In Vitro & In Vivo Studies on Identifying and Designing Temporin-1CEh from the Skin Secretion of Rana chensinensis as the Optimised Antibacterial Prototype Drug
Abstract
:1. Introduction
2. Materials and Methods
2.1. Acquisition of Skin Secretion
2.2. Shot-Gun Cloning of Biosynthetic Precursor
2.3. Solid-Phase Peptide Synthesis
2.4. Liposomes Preparation
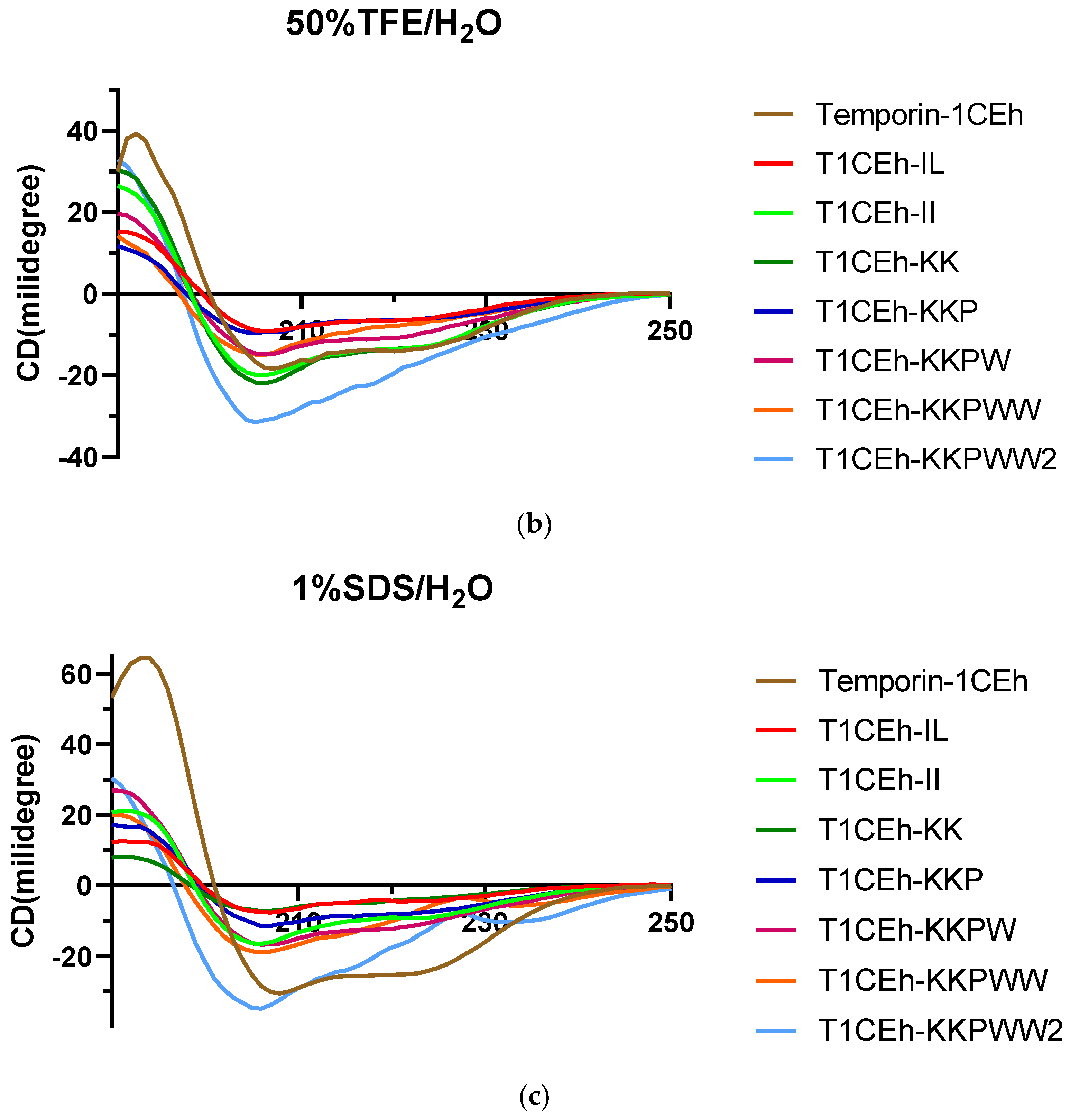
2.5. Secondary Structure Analysis
2.6. Antimicrobial Susceptibility Assay
2.7. Biofilm Susceptibility Assay
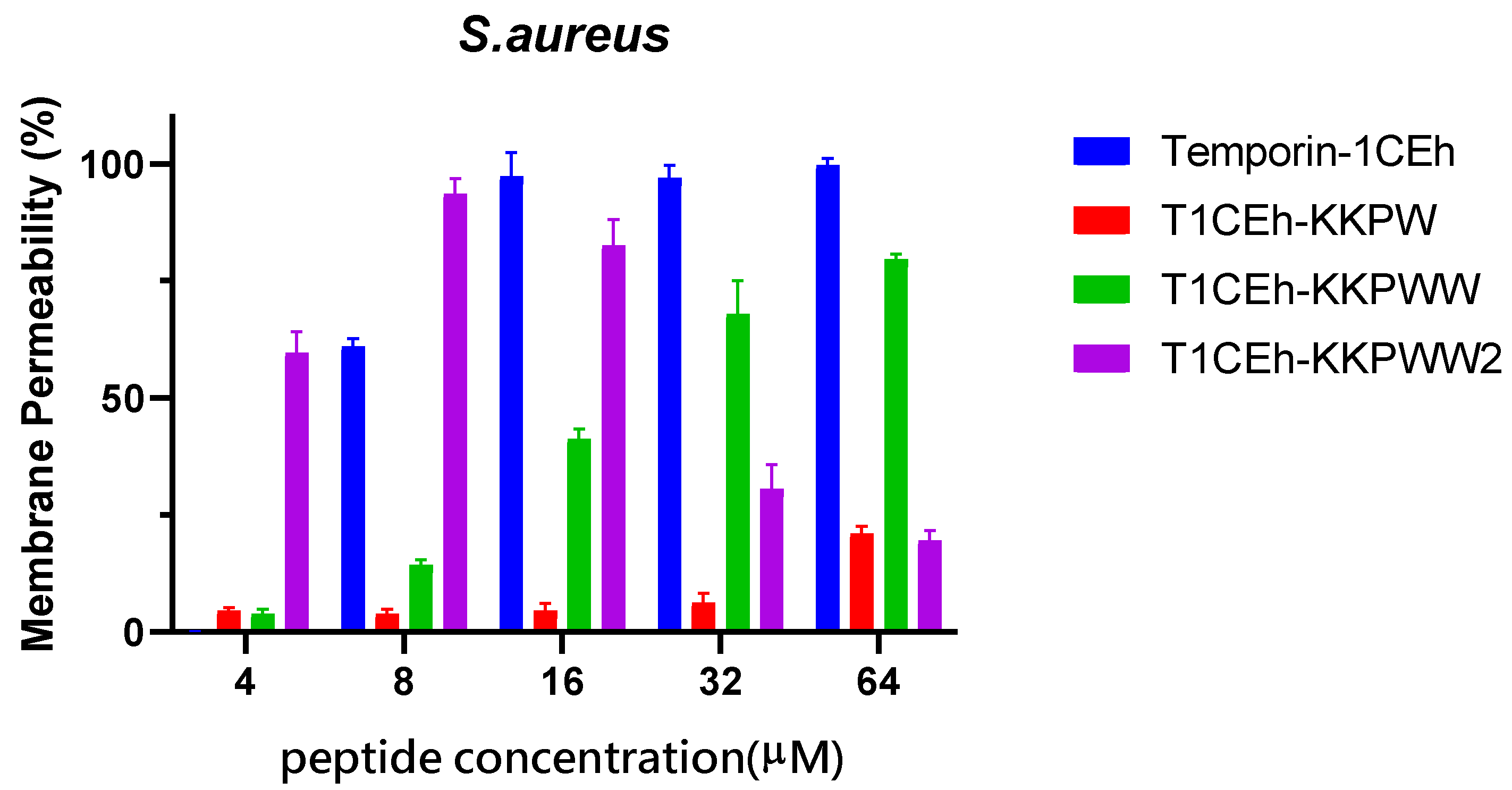
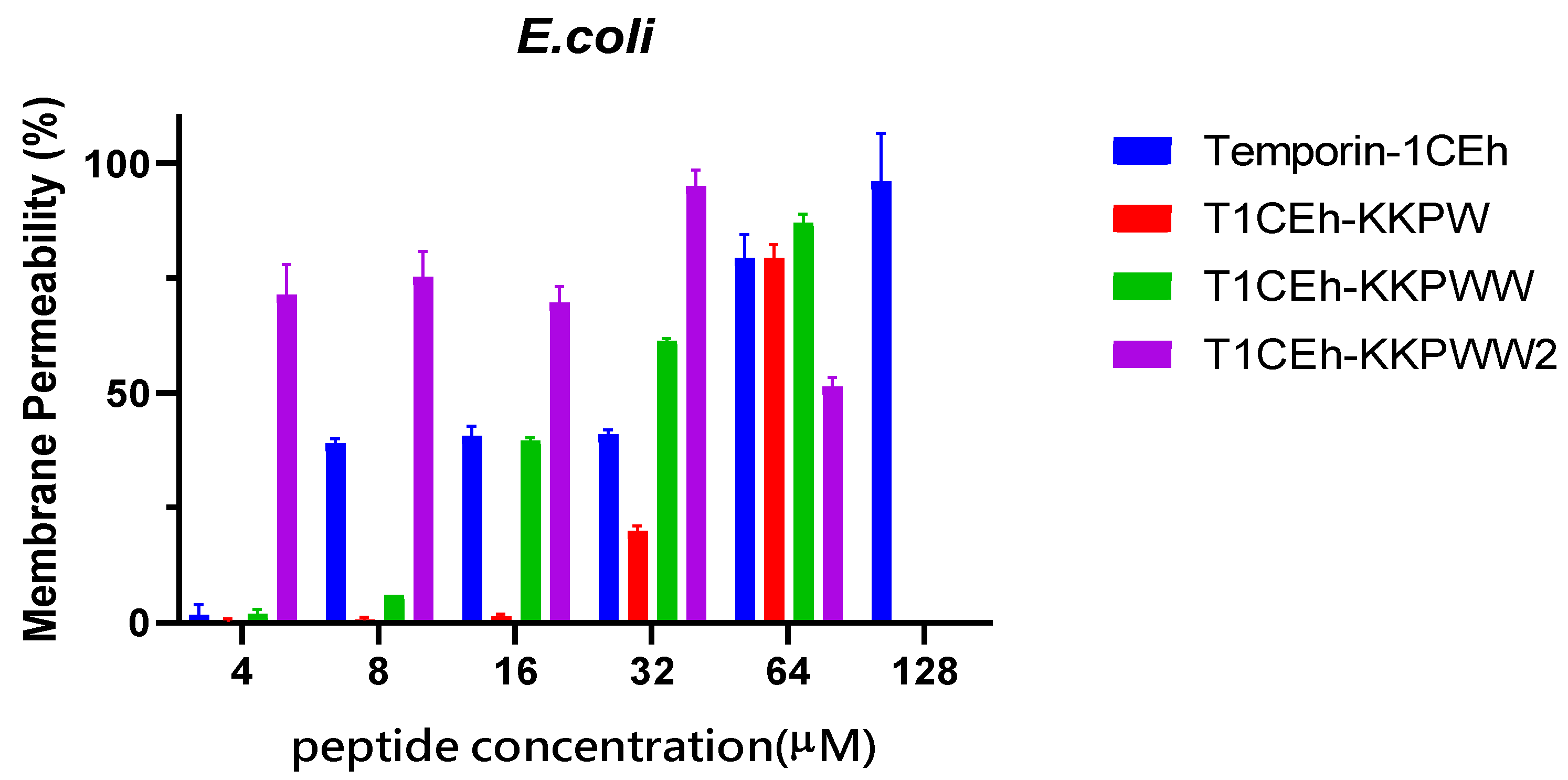
2.8. Membrane Permeability Assay
2.9. Assessing the Impact of Different Environments on Antimicrobial Activity
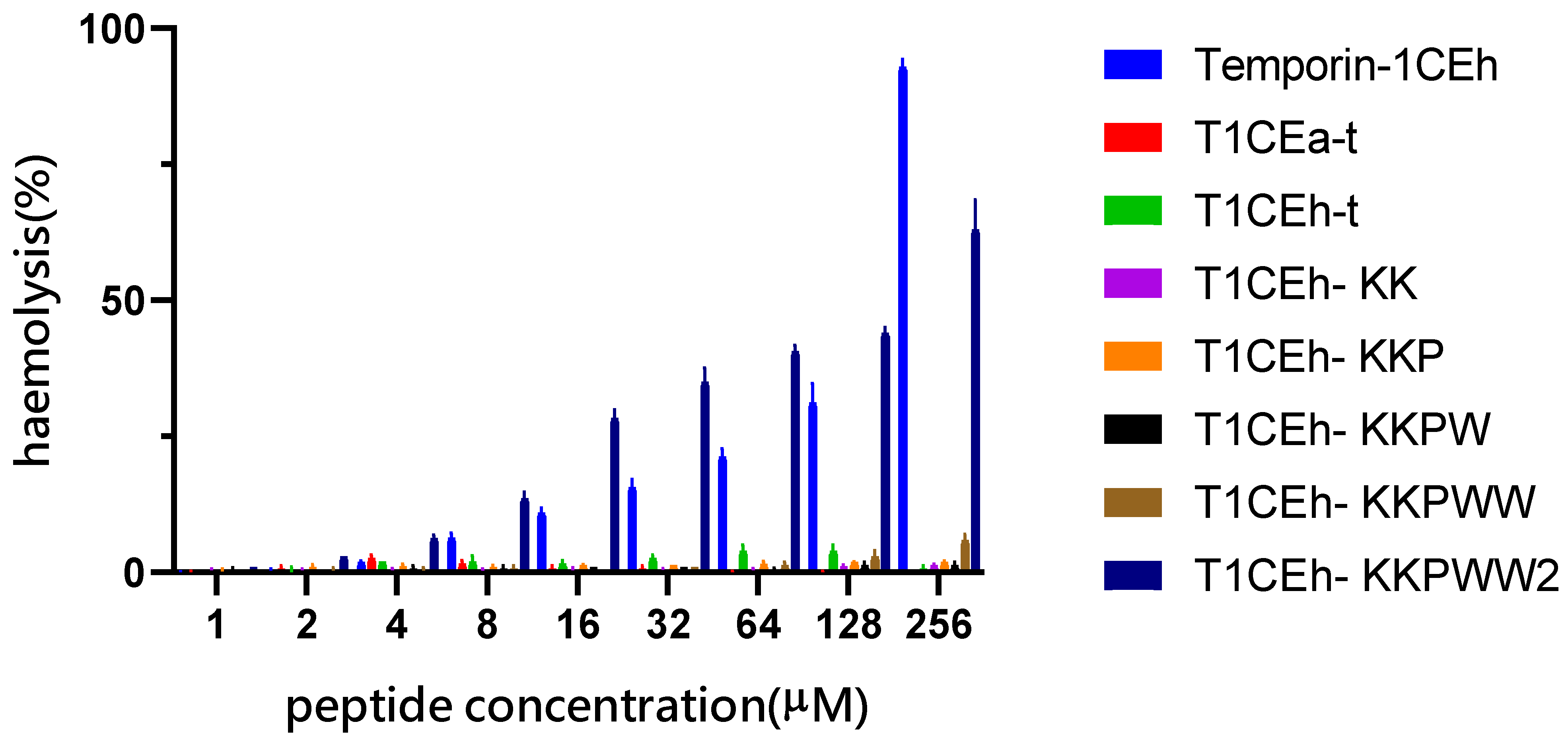
2.10. Haemolysis Assay
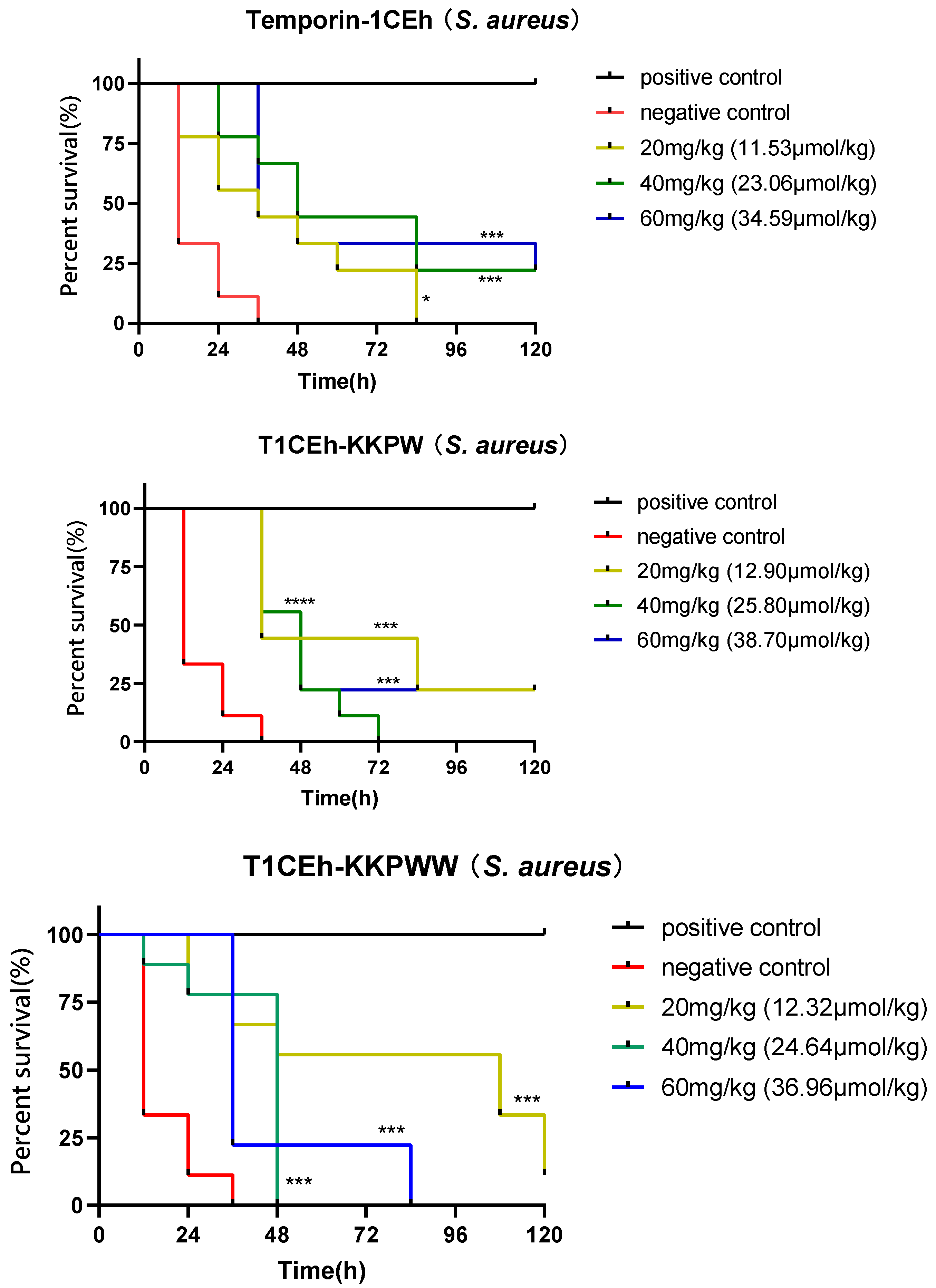
2.11. Assessing the Efficacy of the Peptide Temporin-1CEh, T1CEh-KKPW, T1CEh-KKPWW and T1CEh-KKPWW2 against S. aureus and E. coli In Vivo
2.12. Statistical Analysis
3. Results
3.1. Molecular Cloning of a Novel Peptide Precursor-Encoding cDNA
3.2. Identification of the Peptide from Rana chensinensis Skin Secretion
3.3. Peptide Design following the Tests of Antimicrobial and Haemolytic Activity
3.4. Secondary Structure Analysis
3.5. Membrane Permeabilisation Assays
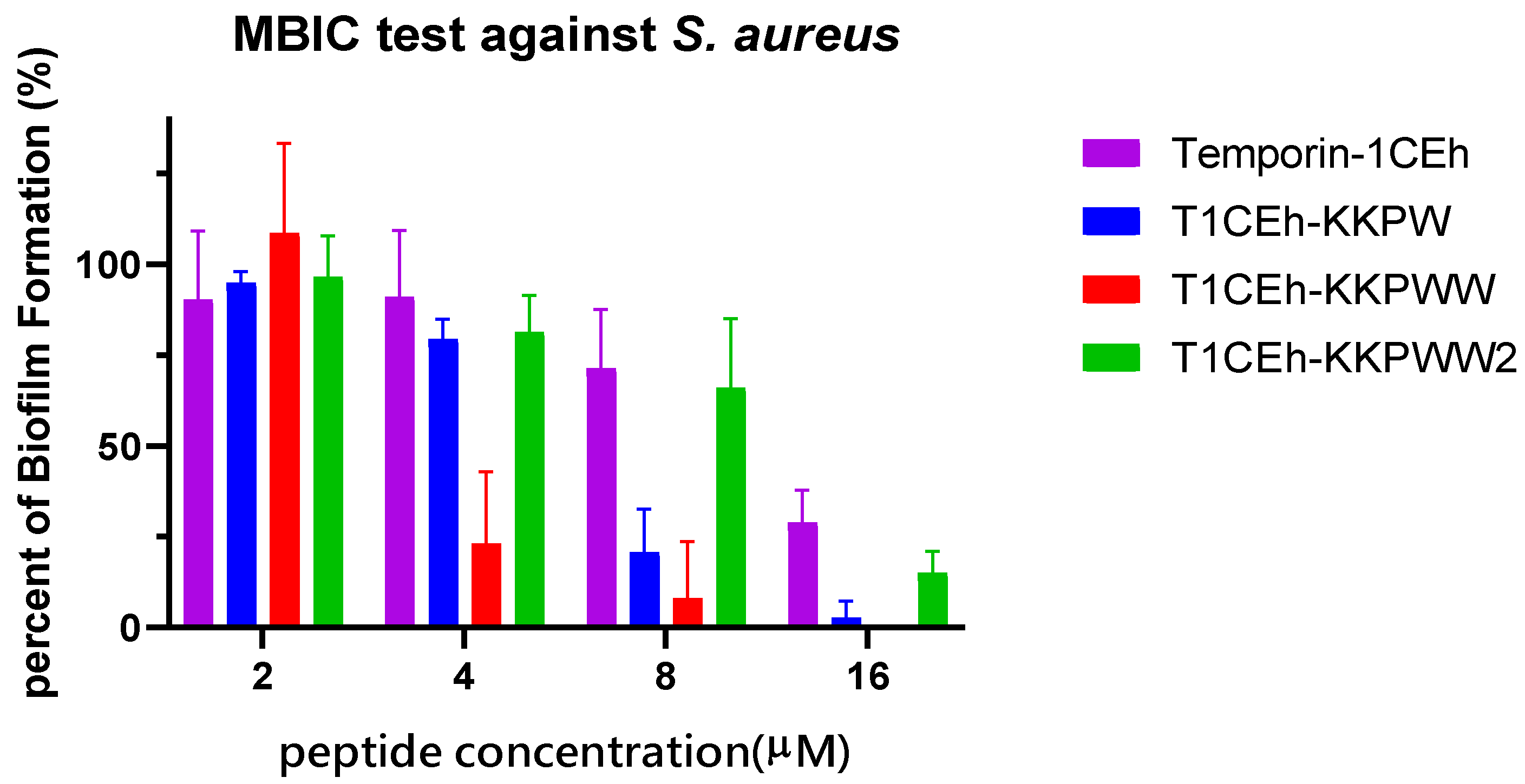
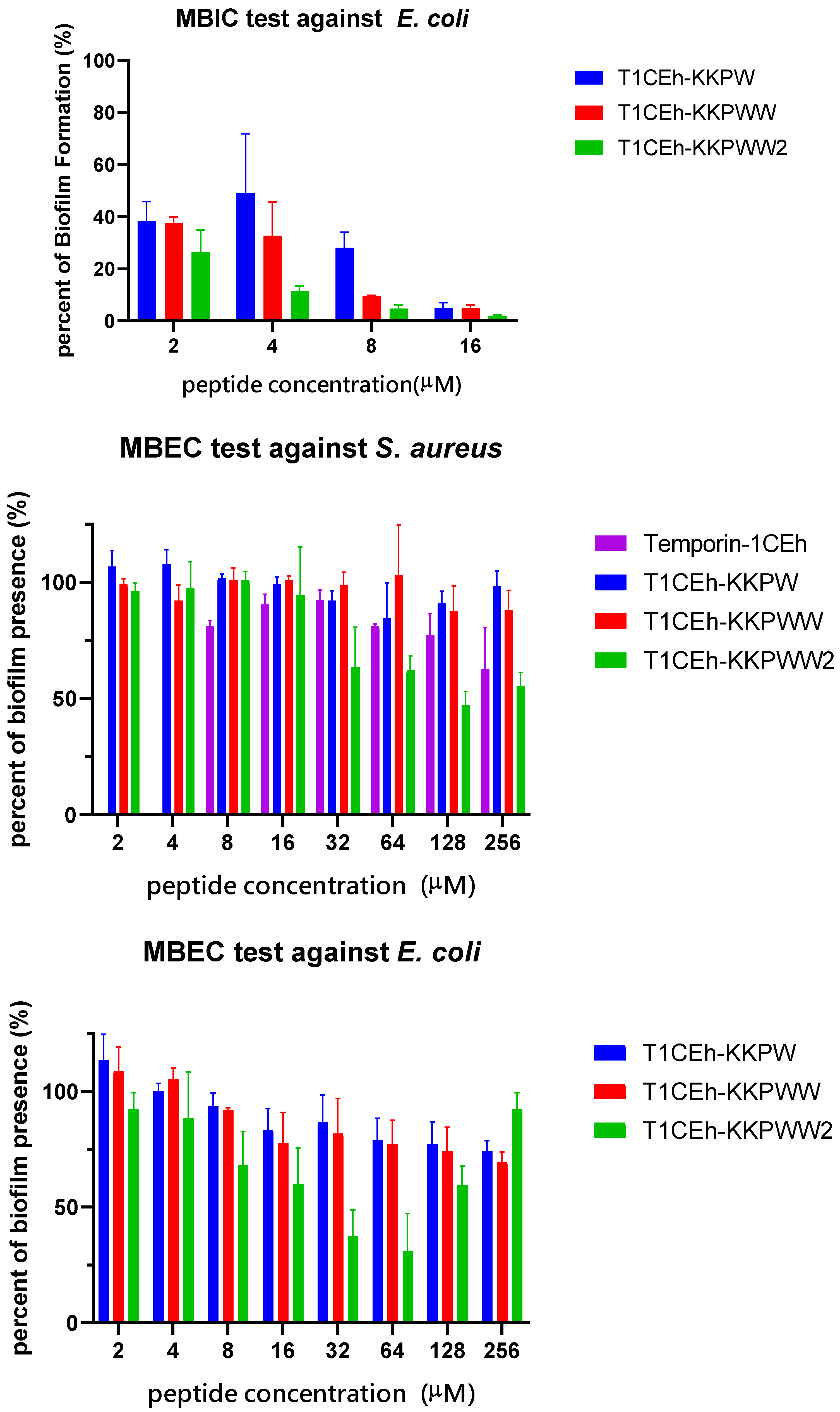
3.6. Antibiofilm Assays
3.7. Assessing the Impact of Different Environments on Antimicrobial Activity
3.8. Assessing the Efficacy of the Peptide against S. aureus and E. coli In Vivo
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Bin Zaman, S.; Hussain, M.A.; Nye, R.; Mehta, V.; Mamun, K.T.; Hossain, N. A Review on Antibiotic Resistance: Alarm Bells Are Ringing. Cureus 2017, 9, e1403. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rotem, S.; Mor, A. Antimicrobial peptide mimics for improved therapeutic properties. Biochim. Biophys. Acta Biomembr. 2009, 1788, 1582–1592. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chung, P.Y.; Khanum, R. Antimicrobial peptides as potential anti-biofilm agents against multidrug-resistant bacteria. J. Microbiol. Immunol. Infect. 2017, 50, 405–410. [Google Scholar] [CrossRef] [PubMed]
- Mishra, B.; Reiling, S.; Zarena, D.; Wang, G. Host defense antimicrobial peptides as antibiotics: Design and application strategies. Curr. Opin. Chem. Biol. 2017, 38, 87–96. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Koh, J.J.; Liu, S.; Lakshminarayanan, R.; Verma, C.S.; Beuerman, R.W. Membrane active antimicrobial peptides: Translating mechanistic insights to design. Front. Neurosci. 2017, 11, 73. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lowe, J. Mechanisms of Antibiotic Resistance. Annu. Rep. Med. Chem. 2016, 17, 119–127. [Google Scholar] [CrossRef]
- Huan, Y.; Kong, Q.; Mou, H.; Yi, H. Antimicrobial Peptides: Classification, Design, Application and Research Progress in Multiple Fields. Front. Microbiol. 2020, 11, 582779. [Google Scholar] [CrossRef]
- Mangoni, M.L.; Carotenuto, A.; Auriemma, L.; Saviello, M.R.; Campiglia, P.; Gomez-Monterrey, I.; Malfi, S.; Marcellini, L.; Barra, D.; Novellino, E.; et al. Structure-activity relationship, conformational and biological studies of temporin l analogues. J. Med. Chem. 2011, 54, 1298–1307. [Google Scholar] [CrossRef]
- Pachón-Ibáñez, M.E.; Smani, Y.; Pachón, J.; Sánchez-Céspedes, J. Perspectives for clinical use of engineered human host defense antimicrobial peptides. FEMS Microbiol. Rev. 2017, 41, 323–342. [Google Scholar] [CrossRef]
- Bhattacharjya, S.; Straus, S.K. Design, engineering and discovery of novel α-helical and β-boomerang antimicrobial peptides against drug resistant bacteria. Int. J. Mol. Sci. 2020, 21, 5773. [Google Scholar] [CrossRef]
- He, H.; Chen, Y.; Ye, Z.; Chen, X.; Ma, C.; Zhou, M.; Xi, X.; Burrows, J.F.; Chen, T.; Wang, L. Modification and targeted design of N-terminal truncates derived from brevinin with improved therapeutic efficacy. Biology 2020, 9, 209. [Google Scholar] [CrossRef] [PubMed]
- Strömstedt, A.A.; Pasupuleti, M.; Schmidtchen, A.; Malmsten, M. Evaluation of strategies for improving proteolytic resistance of antimicrobial peptides by using variants of EFK17, an internal segment of LL-37. Antimicrob. Agents Chemother. 2009, 53, 593–602. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pinilla, C.M.B.; Lopes, N.A.; Brandelli, A. Lipid-Based Nanostructures for the Delivery of Natural Antimicrobials. Molecules 2021, 26, 3587. [Google Scholar] [CrossRef] [PubMed]
- Jin, L.L.; Song, S.S.; Li, Q.; Chen, Y.H.; Wang, Q.Y.; Hou, S.T. Identification and characterisation of a novel antimicrobial polypeptide from the skin secretion of a Chinese frog (Rana chensinensis). Int. J. Antimicrob. Agents 2009, 33, 538–542. [Google Scholar] [CrossRef] [PubMed]
- Ling, X.M.; Zhang, X.H.; Tan, Y.; Yang, J.J.; Ji, B.; Wu, X.R.; Yi, Y.K.; Liang, L. Protective effects of Oviductus Ranae-containing serum on oxidative stress-induced apoptosis in rat ovarian granulosa cells. J. Ethnopharmacol. 2017, 208, 138–148. [Google Scholar] [CrossRef] [PubMed]
- Shang, D.; Sun, Y.; Wang, C.; Wei, S.; Ma, L.; Sun, L. Membrane interaction and antibacterial properties of chensinin-1, an antimicrobial peptide with atypical structural features from the skin of Rana chensinensis. Appl. Microbiol. Biotechnol. 2012, 96, 1551–1560. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Dong, S.; Zhang, L.; Zhao, Y.; Huang, L.; Gong, X.; Wang, H.; Shang, D. Cell surface binding, uptaking and anticancer activity of L-K6, a lysine/leucine-rich peptide, on human breast cancer MCF-7 cells. Sci. Rep. 2017, 7, 8293. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Cheng, Y.; Yang, Y.; Liu, S.; Shi, H.; Lu, C.; Li, S.; Nie, L.; Su, D.; Deng, X.; et al. Polypeptides from the Skin of Rana chensinensis Exert the Antioxidant and Antiapoptotic Activities on HaCaT Cells. ANQ Q. J. Short Artic. Notes Rev. 2017, 30, 1–10. [Google Scholar] [CrossRef]
- Wang, Z.; Zhao, Y.; Su, T.; Zhang, J.; Wang, F. Characterization and antioxidant activity in vitro and in vivo of polysaccharide purified from Rana chensinensis skin. Carbohydr. Polym. 2015, 126, 17–22. [Google Scholar] [CrossRef]
- Langsdorf, M.; Ghassempour, A.; Römpp, A.; Spengler, B. Isolation and sequence analysis of peptides from the skin secretion of the Middle East tree frog Hyla savignyi. Anal. Bioanal. Chem. 2010, 398, 2853–2865. [Google Scholar] [CrossRef]
- Chen, Y.; Xi, X.; Ma, C.; Zhou, M.; Chen, X.; Ye, Z.; Ge, L.; Wu, Q.; Chen, T.; Wang, L.; et al. Structure–activity relationship and molecular docking of a Kunitz-like trypsin inhibitor, Kunitzin-ah, from the skin secretion of Amolops hainanensis. Pharmaceutics 2021, 13, 966. [Google Scholar] [CrossRef] [PubMed]
- Chen, D.; Zhou, X.; Chen, X.; Huang, L.; Xi, X.; Ma, C.; Zhou, M.; Wang, L.; Chen, T. Evaluating the Bioactivity of a Novel Antimicrobial. Molecules 2019, 24, 2974. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hicks, R. Preparation of Membrane Models of Gram-Negative Bacteria and Their Interaction with Antimicrobial Peptides Studied by CD and NMR. Methods Mol. Biol. 2017, 1548, 231–245. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.; Shi, D.; Zhong, R.; Ye, Z.; Ma, C.; Zhou, M.; Xi, X.; Wang, L.; Chen, T.; Kwok, H.F. Bioevaluation of ranatuerin-2Pb from the frog skin secretion of Rana pipiens and its truncated analogues. Biomolecules 2019, 9, 249. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yuan, Y.; Zai, Y.; Xi, X.; Ma, C.; Wang, L.; Zhou, M.; Shaw, C.; Chen, T. A novel membrane-disruptive antimicrobial peptide from frog skin secretion against cystic fibrosis isolates and evaluation of anti-MRSA effect using Galleria mellonella model. Biochim. Biophys. Acta Gen. Subj. 2019, 1863, 849–856. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zai, Y.; Ying, Y.; Ye, Z.; Zhou, M.; Ma, C.; Shi, Z.; Chen, X.; Xi, X.; Chen, T.; Wang, L. Broad-spectrum antimicrobial activity and improved stability of a D-amino acid enantiomer of DMPC-10a, the designed derivative of dermaseptin truncates. Antibiotics 2020, 9, 627. [Google Scholar] [CrossRef]
- Gao, Y.; Wu, D.; Xi, X.; Wu, Y.; Ma, C.; Zhou, M.; Wang, L.; Yang, M.; Chen, T.; Shaw, C. Identification and characterisation of the antimicrobial peptide, phylloseptin-PT, from the skin secretion of Phyllomedusa tarsius, and comparison of activity with designed, cationicity-enhanced analogues and diastereomers. Molecules 2016, 21, 1667. [Google Scholar] [CrossRef] [Green Version]
- Ignasiak, K.; Maxwell, A. Galleria mellonella (greater wax moth) larvae as a model for antibiotic susceptibility testing and acute toxicity trials. BMC Res. Notes 2017, 10, 428. [Google Scholar] [CrossRef] [Green Version]
- Shang, D.; Yu, F.; Li, J.; Zheng, J.; Zhang, L.; Li, Y.; Shang, D.; Yu, F.; Li, J.; Zheng, J.; et al. Molecular Cloning of cDNAs Encoding Antimicrobial Peptide Precursors from the Skin of the Chinese Brown Frog, Rana chensinensis. Zool. Sci. 2009, 26, 220–226. [Google Scholar] [CrossRef] [PubMed]
- Cheng, J.T.J.; Hale, J.D.; Kindrachuk, J.; Jessen, H.; Elliott, M.; Hancock, R.E.W.; Straus, S.K. Importance of residue 13 and the C-terminus for the structure and activity of the antimicrobial peptide aurein 2.2. Biophys. J. 2010, 99, 2926–2935. [Google Scholar] [CrossRef] [Green Version]
- Manzo, G.; Ferguson, P.M.; Gustilo, V.B.; Hind, C.K.; Clifford, M.; Bui, T.T.; Drake, A.F.; Atkinson, R.A.; Sutton, J.M.; Batoni, G.; et al. Minor sequence modifications in temporin B cause drastic changes in antibacterial potency and selectivity by fundamentally altering membrane activity. Sci. Rep. 2019, 9, 1385. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Marimuthu, S.K.; Nagarajan, K.; Perumal, S.K.; Palanisamy, S.; Subbiah, L. Insilico Alpha-Helical Structural Recognition of Temporin Antimicrobial Peptides and Its Interactions with Middle East Respiratory Syndrome-Coronavirus. Int. J. Pept. Res. Ther. 2020, 26, 1473–1483. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Buommino, E.; Carotenuto, A.; Antignano, I.; Bellavita, R.; Casciaro, B.; Loffredo, M.R.; Merlino, F.; Novellino, E.; Mangoni, M.L.; Nocera, F.P.; et al. The Outcomes of Decorated Prolines in the Discovery of Antimicrobial Peptides from Temporin-L. Chem. Med. Chem. 2019, 14, 1283–1290. [Google Scholar] [CrossRef] [PubMed]
- Chou, S.; Li, Q.; Nina, Z.; Shang, L.; Li, J.; Li, J.; Wang, Z.; Shan, A. Peptides with Triplet-Tryptophan-Pivot Promoted Pathogenic Bacteria Membrane Defects. Front. Microbiol. 2020, 11, 537. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Manzo, G.; Scorciapino, M.A.; Wadhwani, P.; Bürck, J.; Montaldo, P.; Pintus, M.; Sanna, R.; Casu, M.; Giuliani, A. Enhanced Amphiphilic Profile of a Short β-Stranded Peptide Improves Its Antimicrobial Activity. Biomolecules 2015, 8, e0116379. [Google Scholar] [CrossRef] [PubMed]
- Micsonai, A.; Wien, F.; Bulyáki, É.; Kun, J.; Moussong, É.; Lee, Y.H.; Goto, Y.; Réfrégiers, M.; Kardos, J. BeStSel: A web server for accurate protein secondary structure prediction and fold recognition from the circular dichroism spectra. Nucleic Acids Res. 2018, 46, W315–W322. [Google Scholar] [CrossRef] [PubMed]
- Sierra, J.M.; Fusté, E.; Rabanal, F.; Vinuesa, T.; Viñas, M. An overview of antimicrobial peptides and the latest advances in their development. Expert Opin. Biol. Ther. 2017, 17, 663–676. [Google Scholar] [CrossRef]
- Abbassi, F.; Humblot, V.; Lequin, O.; Raja, Z.; Andre, S.; Bouceba, T.; Correia, I.; Casale, S.; Foulon, T.; Sereno, D.; et al. Insight into the Mechanism of Action of Temporin-SHa, a New Broad-Spectrum Antiparasitic and Antibacterial Agent. PLoS ONE 2017, 12, e0174024. [Google Scholar] [CrossRef] [Green Version]
- Grohmann, E.; Christie, P.J.; Waksman, G.; Backert, S. Micro Review Type IV secretion in Gram-negative and Gram-positive bacteria. Mol. Microbiol. 2018, 107, 455–471. [Google Scholar] [CrossRef]
- Vanzolini, T.; Bruschi, M.; Rinaldi, A.C.; Magnani, M.; Fraternale, A. Multitalented Synthetic Antimicrobial Peptides and Their Antibacterial, Antifungal and Antiviral Mechanisms. Int. J. Mol. Sci. 2022, 23, 545. [Google Scholar] [CrossRef]
- Hyun, S.; Choi, Y.; Jo, D.; Choo, S.; Park, T.W.; Park, S.J.; Kim, S.; Lee, S.; Park, S.; Jin, S.M.; et al. Proline Hinged Amphipathic α-Helical Peptide Sensitizes Gram-Negative Bacteria to Various Gram-Positive Antibiotics. J. Med. Chem. 2020, 63, 14937–14950. [Google Scholar] [CrossRef] [PubMed]
- Son, M.; Lee, Y.; Hwang, H.; Hyun, S.; Yu, J. Disruption of interactions between hydrophobic residues on nonpolar faces is a key determinant in decreasing hemolysis and increasing antimicrobial activities of α-helical amphipathic peptides. Chem. Med. Chem. 2013, 8, 1638–1642. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rex, S. A Pro→Ala substitution in melittin affects self-association, membrane binding and pore-formation kinetics due to changes in structural and electrostatic properties. Biophys. Chem. 2000, 85, 209–228. [Google Scholar] [CrossRef]
- Park, C.B.; Yi, K.S.; Matsuzaki, K.; Kim, M.S.; Kim, S.C. Structure-activity analysis of buforin II, a histone H2A-derived antimicrobial peptide: The proline hinge is responsible for the cell-penetrating ability of buforin II. Proc. Natl. Acad. Sci. USA 2000, 97, 8245–8250. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, J.K.; Gopal, R.; Park, S.C.; Ko, H.S.; Kim, Y.; Hahm, K.S.; Park, Y. A Proline-Hinge Alters the Characteristics of the Amphipathic α-helical AMPs. PLoS ONE 2013, 8, e67597. [Google Scholar] [CrossRef] [PubMed]
- Di Somma, A.; Moretta, A.; Canè, C.; Cirillo, A.; Duilio, A. Antimicrobial and antibiofilm peptides. Biomolecules 2020, 10, 652. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rohde, H.; Frankenberger, S.; Zähringer, U.; Mack, D. Structure, function and contribution of polysaccharide intercellular adhesin (PIA) to Staphylococcus epidermidis biofilm formation and pathogenesis of biomaterial-associated infections. Eur. J. Cell Biol. 2010, 89, 103–111. [Google Scholar] [CrossRef] [PubMed]
- Vuong, C.; Voyich, J.M.; Fischer, E.R.; Braughton, K.R.; Whitney, A.R.; DeLeo, F.R.; Otto, M. Polysaccharide intercellular adhesin (PIA) protects Staphylococcus epidermidis against major components of the human innate immune system. Cell. Microbiol. 2004, 6, 269–275. [Google Scholar] [CrossRef] [PubMed]
- Mohamed, M.F.; Brezden, A.; Mohammad, H.; Chmielewski, J.; Seleem, M.N. A short D-enantiomeric antimicrobial peptide with potent immunomodulatory and antibiofilm activity against multidrug-resistant Pseudomonas aeruginosa and Acinetobacter baumannii. Sci. Rep. 2017, 7, 6953. [Google Scholar] [CrossRef]
- Zhu, X.; Dong, N.; Wang, Z.; Ma, Z.; Zhang, L.; Ma, Q.; Shan, A. Design of imperfectly amphipathic α-helical antimicrobial peptides with enhanced cell selectivity. Acta Biomater. 2014, 10, 244–257. [Google Scholar] [CrossRef]
- Neumann, A.; Völlger, L.; Berends, E.T.M.; Molhoek, E.M.; Stapels, D.A.C.; Midon, M.; Friães, A.; Pingoud, A.; Rooijakkers, S.H.M.; Gallo, R.L.; et al. Novel role of the antimicrobial peptide LL-37 in the protection of neutrophil extracellular Traps against degradation by bacterial nucleases. J. Innate Immun. 2014, 6, 860–868. [Google Scholar] [CrossRef] [PubMed]
- Kanchi, P.K.; Dasmahapatra, A.K. Enhancing the binding of the β-sheet breaker peptide LPFFD to the amyloid-β fibrils by aromatic modifications: A molecular dynamics simulation study. Comput. Biol. Chem. 2021, 92, 107471. [Google Scholar] [CrossRef] [PubMed]
- Chowdhary, S.; Moschner, J.; Mikolajczak, D.J.; Becker, M.; Thünemann, A.F.; Kästner, C.; Klemczak, D.; Stegemann, A.K.; Böttcher, C.; Metrangolo, P.; et al. The Impact of Halogenated Phenylalanine Derivatives on NFGAIL Amyloid Formation. ChemBioChem 2020, 21, 3544–3554. [Google Scholar] [CrossRef] [PubMed]
- Luo, R.; Wan, Y.; Luo, X.; Liu, G.; Li, Z.; Chen, J.; Su, D.; Lu, N.; Luo, Z. A Rapid Self-Assembly Peptide Hydrogel for Recruitment and Activation of Immune Cells. Molecules 2022, 27, 419. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Qian, Y.; Xu, L.; Shao, Y.; Zhang, H.; Shi, F.; Chen, J.; Cui, S.; Chen, X.; Zhu, D.; et al. Hyaluronic acid-shelled, peptide drug conjugate-cored nanomedicine for the treatment of hepatocellular carcinoma. Mater. Sci. Eng. C 2020, 117, 111261. [Google Scholar] [CrossRef] [PubMed]
- Zai, Y.; Xi, X.; Ye, Z.; Ma, C.; Zhou, M.; Chen, X.; Siu, S.W.I.; Chen, T.; Wang, L.; Kwok, H.F. Aggregation and its influence on the bioactivities of a novel antimicrobial peptide, temporin-pf, and its analogues. Int. J. Mol. Sci. 2021, 22, 4509. [Google Scholar] [CrossRef] [PubMed]
- Sarig, H.; Rotem, S.; Ziserman, L.; Danino, D.; Mor, A. Impact of self-assembly properties on antibacterial activity of short acyl-lysine oligomers. Antimicrob. Agents Chemother. 2008, 52, 4308–4314. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Haney, E.F.; Wu, B.; Lee, K.; Hilchie, A.L.; Hancock, R.E.W. Aggregation and Its Influence on the Immunomodulatory Activity of Synthetic Innate Defense Regulator Peptides. Cell Chem. Biol. 2017, 24, 969–980.e4. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Batoni, G.; Casu, M.; Giuliani, A.; Luca, V.; Maisetta, G.; Mangoni, M.L.; Manzo, G.; Pintus, M.; Pirri, G.; Rinaldi, A.C.; et al. Rational modification of a dendrimeric peptide with antimicrobial activity: Consequences on membrane-binding and biological properties. Amino Acids 2016, 48, 887–900. [Google Scholar] [CrossRef] [PubMed]
- Tsai, C.J.Y.; Loh, J.M.S.; Proft, T. Galleria mellonella infection models for the study of bacterial diseases and for antimicrobial drug testing. Virulence 2016, 7, 214–229. [Google Scholar] [CrossRef] [Green Version]






| Peptide | Sequence |
|---|---|
| Temporin-1CEh | FVDLKKIANILNSIF-NH2 |
| Temporin-1CEa | FVDLKKIANIINSIF-NH2 |
| T1CEh-t | FVDLKKIANIL-NH2 |
| T1CEa-t | FVDLKKIANII-NH2 |
| T1CEh-KK | KKFVDLKKIANIL-NH2 |
| T1CEh-KKP | KKFVPLKKIANIL-NH2 |
| T1CEh-KKPW | KKWVPLKKIANIL-NH2 |
| T1CEh-KKPWW | KKWVPWKKIANIL-NH2 |
| T1CEh-KKPWW2 | (KKWVPWK)2KIANIL-NH2 |
| Bacteria Strains | MIC/MBC (μM) | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Temporin-1CEh | Temporin-1CEa | T1CEh-t | T1CEa-t | T1CEh-KK | T1CEh-KKP | T1CEh-KKPW | T1CEh-KKPWW | T1CEh-KKPWW2 | |
| S. aureus | 8/16 | 13.1 | >512/>512 | >512/>512 | 256/>512 | 32/64 | 8/16 | 4/4 | 4/8 |
| MRSA | 16/16 | - | >512/>512 | >512/>512 | >512/>512 | 256/256 | 16/64 | 32/256 | 8/16 |
| E. faecalis | 32/32 | - | >512/>512 | >512/>512 | 64/>512 | >512/>512 | >512/>512 | 128/128 | 32/64 |
| K. pneumoniae | 256/256 | - | >512/>512 | >512/>512 | 128/128 | >512/>512 | 16/16 | 4/4 | 4/8 |
| P. aeruginosa | 128/256 | - | >512/>512 | >512/>512 | 256/256 | 512/>512 | 32/64 | 16/16 | 8/16 |
| E. coli | 128/128 | >100 | >512/>512 | >512/>512 | 8/16 | 128/128 | 8/16 | 4/4 | 2/2 |
| Peptide | Hydrophobicity (H) | Hydrophobic Moment (μH) | Net Charge (z) | α-Helicity (%) |
|---|---|---|---|---|
| Temporin-1CEh | 0.661 | 0.518 | 1 | 47.8 |
| Temporin-1CEa | 0.668 | 0.524 | 1 | - |
| T1CEh-t | 0.634 | 0.481 | 1 | 33 |
| T1CEa-t | 0.643 | 0.490 | 1 | 40.2 |
| T1CEh-KK | 0.384 | 0.385 | 3 | 46.2 |
| T1CEh-KKP | 0.498 | 0.394 | 4 | 28.8 |
| T1CEh-KKPW | 0.534 | 0.400 | 4 | 40.4 |
| T1CEh-KKPWW | 0.576 | 0.440 | 4 | 30.2 |
| Bacteria Strains | MBIC/MBEC (µM) | |||
|---|---|---|---|---|
| Temporin-1CEh | T1CEh-KKPW | T1CEh-KKPWW | T1CEh-KKPWW2 | |
| S. aureus | 64/>256 | 16/>256 | 8/>256 | 32/>256 |
| E. coli | - | 16/>256 | 8/>256 | 8/>256 |
| Peptide | Bacteria | MIC (μM) | ||||||
|---|---|---|---|---|---|---|---|---|
| pH 6.0 | pH 7.0 | pH8.0 | MHB + 10% FBS | MHB + 150 mM NaCl | MHB + 1 mM MgCl2 | MHB + 4 µM FeCl3 | ||
| Temporin-1CEh | S. aureus | 128 | 8 | 128 | 128 | 128 | 128 | 128 |
| MRSA | 256 | 16 | 256 | 256 | 256 | 128 | 128 | |
| E. faecalis | >512 | 32 | >512 | >512 | >512 | 128 | 128 | |
| K. pneumoniae | >512 | 256 | >512 | >512 | >512 | >512 | >512 | |
| P. aeruginosa | >512 | 128 | >512 | >512 | >512 | >512 | >512 | |
| E. coli | >512 | 128 | >512 | >512 | >512 | >512 | >512 | |
| T1CEh-KKPW | S. aureus | >512 | 8 | 32 | >512 | >512 | 256 | >512 |
| MRSA | >512 | 16 | 128 | >512 | >512 | >512 | >512 | |
| E. faecalis | >512 | >512 | 256 | 512 | >512 | >512 | >512 | |
| K. pneumoniae | 256 | 16 | 128 | 256 | >512 | 16 | 16 | |
| P. aeruginosa | 64 | 32 | 32 | 64 | >512 | >512 | >512 | |
| E. coli | 16 | 8 | 8 | 8 | 128 | 32 | 64 | |
| T1CEh-KKPWW | S. aureus | 32 | 4 | 4 | 128 | 16 | 32 | 64 |
| MRSA | 256 | 32 | 32 | 64 | >512 | >512 | 512 | |
| E. faecalis | >512 | 128 | 64 | 128 | >512 | 512 | 512 | |
| K. pneumoniae | 32 | 4 | 8 | 64 | 256 | 16 | 8 | |
| P. aeruginosa | 16 | 16 | 4 | 32 | >512 | >512 | >512 | |
| E. coli | 4 | 4 | 4 | 4 | 8 | 8 | 8 | |
| T1CEh-KKPWW2 | S. aureus | 8 | 4 | 2 | 4 | 16 | 8 | 4 |
| MRSA | 16 | 8 | 2 | 8 | 8 | 8 | 4 | |
| E. faecalis | 128 | 32 | 64 | 64 | 128 | 64 | 64 | |
| K. pneumoniae | 8 | 4 | 4 | 16 | 16 | 8 | 8 | |
| P. aeruginosa | 32 | 8 | 8 | 32 | 32 | 16 | 16 | |
| E. coli | 2 | 2 | 2 | 4 | 2 | 4 | 2 | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ye, Z.; Zhou, X.; Xi, X.; Zai, Y.; Zhou, M.; Chen, X.; Ma, C.; Chen, T.; Wang, L.; Kwok, H.F. In Vitro & In Vivo Studies on Identifying and Designing Temporin-1CEh from the Skin Secretion of Rana chensinensis as the Optimised Antibacterial Prototype Drug. Pharmaceutics 2022, 14, 604. https://doi.org/10.3390/pharmaceutics14030604
Ye Z, Zhou X, Xi X, Zai Y, Zhou M, Chen X, Ma C, Chen T, Wang L, Kwok HF. In Vitro & In Vivo Studies on Identifying and Designing Temporin-1CEh from the Skin Secretion of Rana chensinensis as the Optimised Antibacterial Prototype Drug. Pharmaceutics. 2022; 14(3):604. https://doi.org/10.3390/pharmaceutics14030604
Chicago/Turabian StyleYe, Zhuming, Xiaowei Zhou, Xinping Xi, Yu Zai, Mei Zhou, Xiaoling Chen, Chengbang Ma, Tianbao Chen, Lei Wang, and Hang Fai Kwok. 2022. "In Vitro & In Vivo Studies on Identifying and Designing Temporin-1CEh from the Skin Secretion of Rana chensinensis as the Optimised Antibacterial Prototype Drug" Pharmaceutics 14, no. 3: 604. https://doi.org/10.3390/pharmaceutics14030604
APA StyleYe, Z., Zhou, X., Xi, X., Zai, Y., Zhou, M., Chen, X., Ma, C., Chen, T., Wang, L., & Kwok, H. F. (2022). In Vitro & In Vivo Studies on Identifying and Designing Temporin-1CEh from the Skin Secretion of Rana chensinensis as the Optimised Antibacterial Prototype Drug. Pharmaceutics, 14(3), 604. https://doi.org/10.3390/pharmaceutics14030604







_Kwok.png)

