An Evaluation of CXCR4 Targeting with PAMAM Dendrimer Conjugates for Oncologic Applications
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Modification of POL3026 with the Linker
2.3. Synthesis of CXCR4-Targeted G5-X4 and Control G5-Ctrl Dendrimers
2.4. Matrix-Assisted Laser Desorption Ionization-Time-of-Flight
2.5. Dynamic Light Scattering (DLS) and Zeta Potential (ZP)
2.6. Cell Lines
2.7. Competitive Binding Assays
2.8. Flow Cytometry
2.9. Inhibition of Chemotaxis
2.10. Radiolabeling
2.11. Animal Models
2.12. Ex Vivo Biodistribution
2.13. SPECT/CT Imaging and Analysis
2.14. Data Analysis
3. Results
3.1. Synthesis and Physicochemical Characterization of G5-Ctrl and G5-X4
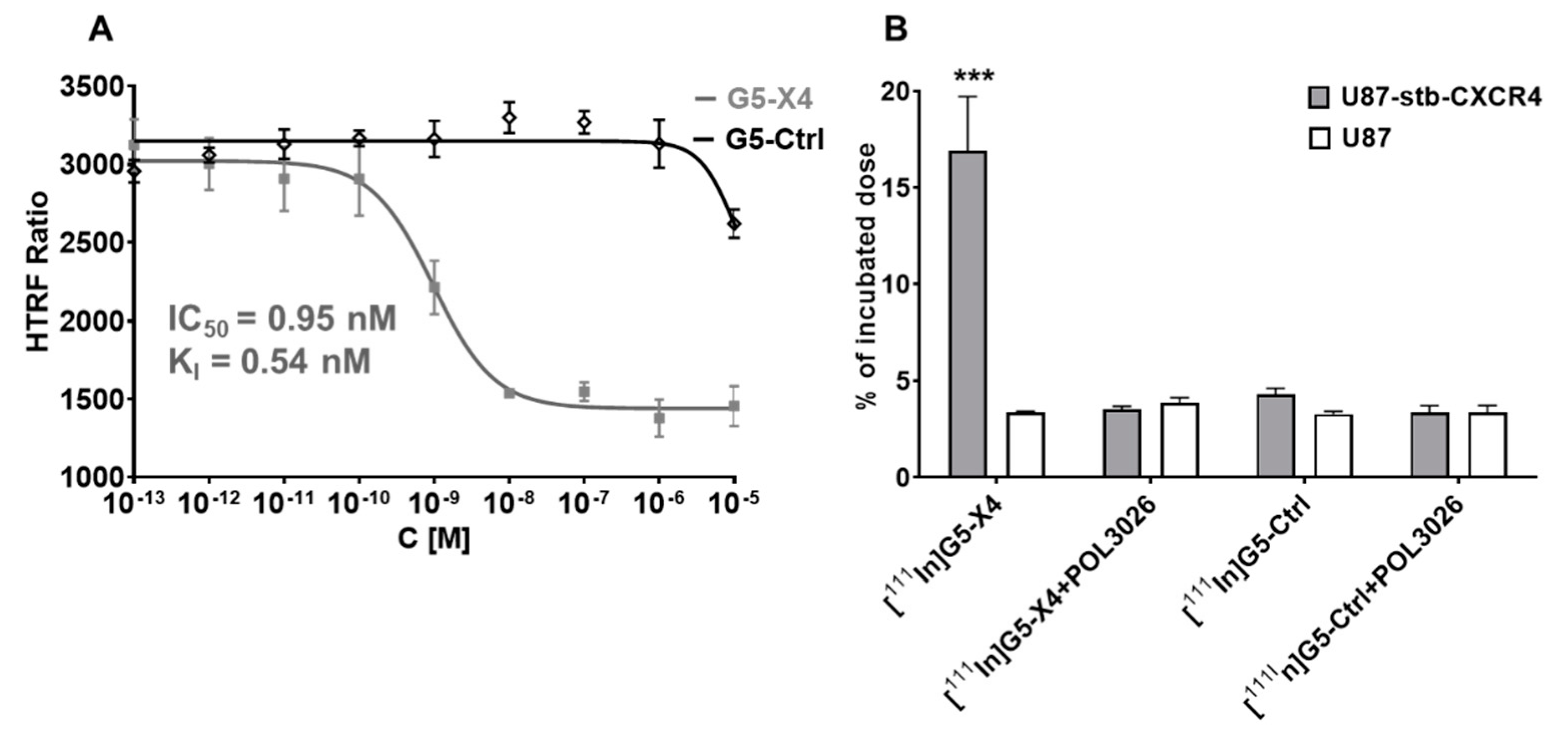
3.2. Evaluation of In Vitro CXCR4 Specificity
3.3. Radiolabeling of G5-Ctrl and G5-X4
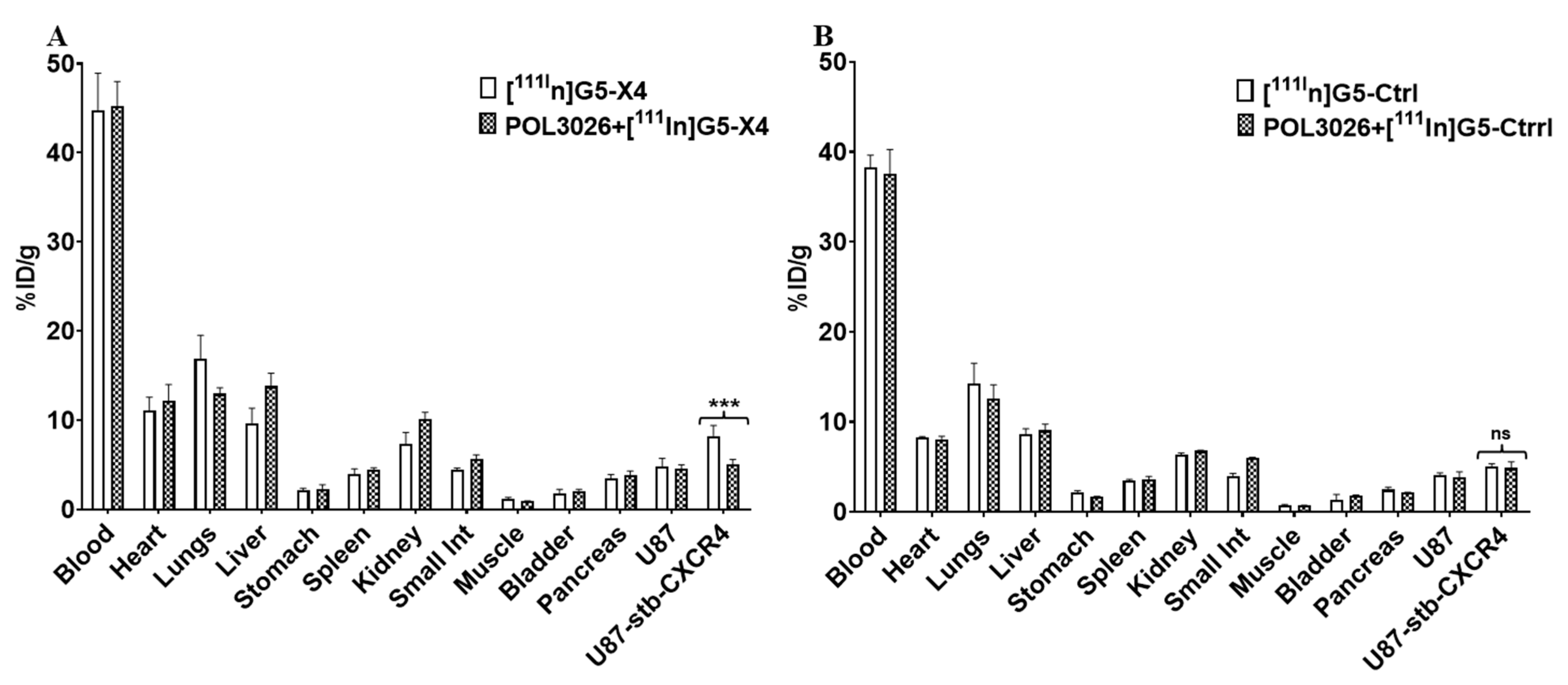
3.4. Evaluation of In Vitro Specificity
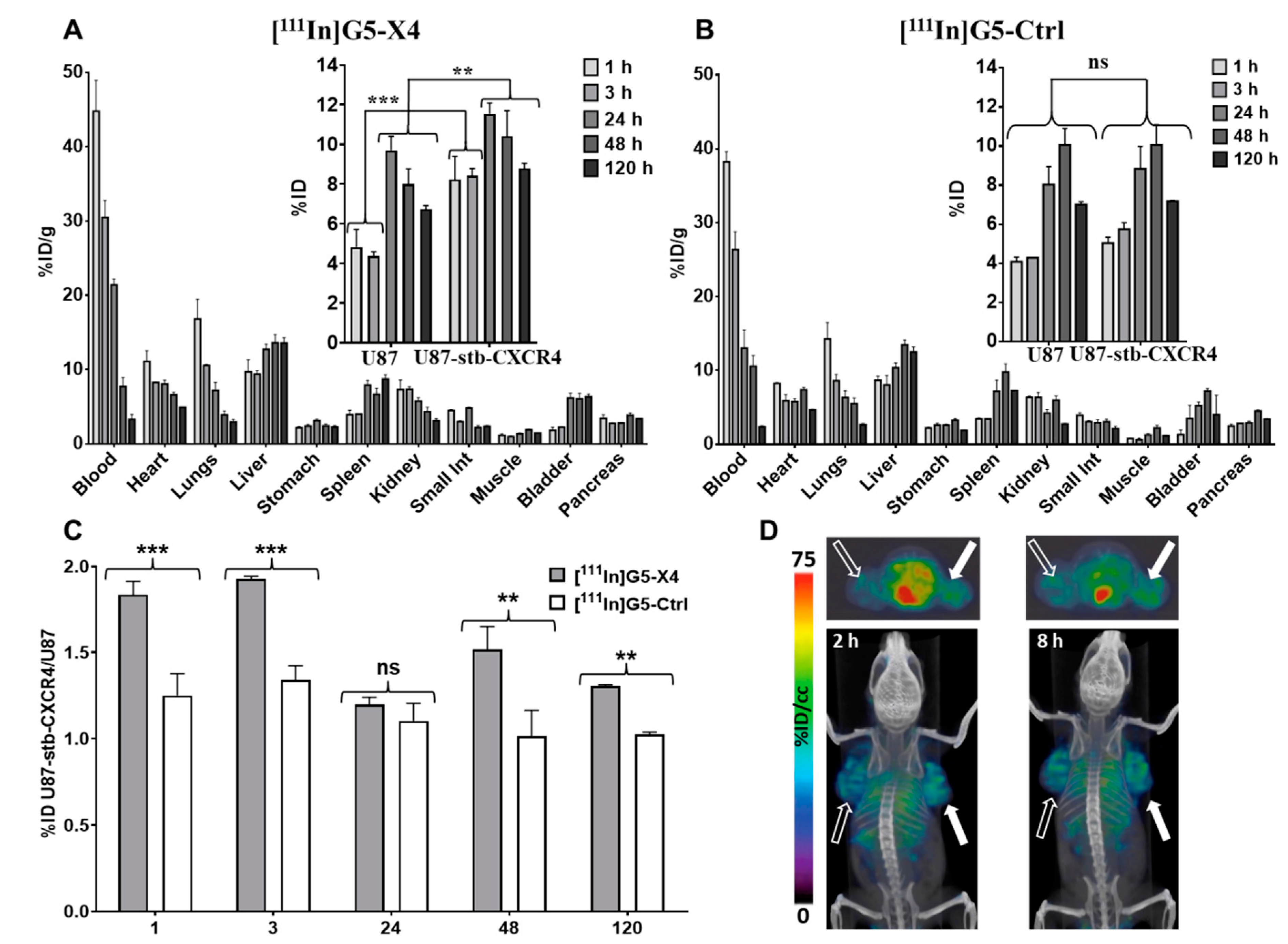
3.5. Biodistribution of [111In]G5-X4 and [111In]G5-Ctrl
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Teicher, B.A.; Fricker, S.P. CXCL12 (SDF-1)/CXCR4 pathway in cancer. Clin. Cancer Res. 2010, 16, 2927–2931. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Z.; Ni, C.; Chen, W.; Wu, P.; Wang, Z.; Yin, J.; Huang, J.; Qiu, F. Expression of CXCR4 and breast cancer prognosis: A systematic review and meta-analysis. BMC Cancer 2014, 14, 49. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Müller, A.; Homey, B.; Soto, H.; Ge, N.; Catron, D.; Buchanan, M.E.; McClanahan, T.; Murphy, E.; Yuan, W.; Wagner, S.N.; et al. Involvement of chemokine receptors in breast cancer metastasis. Nature 2001, 410, 50–56. [Google Scholar] [CrossRef] [PubMed]
- Wald, O.; Shapira, O.M.; Izhar, U. CXCR4/CXCL12 axis in non small cell lung cancer (NSCLC) pathologic roles and therapeutic potential. Theranostics 2013, 3, 26–33. [Google Scholar] [CrossRef] [Green Version]
- Burger, J.A.; Stewart, D.J.; Wald, O.; Peled, A. Potential of CXCR4 antagonists for the treatment of metastatic lung cancer. Expert Rev. Anticancer Ther. 2011, 11, 621–630. [Google Scholar] [CrossRef]
- Bachelerie, F.; Ben-Baruch, A.; Burkhardt, A.M.; Combadiere, C.; Farber, J.M.; Graham, G.; Horuk, R.; Sparre-Ulrich, A.H.; Locati, M.; Luster, A.D.; et al. International Union of Basic and Clinical Pharmacology. LXXXIX. Update on the Extended Family of Chemokine Receptors and Introducing a New Nomenclature for Atypical Chemokine Receptors. Pharmacol. Rev. 2013, 66, 1–79. [Google Scholar] [CrossRef] [Green Version]
- Chatterjee, S.; Azad, B.B.; Nimmagadda, S. The intricate role of CXCR4 in cancer. Adv. Cancer Res. 2014, 124, 31–82. [Google Scholar] [CrossRef] [Green Version]
- Feig, C.; Jones, J.O.; Kraman, M.; Wells, R.J.; Deonarine, A.; Chan, D.S.; Connell, C.M.; Roberts, E.W.; Zhao, Q.; Caballero, O.L.; et al. Targeting CXCL12 from FAP-expressing carcinoma-associated fibroblasts synergizes with anti-PD-L1 immunotherapy in pancreatic cancer. Proc. Natl. Acad. Sci. USA 2013, 110, 20212–20217. [Google Scholar] [CrossRef] [Green Version]
- Hassan, S.; Buchanan, M.; Jahan, K.; Aguilar-Mahecha, A.; Gaboury, L.; Muller, W.J.; Alsawafi, Y.; Mourskaia, A.A.; Siegel, P.M.; Salvucci, O.; et al. CXCR4 peptide antagonist inhibits primary breast tumor growth, metastasis and enhances the efficacy of anti-VEGF treatment or docetaxel in a transgenic mouse model. Int. J. Cancer 2010, 129, 225–232. [Google Scholar] [CrossRef]
- Kioi, M.; Vogel, H.; Schultz, G.; Hoffman, R.M.; Harsh, G.R.; Brown, J.M. Inhibition of vasculogenesis, but not angiogenesis, prevents the recurrence of glioblastoma after irradiation in mice. J. Clin. Investig. 2010, 120, 694–705. [Google Scholar] [CrossRef]
- Liang, Z.; Wu, T.; Lou, H.; Yu, X.; Taichman, R.S.; Lau, S.K.; Nie, S.; Umbreit, J.; Shim, H. Inhibition of breast cancer metastasis by selective synthetic polypeptide against CXCR4. Cancer Res. 2004, 64, 4302–4308. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Domanska, U.M.; Kruizinga, R.C.; Nagengast, W.B.; Timmer-Bosscha, H.; Huls, G.; de Vries, E.G.E.; Walenkamp, A.M.E. A review on CXCR4/CXCL12 axis in oncology: No place to hide. Eur. J. Cancer 2013, 49, 219–230. [Google Scholar] [CrossRef] [PubMed]
- Ghobrial, I.M.; Liu, C.-J.; Redd, R.A.; Perez, R.P.; Baz, R.; Zavidij, O.; Sklavenitis-Pistofidis, R.; Richardson, P.G.; Anderson, K.C.; Laubach, J.P.; et al. A Phase Ib/II Trial of the First-in-Class Anti-CXCR4 Antibody Ulocuplumab in Combination with Lenalidomide or Bortezomib Plus Dexamethasone in Relapsed Multiple Myeloma. Clin. Cancer Res. 2020, 26, 344–353. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Buck, A.K.; Stolzenburg, A.; Hänscheid, H.; Schirbel, A.; Lückerath, K.; Schottelius, M.; Wester, H.-J.; Lapa, C. Chemokine receptor—Directed imaging and therapy. Methods 2017, 130, 63–71. [Google Scholar] [CrossRef]
- Gallo, J.; Kamaly, N.; Lavdas, I.; Stevens, E.; Nguyen, Q.-D.; Wylezinska-Arridge, M.; Aboagye, E.O.; Long, N.J. CXCR4-targeted and MMP-responsive iron oxide nanoparticles for enhanced magnetic resonance imaging. Angew. Chem. Int. Ed. 2014, 53, 9550–9554. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.; Kumar, S.; Rachagani, S.; Sajja, B.R.; Xie, Y.; Hang, Y.; Jain, M.; Li, J.; Boska, M.D.; Batra, S.K.; et al. Polyplex-mediated inhibition of chemokine receptor CXCR4 and chromatin-remodeling enzyme NCOA3 impedes pancreatic cancer progression and metastasis. Biomaterials 2016, 101, 108–120. [Google Scholar] [CrossRef] [Green Version]
- Asaftei, S.; Huskens, D.; Schols, D. HIV-1 X4 Activities of Polycationic “Viologen” based dendrimers by interaction with the chemokine receptor CXCR4: Study of structure–activity relationship. J. Med. Chem. 2012, 55, 10405–10413. [Google Scholar] [CrossRef]
- Pisani, A.; Donno, R.; Gennari, A.; Cibecchini, G.; Catalano, F.; Marotta, R.; Pompa, P.P.; Tirelli, N.; Bardi, G. CXCL12-PLGA/Pluronic Nanoparticle Internalization Abrogates CXCR4-Mediated Cell Migration. Nanomaterials 2020, 10, 2304. [Google Scholar] [CrossRef]
- De Araujo, R.V.; da Silva Santos, S.; Ferreira, F.I.; Giarolla, J. New Advances in General Biomedical Applications of PAMAM Dendrimers. Molecules 2018, 23, 2849. [Google Scholar] [CrossRef] [Green Version]
- Kannan, R.M.; Nance, E.; Kannan, S.; Tomalia, D.A. Emerging concepts in dendrimer-based nanomedicine: From design principles to clinical applications. J. Intern. Med. 2014, 276, 579–617. [Google Scholar] [CrossRef]
- Kim, Y.; Park, E.J.; Na, D.H. Recent progress in dendrimer-based nanomedicine development. Arch. Pharm. Res. 2018, 41, 571–582. [Google Scholar] [CrossRef] [PubMed]
- Menjoge, A.R.; Kannan, R.M.; Tomalia, D.A. Dendrimer-based drug and imaging conjugates: Design considerations for nanomedical applications. Drug Discov. Today 2010, 15, 171–185. [Google Scholar] [CrossRef] [PubMed]
- Tomalia, D.; Reyna, L.; Svenson, S. Dendrimers as multi-purpose nanodevices for oncology drug delivery and diagnostic imaging. Biochem. Soc. Trans. 2007, 35, 61–67. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, H. Targeted nanosystems: Advances in targeted dendrimers for cancer therapy. Nanomed. Nanotechnol. Biol. Med. 2015, 12, 309–316. [Google Scholar] [CrossRef] [Green Version]
- Chittasupho, C.; Anuchapreeda, S.; Sarisuta, N. CXCR4 targeted dendrimer for anti-cancer drug delivery and breast cancer cell migration inhibition. Eur. J. Pharm. Biopharm. 2017, 119, 310–321. [Google Scholar] [CrossRef]
- Chittasupho, C.; Aonsri, C.; Imaram, W. Targeted dendrimers for antagonizing the migration and viability of NALM-6 lymphoblastic leukemia cells. Bioorganic Chem. 2021, 107, 104601. [Google Scholar] [CrossRef]
- Liu, C.L.; Duan, H.; Zhao, Z.; Li, W.; Ma, L.; Fang, X.; Wang, C.; Yang, Y. Improving the inhibitory effect of CXCR4 peptide antagonist in tumor metastasis with an acetylated PAMAM dendrimer. RSC Adv. 2018, 8, 42253. [Google Scholar] [CrossRef] [Green Version]
- Lesniak, W.G.; Sikorska, E.; Shallal, H.; Aza, B.B.; Lisok, A.; Pullambhatla, M.; Pomper, M.G. Structural Characterization and In Vivo Evaluation of Beta-Hairpin Peptidomimetics as Specific CXCR4 Imaging Agents. Mol. Pharm. 2015, 12, 941–953. [Google Scholar] [CrossRef]
- Björndal, A.; Deng, H.; Jansson, M.; Fiore, J.R.; Colognesi, C.; Karlsson, A.; Albert, J.; Scarlatti, G.; Littman, D.R.; Fenyö, E.M. Coreceptor usage of primary human immunodeficiency virus type 1 isolates varies according to biological phenotype. J. Virol. 1997, 71, 7478–7487. [Google Scholar] [CrossRef] [Green Version]
- Woodard, L.E.; De Silva, R.A.; Azad, B.B.; Lisok, A.; Pullambhatla, M.; Lesniak, W.G.; Mease, R.C.; Pomper, M.G.; Nimmagadda, S. Bridged cyclams as imaging agents for chemokine receptor 4 (CXCR4). Nucl. Med. Biol. 2014, 41, 552–561. [Google Scholar] [CrossRef] [Green Version]
- Burger, M.; Glodek, A.; Hartmann, T.N.; Schmitt-Gräff, A.; Silberstein, L.E.; Fujii, N.; Kipps, T.J.; Burger, J.A. Functional expression of CXCR4 (CD184) on small-cell lung cancer cells mediates migration, integrin activation, and adhesion to stromal cells. Oncogene 2003, 22, 8093–8101. [Google Scholar] [CrossRef] [Green Version]
- Nimmagadda, S.; Pullambhatla, M.; Pomper, M.G. Immunoimaging of CXCR4 Expression in Brain Tumor Xenografts Using SPECT/CT. J. Nucl. Med. 2009, 50, 1124–1130. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lesniak, W.G.; Kariapper, M.S.T.; Nair, B.M.; Tan, W.; Hutson, A.; Balogh, L.P.; Khan, M.K. Synthesis and characterization of PAMAM dendrimer-based multifunctional nanodevices for targeting alpha(v)beta(3) integrins. Bioconj. Chem. 2007, 18, 1148–1154. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shi, X.; Bányai, I.; Islam, M.T.; Lesniak, W.; Davis, D.Z.; Baker, J.R.; Balogh, L.P. Generational, skeletal and substitutional diversities in generation one poly(amidoamine) dendrimers. Polymer 2005, 46, 3022–3034. [Google Scholar] [CrossRef]
- Lesniak, W.G.; Boinapally, S.; Banerjee, S.R.; Azad, B.B.; Foss, C.A.; Shen, C.; Lisok, A.; Wharram, B.; Nimmagadda, S.; Pomper, M.G. Evaluation of PSMA-Targeted PAMAM Dendrimer Nanoparticles in a Murine Model of Prostate Cancer. Mol. Pharm. 2019, 16, 2590–2604. [Google Scholar] [CrossRef]
- Villaraza, A.J.L.; Bumb, A.; Brechbiel, M.W. Macromolecules, Dendrimers, and Nanomaterials in Magnetic Resonance Imaging: The Interplay between Size, Function, and Pharmacokinetics. Chem. Rev. 2010, 110, 2921–2959. [Google Scholar] [CrossRef] [Green Version]
- Kobayashi, H.; Sato, N.; Hiraga, A.; Saga, T.; Nakamoto, Y.; Ueda, H.; Konishi, J.; Togashi, K.; Brechbiel, M.W. 3D-micro-MR angiography of mice using macromolecular MR contrast agents with polyamidoamine dendrimer core with reference to their pharmacokinetic properties. Magn. Reson. Med. 2001, 45, 454–460. [Google Scholar] [CrossRef]
- Lesniak, W.G.; Mishra, M.K.; Jyoti, A.; Balakrishnan, B.; Zhang, F.; Nance, E.; Romero, R.; Kannan, S.; Kannan, R.M. Biodistribution of Fluorescently Labeled PAMAM Dendrimers in Neonatal Rabbits: Effect of Neuroinflammation. Mol. Pharm. 2013, 10, 4560–4571. [Google Scholar] [CrossRef]
- Sadekar, S.; Ray, A.; Janàt-Amsbury, M.; Peterson, C.M.; Ghandehari, H. Comparative Biodistribution of PAMAM Dendrimers and HPMA Copolymers in Ovarian-Tumor-Bearing Mice. Biomacromolecules 2010, 12, 88–96. [Google Scholar] [CrossRef] [Green Version]
- Nel, A.; Ruoslahti, E.; Meng, H. New Insights into “Permeability” as in the Enhanced Permeability and Retention Effect of Cancer Nanotherapeutics. ACS Nano 2017, 11, 9567–9569. [Google Scholar] [CrossRef]
- Banerjee, S.R.; Foss, C.A.; Horhota, A.; Pullambhatla, M.; McDonnell, K.; Zale, S.; Pomper, M.G. 111In- and IRDye800CW-Labeled PLA–PEG Nanoparticle for Imaging Prostate-Specific Membrane Antigen-Expressing Tissues. Biomacromolecules 2016, 18, 201–209. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ishida, O.; Maruyama, K.; Sasaki, K.; Iwatsuru, M. Size-dependent extravasation and interstitial localization of polyethyleneglycol liposomes in solid tumor-bearing mice. Int. J. Pharm. 1999, 190, 49–56. [Google Scholar] [CrossRef]

| DC | # of NH2 | # of DOTA | # of PEG2000 | # of PEG2 | # of SAPOL3026 | MW [m/z] | Size [nm] | ZP [mV] |
|---|---|---|---|---|---|---|---|---|
| 1 | 128 | 0 | 0 | 0 | 0 | 26,111 | 5.57 ± 0.84 | 45.52 ± 0.74 |
| 2 | 124 | 4 | 0 | 0 | 0 | 27,661 | 4.77 ± 0.44 | 38.15 ± 024 |
| 3 | 116 | 4 | 8 | 0 | 0 | 44,027 | 8.99 ± 0.76 | 20.10 ± 0.89 |
| 4 | 54 | 4 | 8 | 62 | 0 | 52,250 | 8.78 ± 0.37 | 10.04 ± 0.65 |
| G5-Ctrl | 54 | 4 | 8 | 62 | 0 | 50,666 | 7.81 ± 0.85 | 12.41 ± 1.4 |
| G5-X4 | 54 | 4 | 8 | 62 | 3 | 57,250 | 9.11 ± 0.92 | 14.9 ± 0.51 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lesniak, W.G.; Azad, B.B.; Chatterjee, S.; Lisok, A.; Pomper, M.G. An Evaluation of CXCR4 Targeting with PAMAM Dendrimer Conjugates for Oncologic Applications. Pharmaceutics 2022, 14, 655. https://doi.org/10.3390/pharmaceutics14030655
Lesniak WG, Azad BB, Chatterjee S, Lisok A, Pomper MG. An Evaluation of CXCR4 Targeting with PAMAM Dendrimer Conjugates for Oncologic Applications. Pharmaceutics. 2022; 14(3):655. https://doi.org/10.3390/pharmaceutics14030655
Chicago/Turabian StyleLesniak, Wojciech G., Babak Behnam Azad, Samit Chatterjee, Ala Lisok, and Martin G. Pomper. 2022. "An Evaluation of CXCR4 Targeting with PAMAM Dendrimer Conjugates for Oncologic Applications" Pharmaceutics 14, no. 3: 655. https://doi.org/10.3390/pharmaceutics14030655







