivin Silencing siRNA in Resistant Non-Small Cell Lung Cancer. Pharm. Res. 2016, 33, 2943–2953. [Google Scholar] [CrossRef]
- Yang, Y.; Huang, Z.; Pu, X.; Yin, G.; Wang, L.; Gao, F. Fabrication of magnetic nanochains linked with CTX and curcumin for dual modal imaging detection and limitation of early tumour. Cell Prolif. 2018, 51, e12486. [Google Scholar] [CrossRef]
- Qin, C.; He, B.; Dai, W.; Zhang, H.; Wang, X.; Wang, J.; Zhang, X.; Wang, G.; Yin, L.; Zhang, Q. Inhibition of metastatic tumor growth and metastasis via targeting metastatic breast cancer by chlorotoxin-modified liposomes. Mol. Pharm. 2014, 11, 3233–3241. [Google Scholar] [CrossRef] [PubMed]
- Maertens, C.; Wei, L.; Tytgat, J.; Droogmans, G.; Nilius, B. Chlorotoxin does not inhibit volume-regulated, calcium-activated and cyclic AMP-activated chloride channels. Br. J. Pharmacol. 2000, 129, 791–801. [Google Scholar] [CrossRef]
- Pierschbacher, M.D.; Ruoslahti, E. Cell attachment activity of fibronectin can be duplicated by small synthetic fragments of the molecule. Nature 1984, 309, 30–33. [Google Scholar] [CrossRef] [PubMed]
- Sawyers, C.L. Herceptin: A First Assault on Oncogenes that Launched a Revolution. Cell 2019, 179, 8–12. [Google Scholar] [CrossRef]
- Young Kim, H.; Young Yum, S.; Jang, G.; Ahn, D.R. Discovery of a non-cationic cell penetrating peptide derived from membrane-interacting human proteins and its potential as a protein delivery carrier. Sci. Rep. 2015, 5, 11719. [Google Scholar] [CrossRef]
- Gry, M.; Rimini, R.; Stromberg, S.; Asplund, A.; Ponten, F.; Uhlen, M.; Nilsson, P. Correlations between RNA and protein expression profiles in 23 human cell lines. BMC Genom. 2009, 10, 365. [Google Scholar] [CrossRef]
- Koussounadis, A.; Langdon, S.P.; Um, I.H.; Harrison, D.J.; Smith, V.A. Relationship between differentially expressed mRNA and mRNA-protein correlations in a xenograft model system. Sci. Rep. 2015, 5, 10775. [Google Scholar] [CrossRef]
- Carlson, M.W.; Iyer, V.R.; Marcotte, E.M. Quantitative gene expression assessment identifies appropriate cell line models for individual cervical cancer pathways. BMC Genom. 2007, 8, 117. [Google Scholar] [CrossRef] [PubMed]
- Deshane, J.; Garner, C.C.; Sontheimer, H. Chlorotoxin inhibits glioma cell invasion via matrix metalloproteinase-2. J. Biol. Chem. 2003, 278, 4135–4144. [Google Scholar] [CrossRef]
- Patel, D.; Lahiji, A.; Patel, S.; Franklin, M.; Jimenez, X.; Hicklin, D.J.; Kang, X. Monoclonal antibody cetuximab binds to and down-regulates constitutively activated epidermal growth factor receptor vIII on the cell surface. Anticancer Res. 2007, 27, 3355–3366. [Google Scholar]
- Costantini, D.L.; Chan, C.; Cai, Z.; Vallis, K.A.; Reilly, R.M. (111)In-labeled trastuzumab (Herceptin) modified with nuclear localization sequences (NLS): An Auger electron-emitting radiotherapeutic agent for HER2/neu-amplified breast cancer. J. Nucl. Med. 2007, 48, 1357–1368. [Google Scholar] [CrossRef]
- Yi, Y.S. Folate Receptor-Targeted Diagnostics and Therapeutics for Inflammatory Diseases. Immune Netw. 2016, 16, 337–343. [Google Scholar] [CrossRef]
- Doucette, M.M.; Stevens, V.L. Folate receptor function is regulated in response to different cellular growth rates in cultured mammalian cells. J. Nutr. 2001, 131, 2819–2825. [Google Scholar] [CrossRef] [PubMed]
- Erijman, A.; Rosenthal, E.; Shifman, J.M. How structure defines affinity in protein-protein interactions. PLoS ONE 2014, 9, e110085. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Zhao, R.; Wu, X.; Sun, Y.; Yao, M.; Li, J.; Xu, Y.; Gu, J. Identification and characterization of a novel peptide ligand of epidermal growth factor receptor for targeted delivery of therapeutics. FASEB J. 2005, 19, 1978–1985. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Joshi, B.P.; Duan, X.; Pant, A.; Qiu, Z.; Kuick, R.; Owens, S.R.; Wang, T.D. EGFR Overexpressed in Colonic Neoplasia Can be Detected on Wide-Field Endoscopic Imaging. Clin. Transl. Gastroenterol. 2015, 6, e101. [Google Scholar] [CrossRef]
- Toporkiewicz, M.; Meissner, J.; Matusewicz, L.; Czogalla, A.; Sikorski, A.F. Toward a magic or imaginary bullet? Ligands for drug targeting to cancer cells: Principles, hopes, and challenges. Int. J. Nanomed. 2015, 10, 1399–1414. [Google Scholar] [CrossRef]
- Kumar Kulabhusan, P.; Hussain, B.; Yuce, M. Current Perspectives on Aptamers as Diagnostic Tools and Therapeutic Agents. Pharmaceutics 2020, 12, 646. [Google Scholar] [CrossRef]
- Mirian, M.; Khanahmad, H.; Darzi, L.; Salehi, M.; Sadeghi-Aliabadi, H. Oligonucleotide aptamers: Potential novel molecules against viral hepatitis. Res. Pharm. Sci. 2017, 12, 88–98. [Google Scholar] [CrossRef]
- Goddard, Z.R.; Marin, M.J.; Russell, D.A.; Searcey, M. Active targeting of gold nanoparticles as cancer therapeutics. Chem. Soc. Rev. 2020, 49, 8774–8789. [Google Scholar] [CrossRef]
- Fuchs, H.; Gessner, R. Iodination significantly influences the binding of human transferrin to the transferrin receptor. Biochim. Biophys. Acta 2002, 1570, 19–26. [Google Scholar] [CrossRef] [PubMed]
- Fosgerau, K.; Hoffmann, T. Peptide therapeutics: Current status and future directions. Drug Discov. Today 2015, 20, 122–128. [Google Scholar] [CrossRef] [PubMed]
- Erazo-Oliveras, A.; Muthukrishnan, N.; Baker, R.; Wang, T.Y.; Pellois, J.P. Improving the endosomal escape of cell-penetrating peptides and their cargos: Strategies and challenges. Pharmaceuticals 2012, 5, 1177–1209. [Google Scholar] [CrossRef]
- Ayed, A.S.; Omran, M.A.A.A.; Nabil, Z.I.; Strong, P.N.; Newton, K.A.; Abdel-Rahman, M.A. C-Terminal Amidation of Chlorotoxin Does Not Affect Tumour Cell Proliferation and Has No Effect on Toxin Cytotoxicity. Int. J. Pept. Res. Ther. 2021, 27, 659–667. [Google Scholar] [CrossRef]
- Lodeserto, P.; Rossi, M.; Blasi, P.; Farruggia, G.; Orienti, I. Nanospermidine in Combination with Nanofenretinide Induces Cell Death in Neuroblastoma Cell Lines. Pharmaceutics 2022, 14, 1215. [Google Scholar] [CrossRef] [PubMed]


| Targeting Moiety | Molecule Type | Target | Kd (nM) | Ref. |
|---|---|---|---|---|
| GE11 | Peptide | EGFR | 22.28 | [66] |
| QRHKPRE | Peptide | EGFR | 50 | [67] |
| RGD | Peptide | Integrins ανβ3 and ανβ5 | 3.2–100 | [68] |
| Trastuzumab | Antibody | HER2 | 5–9 | [62] |
| Bevacizumab | Antibody | VEGF | 0.5 | [68] |
| NOX-E36 | Aptamer | MCP-1 | 1.32 | [69] |
| NU172 | Aptamer | Thrombin | 0.3–0.5 | [69] |
| AB3 | Aptamer | OFA/iLRP | 101 | [69] |
| AO-01 | Aptamer | Matrix-binding domain | 180 | [70] |
| Hyaluronic acid | Poly saccharide | CD44 | 0.3 | [71] |
| Transferrin | Glycoprotein | Transferrin Receptor | 0.23 | [72] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shrestha, A.; Lahooti, B.; Mikelis, C.M.; Mattheolabakis, G. Chlorotoxin and Lung Cancer: A Targeting Perspective for Drug Delivery. Pharmaceutics 2022, 14, 2613. https://doi.org/10.3390/pharmaceutics14122613
Shrestha A, Lahooti B, Mikelis CM, Mattheolabakis G. Chlorotoxin and Lung Cancer: A Targeting Perspective for Drug Delivery. Pharmaceutics. 2022; 14(12):2613. https://doi.org/10.3390/pharmaceutics14122613
Chicago/Turabian StyleShrestha, Archana, Behnaz Lahooti, Constantinos M. Mikelis, and George Mattheolabakis. 2022. "Chlorotoxin and Lung Cancer: A Targeting Perspective for Drug Delivery" Pharmaceutics 14, no. 12: 2613. https://doi.org/10.3390/pharmaceutics14122613
APA StyleShrestha, A., Lahooti, B., Mikelis, C. M., & Mattheolabakis, G. (2022). Chlorotoxin and Lung Cancer: A Targeting Perspective for Drug Delivery. Pharmaceutics, 14(12), 2613. https://doi.org/10.3390/pharmaceutics14122613







