Antimicrobial Properties of Amino-Acid-Derived N-Heterocyclic Carbene Silver Complexes
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials
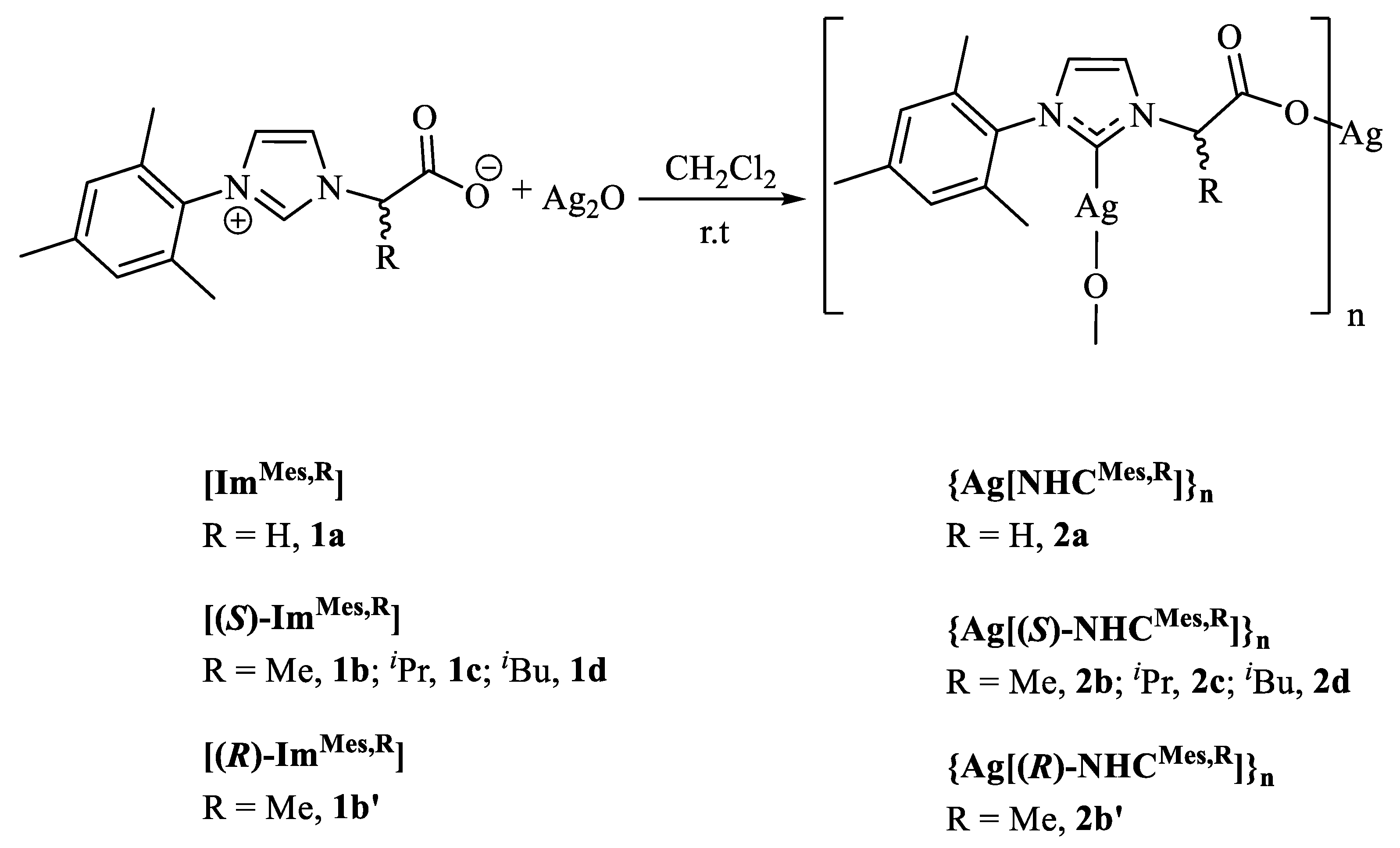
2.2. Synthesis of the {Ag[NHCMes,R]}n Complexes
2.2.1. Complex {Ag[NHCMes,H]}n (2a)
2.2.2. Complexes {Ag[(S)-NHCMes,Me]}n (2b) and {Ag[(R)-NHCMes,Me]}n (2b’)
2.2.3. Complex {Ag[(S)-NHCMes,iPr]}n (2c)
2.2.4. Complex {Ag[(S)-NHCMes,iBu]}n (2d)
2.3. Single-Crystal X-ray Crystallography
2.4. Antimicrobial Studies and Microbiological Assays
2.4.1. Bacterial Strains and Culture Conditions
2.4.2. Determination of Minimal Inhibitory Growth Concentrations (MIC) and Minimal Bactericidal Concentrations (MBC)
2.4.3. Determination of Antioxidant Enzymes and Thiobarbituric Acid Reactive Substances (TBARs)
2.4.4. Effects on Biofilm Formation: Evaluation by Colorimetric Technique and by Scanning Electron Microscopy (SEM)
2.5. Computational Details
3. Results and Discussion
3.1. Synthesis and Characterisation of the {Ag[NHCMes,R]}n Complexes
3.2. X-ray Characterisation of Complex {Ag[NHCMes,H]}n, 2a
3.3. Solution Behaviour of {Ag[NHCMes,R]}n Compounds
3.4. Antimicrobial Properties: Determination of Minimal Inhibitory Growth Concentrations (MIC) and Minimal Bactericidal Concentrations (MBC)
3.5. Antimicrobial Properties: Determination of Antioxidant Enzymes and Thiobarbituric Acid Reactive Substances (TBARs)
3.6. Antimicrobial Properties’ Effects on Biofilm Formation: Evaluation by Colorimetric Technique and by Scanning Electron Microscopy (SEM)
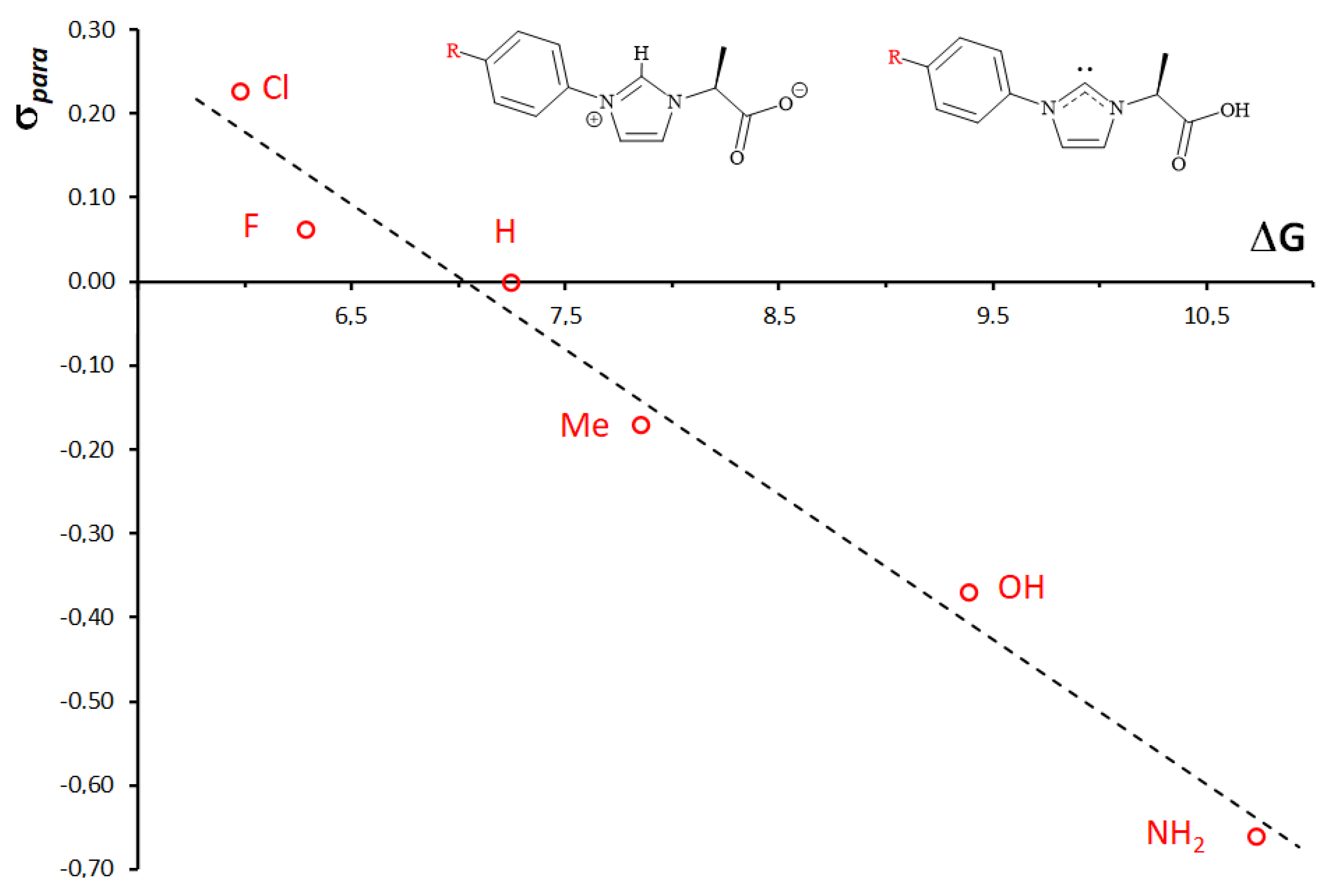
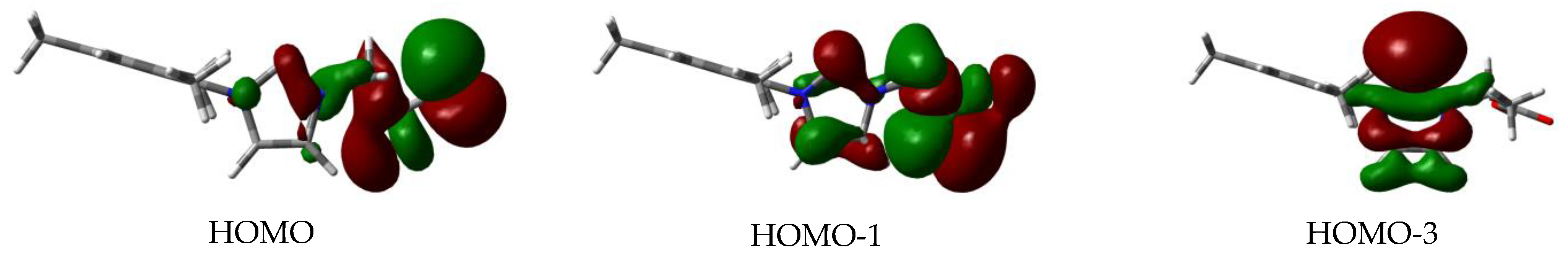
3.7. Computational Studies
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Betts, H.D.; Whitehead, C.; Harris, H.H. Silver in biology and medicine: Opportunities for metallomics researchers. Metallomics 2021, 13, mfaa001. [Google Scholar] [CrossRef] [PubMed]
- Klasen, H.J. Historical review of the use of silver in the treatment of burns. I. Early uses. Burns 2000, 26, 117–130. [Google Scholar] [CrossRef]
- Klasen, H. A historical review of the use of silver in the treatment of burns. II. Renewed interest for silver. Burns 2000, 26, 131–138. [Google Scholar] [CrossRef]
- Thornton, L.; Dixit, V.; Assad, L.O.N.; Ribeiro, T.P.; Queiroz, D.D.; Kellett, A.; Casey, A.; Colleran, J.; Pereira, M.D.; Rochford, G.; et al. Water-soluble and photo-stable silver(I) dicarboxylate complexes containing 1,10-phenanthroline ligands: Antimicrobial and anticancer chemotherapeutic potential, DNA interactions and antioxidant activity. J. Inorg. Biochem. 2016, 159, 120–132. [Google Scholar] [CrossRef]
- Lemire, J.A.; Harrison, J.J.; Turner, R.J. Antimicrobial activity of metals: Mechanisms, molecular targets and applications. Nat. Rev. Microbiol. 2013, 11, 371–384. [Google Scholar] [CrossRef] [PubMed]
- Jung, W.K.; Koo, H.C.; Kim, K.W.; Shin, S.; Kim, S.H.; Park, Y.H. Antibacterial Activity and Mechanism of Action of the Silver Ion in Staphylococcus aureus and Escherichia coli. Appl. Environ. Microbiol. 2008, 74, 2171–2178. [Google Scholar] [CrossRef] [Green Version]
- Patil, S.; Deally, A.; Gleeson, B.; Müller-Bunz, H.; Paradisi, F.; Tacke, M. Novel benzyl-substituted N-heterocyclic carbene-silver acetate complexes: Synthesis, cytotoxicity and antibacterial studies. Metallomics 2011, 3, 74–88. [Google Scholar] [CrossRef]
- Hindi, K.M.; Siciliano, T.J.; Durmus, S.; Panzner, M.J.; Medvetz, D.A.; Reddy, D.V.; Hogue, L.A.; Hovis, C.E.; Hilliard, J.K.; Mallet, R.J.; et al. Synthesis, Stability, and Antimicrobial Studies of Electronically Tuned Silver Acetate N -Heterocyclic Carbenes. J. Med. Chem. 2008, 51, 1577–1583. [Google Scholar] [CrossRef]
- Kascatan-Nebioglu, A.; Panzner, M.J.; Tessier, C.A.; Cannon, C.L.; Youngs, W.J. N-Heterocyclic carbene–silver complexes: A new class of antibiotics. Coord. Chem. Rev. 2007, 251, 884–895. [Google Scholar] [CrossRef]
- Patil, S.; Deally, A.; Gleeson, B.; Hackenberg, F.; Müller-Bunz, H.; Paradisi, F.; Tacke, M. Synthesis, cytotoxicity and antibacterial studies of novel symmetrically and non-symmetrically p-nitrobenzyl-substituted N-heterocyclic carbene-silver(i) acetate complexes. Z. Fur Anorg. Und Allg. Chem. 2011, 637, 386–396. [Google Scholar] [CrossRef]
- Kascatan-Nebioglu, A.; Melaiye, A.; Hindi, K.; Durmus, S.; Panzner, M.J.; Hogue, L.A.; Mallett, R.J.; Hovis, C.E.; Coughenour, M.; Crosby, S.D.; et al. Synthesis from Caffeine of a Mixed N-Heterocyclic Carbene−Silver Acetate Complex Active against Resistant Respiratory Pathogens. J. Med. Chem. 2006, 49, 6811–6818. [Google Scholar] [CrossRef] [PubMed]
- Roland, S.; Jolivalt, C.; Cresteil, T.; Eloy, L.; Bouhours, P.; Hequet, A.; Mansuy, V.; Vanucci, C.; Paris, J.-M. Investigation of a Series of Silver-N-Heterocyclic Carbenes as Antibacterial Agents: Activity, Synergistic Effects, and Cytotoxicity. Chem. A Eur. J. 2011, 17, 1442–1446. [Google Scholar] [CrossRef] [PubMed]
- Sarı, Y.; Akkoç, S.; Gök, Y.; Sifniotis, V.; Özdemir, İ.; Günal, S.; Kayser, V. Benzimidazolium-based novel silver N -heterocyclic carbene complexes: Synthesis, characterisation and in vitro antimicrobial activity. J. Enzyme Inhib. Med. Chem. 2016, 31, 1527–1530. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sakamoto, R.; Morozumi, S.; Yanagawa, Y.; Toyama, M.; Takayama, A.; Kasuga, N.C.; Nomiya, K. Synthesis, characterization, and structure–activity relationship of the antimicrobial activities of dinuclear N-heterocyclic carbene (NHC)-silver(I) complexes. J. Inorg. Biochem. 2016, 163, 110–117. [Google Scholar] [CrossRef] [PubMed]
- O’Beirne, C.; Piatek, M.E.; Fossen, J.; Müller-Bunz, H.; Andes, D.R.; Kavanagh, K.; Patil, S.A.; Baumann, M.; Tacke, M. Continuous flow synthesis and antimicrobial evaluation of NHC* silver carboxylate derivatives of SBC3 in vitro and in vivo. Metallomics 2021, 13, mfaa011. [Google Scholar] [CrossRef] [PubMed]
- Imaz, I.; Rubio-Martínez, M.; An, J.; Solé-Font, I.; Rosi, N.L.; Maspoch, D. Metal–biomolecule frameworks (MBioFs). Chem. Commun. 2011, 47, 7287–7302. [Google Scholar] [CrossRef] [PubMed]
- Severin, K.; Bergs, R.; Beck, W. Bioorganometallic Chemistry—Transition Metal Complexes withα-Amino Acids and Peptides. Angew. Chem. Int. Ed. 1998, 37, 1634–1654. [Google Scholar] [CrossRef]
- Esposito, D.; Kirchhecker, S.; Antonietti, M. A sustainable route towards imidazolium building blocks based on biomass molecules. Chem. A Eur. J. 2013, 19, 15097–15100. [Google Scholar] [CrossRef]
- Kühl, O.; Palm, G. Imidazolium salts from amino acids—A new route to chiral zwitterionic carbene precursors? Tetrahedron Asymmetry 2010, 21, 393–397. [Google Scholar] [CrossRef]
- Jahier-Diallo, C.; Morin, M.S.T.; Queval, P.; Rouen, M.; Artur, I.; Querard, P.; Toupet, L.; Crévisy, C.; Baslé, O.; Mauduit, M. Multicomponent Synthesis of Chiral Bidentate Unsymmetrical Unsaturated N -Heterocyclic Carbenes: Copper-Catalyzed Asymmetric C-C Bond Formation. Chem. A Eur. J. 2015, 21, 993–997. [Google Scholar] [CrossRef]
- Kirchhecker, S.; Antonietti, M.; Esposito, D. Hydrothermal decarboxylation of amino acid derived imidazolium zwitterions: A sustainable approach towards ionic liquids. Green Chem. 2014, 16, 3705–3709. [Google Scholar] [CrossRef] [Green Version]
- Ferry, A.; Schaepe, K.; Tegeder, P.; Richter, C.; Chepiga, K.M.; Ravoo, B.J.; Glorius, F. Negatively Charged N-Heterocyclic Carbene-Stabilized Pd and Au Nanoparticles and Efficient Catalysis in Water. ACS Catal. 2015, 5, 5414–5420. [Google Scholar] [CrossRef]
- Reynoso-Esparza, M.A.; Rangel-Salas, I.I.; Peregrina-Lucano, A.A.; Alvarado-Rodríguez, J.G.; López-Dellamary-Toral, F.A.; Manríquez-González, R.; Espinosa-Macías, M.L.; Cortes-Llamas, S.A. Synthesis and characterization of Au(I) and Au(III) complexes containing N-heterocyclic ligands derived from amino acids. Polyhedron 2014, 81, 564–571. [Google Scholar] [CrossRef]
- Lomelí-Rosales, D.A.; Rangel-Salas, I.I.; Zamudio-Ojeda, A.; Carbajal-Arízaga, G.G.; Godoy-Alcántar, C.; Manríquez-González, R.; Alvarado-Rodríguez, J.G.; Martínez-Otero, D.; Cortes-Llamas, S.A. Chiral Imidazolium-Functionalized Au Nanoparticles: Reversible Aggregation and Molecular Recognition. ACS Omega 2016, 1, 876–885. [Google Scholar] [CrossRef] [PubMed]
- Steeples, E.; Kelling, A.; Schilde, U.; Esposito, D. Amino acid-derived N-heterocyclic carbene palladium complexes for aqueous phase Suzuki–Miyaura couplings. New J. Chem. 2016, 40, 4922–4930. [Google Scholar] [CrossRef] [Green Version]
- DePasquale, J.; White, N.J.; Ennis, E.J.; Zeller, M.; Foley, J.P.; Papish, E.T. Synthesis of chiral N-heterocyclic carbene (NHC) ligand precursors and formation of ruthenium(II) complexes for transfer hydrogenation catalysts. Polyhedron 2013, 58, 162–170. [Google Scholar] [CrossRef]
- Meyer, A.; Taige, M.A.; Strassner, T. Chiral bisimidazolium salts derived from amino acids and their palladium(II)- and platinum(II)-biscarbene complexes. J. Organomet. Chem. 2009, 694, 1861–1868. [Google Scholar] [CrossRef]
- Wang, X.; Li, X.B.; Yan, R.H.; Wang, Y.Q.; Gao, E.Q. Diverse manganese(ii) coordination polymers derived from achiral/chiral imidazolium-carboxylate zwitterions and azide: Structure and magnetic properties. Dalton Trans. 2013, 42, 10000–10010. [Google Scholar] [CrossRef]
- Babu, C.N.; Sathyanarayana, A.; Mobin, S.M.; Prabusankar, G. Structurally characterized zwitterionic chiral zinc imidazolium [4,4] grid. Inorg. Chem. Commun. 2013, 37, 222–224. [Google Scholar] [CrossRef]
- Nicasio, A.I.; Montilla, F.; Álvarez, E.; Colodrero, R.P.; Galindo, A. Synthesis and structural characterization of homochiral 2D coordination polymers of zinc and copper with conformationally flexible ditopic imidazolium-based dicarboxylate ligands. Dalton Trans. 2017, 46, 471–482. [Google Scholar] [CrossRef]
- Caballero, P.; Colodrero, R.M.P.; del Mar Conejo, M.; Pastor, A.; Álvarez, E.; Montilla, F.; Carrasco, C.J.; Nicasio, A.I.; Galindo, A. Homochiral imidazolium-based dicarboxylate compounds: Structure and solution behaviour. Inorg. Chim. Acta 2020, 513, 119923. [Google Scholar] [CrossRef]
- Borrego-Blanco, E.; Nicasio, A.I.; Alvarez González, E.; Montilla, F.; Córdoba, J.M.; Galindo, A. Synthesis and structural characterization of homochiral coordination polymers with imidazole-based monocarboxylate ligands. Dalton Trans. 2019, 48, 8731–8739. [Google Scholar] [CrossRef] [PubMed]
- Carrasco, C.J.; Montilla, F.; Galindo, A. Molybdenum-catalyzed asymmetric sulfoxidation with hydrogen peroxide and subsequent kinetic resolution, using an imidazolium-based dicarboxylate compound as chiral inductor. Catal. Commun. 2016, 84, 134–136. [Google Scholar] [CrossRef]
- Carrasco, C.; Montilla, F.; Galindo, A. Molybdenum-Catalyzed Enantioselective Sulfoxidation Controlled by a Nonclassical Hydrogen Bond between Coordinated Chiral Imidazolium-Based Dicarboxylate and Peroxido Ligands. Molecules 2018, 23, 1595. [Google Scholar] [CrossRef] [Green Version]
- Tarrieu, R.; Dumas, A.; Thongpaen, J.; Vives, T.; Roisnel, T.; Dorcet, V.; Crévisy, C.; Baslé, O.; Mauduit, M. Readily Accessible Unsymmetrical Unsaturated 2,6-Diisopropylphenyl N-Heterocyclic Carbene Ligands. Applications in Enantioselective Catalysis. J. Org. Chem. 2017, 82, 1880–1887. [Google Scholar] [CrossRef]
- Carrasco, C.J.; Montilla, F.; Alvarez, E.; Galindo, A.; Perez-Aranda, M.; Pajuelo, E.; Alcudia, A. Homochiral imidazolium based dicarboxylate silver(I) compounds: Synthesis, characterisation and antimicrobial properties. Dalton Trans. 2022. [Google Scholar] [CrossRef]
- Thongpaen, J.; Schmid, T.E.; Toupet, L.; Dorcet, V.; Mauduit, M.; Baslé, O. Directed ortho C–H borylation catalyzed using Cp*Rh(iii)–NHC complexes. Chem. Commun. 2018, 54, 8202–8205. [Google Scholar] [CrossRef] [Green Version]
- Manguin, R.; Pichon, D.; Tarrieu, R.; Vives, T.; Roisnel, T.; Dorcet, V.; Crévisy, C.; Miqueu, K.; Favereau, L.; Crassous, J.; et al. A kinetic resolution strategy for the synthesis of chiral octahedral NHC–iridium(iii) catalysts. Chem. Commun. 2019, 55, 6058–6061. [Google Scholar] [CrossRef]
- Thongpaen, J.; Manguin, R.; Dorcet, V.; Vives, T.; Duhayon, C.; Mauduit, M.; Baslé, O. Visible Light Induced Rhodium(I)-Catalyzed C−H Borylation. Angew. Chem. Int. Ed. 2019, 58, 15244–15248. [Google Scholar] [CrossRef]
- Bowler, P.; Murphy, C.; Wolcott, R. Biofilm exacerbates antibiotic resistance: Is this a current oversight in antimicrobial stewardship? Antimicrob. Resist. Infect. Control 2020, 9, 162. [Google Scholar] [CrossRef]
- Bruker SAINT+. SAINT+; Bruker AXS Inc.: Madison, WI, USA, 2007. [Google Scholar]
- Sheldrick, G.M. SADABS, Programs for Scaling and Absorption Correction of Area Detector Data; University of Göttingen: Göttingen, Germany, 1997. [Google Scholar]
- Burla, M.C.; Camalli, M.; Carrozzini, B.; Cascarano, G.L.; Giacovazzo, C.; Polidori, G.; Spagna, R. SIR2002: The program. J. Appl. Crystallogr. 2003, 36, 1103. [Google Scholar] [CrossRef] [Green Version]
- Sheldrick, G.M. A short history of SHELX. Acta Crystallogr. Sect. A Found. Crystallogr. 2008, 64, 112–122. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Van Der Sluis, P.; Spek, A.L. BYPASS: An effective method for the refinement of crystal structures containing disordered solvent regions. Acta Crystallogr. Sect. A 1990, 46, 194–201. [Google Scholar] [CrossRef]
- Cavalier, S.J.; Harbeck, R.J.; McCarter, Y.S.; Ortez, J.H.; Rankin, I.D.; Sautter, R.L.; Sharp, S.E.; Spiegel, C.A. MIC Testing. Man. Antimicrob. Susceptibility Test. 2005, 32, 53–62. [Google Scholar]
- Pine, L.; Hoffman, P.S.; Malcolm, G.B.; Benson, R.F.; Keen, M.G. Determination of catalase, peroxidase, and superoxide dismutase within the genus Legionella. J. Clin. Microbiol. 1984, 20, 421–429. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chance, B.; Maehly, A.C. [136] Assay of catalases and peroxidases. Methods Enzymol. 1955, 2, 764–775. [Google Scholar] [CrossRef]
- Beyer, W.F.; Fridovich, I. Assaying for superoxide dismutase activity: Some large consequences of minor changes in conditions. Anal. Biochem. 1987, 161, 559–566. [Google Scholar] [CrossRef]
- Ong, K.S.; Cheow, Y.L.; Lee, S.M. The role of reactive oxygen species in the antimicrobial activity of pyochelin. J. Adv. Res. 2017, 8, 393–398. [Google Scholar] [CrossRef]
- Bradford, M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- O’Toole, G.A. Microtiter Dish Biofilm Formation Assay. J. Vis. Exp. 2011, 9, e2437. [Google Scholar] [CrossRef]
- Becke, A.D. Density-functional thermochemistry. III. The role of exact exchange. J. Chem. Phys. 1993, 98, 5648–5652. [Google Scholar] [CrossRef] [Green Version]
- Lee, C.; Yang, W.; Parr, R.G. Development of the Colle-Salvetti correlation-energy formula into a functional of the electron density. Phys. Rev. B 1988, 37, 785–789. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hay, P.J.; Wadt, W.R. Ab initio effective core potentials for molecular calculations. Potentials for K to Au including the outermost core orbitale. J. Chem. Phys. 1985, 82, 299–310. [Google Scholar] [CrossRef]
- Frisch, M.J.; Trucks, G.W.; Schlegel, H.B.; Scuseria, G.E.; Robb, M.A.; Cheeseman, J.R.; Scalmani, G.; Barone, V.; Petersson, G.A.; Nakatsuji, H.; et al. Gaussian 09; Revision B.01; Gaussian, Inc.: Wallingford, CT, USA, 2016.
- Groom, C.R.; Bruno, I.J.; Lightfoot, M.P.; Ward, S.C. The Cambridge structural database. Acta Crystallogr. Sect. B Struct. Sci. Cryst. Eng. Mater. 2016, 72, 171–179. [Google Scholar] [CrossRef] [PubMed]
- Udvardy, A.; Szolnoki, C.T.; Kováts, É.; Nyul, D.; Gál, G.T.; Papp, G.; Joó, F.; Kathó, Á. Water-soluble Ag(I)-based coordination polymers obtained by anion-directed self-assembly of various AgX salts and a phosphabetaine derived from 1,3,5-triaza-7-phophaadamantane. Inorg. Chim. Acta 2021, 520, 120299. [Google Scholar] [CrossRef]
- Macchioni, A.; Ciancaleoni, G.; Zuccaccia, C.; Zuccaccia, D. Determining accurate molecular sizes in solution through NMR diffusion spectroscopy. Chem. Soc. Rev. 2008, 37, 479–489. [Google Scholar] [CrossRef]
- Thurman, R.B.; Gerba, C.P. The molecular mechanisms of copper and silver ion disinfection of bacteria and viruses. Crit. Rev. Environ. Control 1989, 18, 295–315. [Google Scholar] [CrossRef]
- Nomiya, K.; Tsuda, K.; Sudoh, T.; Oda, M. Ag(I)-N bond-containing compound showing wide spectra in effective antimicrobial activities: Polymeric silver(I) imidazolate. J. Inorg. Biochem. 1997, 68, 39–44. [Google Scholar] [CrossRef]
- Asaad, K.; Mashhadi, S. Topical Application of Silver Nitrate. Int. J. Low. Extrem. Wounds 2013, 12, 324. [Google Scholar] [CrossRef]
- Cava, F.; Lam, H.; de Pedro, M.A.; Waldor, M.K. Emerging knowledge of regulatory roles of d-amino acids in bacteria. Cell. Mol. Life Sci. 2011, 68, 817–831. [Google Scholar] [CrossRef] [Green Version]
- Trivedi, R.R.; Crooks, J.A.; Auer, G.K.; Pendry, J.; Foik, I.P.; Siryaporn, A.; Abbott, N.L.; Gitai, Z.; Weibel, D.B. Mechanical Genomic Studies Reveal the Role of D-Alanine Metabolism in Pseudomonas aeruginosa Cell Stiffness. MBio 2018, 9, e01340-18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cherian, D.; Peter, T.; Narayanan, A.; Madhavan, S.; Achammada, S.; Vynat, G. Malondialdehyde as a marker of oxidative stress in periodontitis patients. J. Pharm. Bioallied Sci. 2019, 11, 297–300. [Google Scholar] [CrossRef] [PubMed]
- Lopez, D.; Vlamakis, H.; Kolter, R. Biofilms. Cold Spring Harb. Perspect. Biol. 2010, 2, a000398. [Google Scholar] [CrossRef] [PubMed]
- Fu, J.; Zhang, Y.; Lin, S.; Zhang, W.; Shu, G.; Lin, J.; Li, H.; Xu, F.; Tang, H.; Peng, G.; et al. Strategies for Interfering With Bacterial Early Stage Biofilms. Front. Microbiol. 2021, 12, 1339. [Google Scholar] [CrossRef]


| E. coli | P. aeruginosa | |||
|---|---|---|---|---|
| Complex | MIC | MBC | MIC | MBC |
| 2a | 0.156 | 0.625 | 0.078 | 0.3125 |
| 2b | 0.156 | 0.3125 | 0.156 | 0.625 |
| 2b’ | 0.078 | 0.156 | 0.039 | 0.156 |
| 2c | 0.3125 | 1.250 | 0.3125 | 2.500 |
| 2d | 1.250 | 2.500 | 1.250 | 2.500 |
| AgNO3 | 0.156 | 0.3125 | 0.156 | 0.3125 |
| E. coli | P. aeruginosa | ||||||
|---|---|---|---|---|---|---|---|
| Complex | Enzyme | Control | MIC | MBC | Control | MIC | MBC |
| 2b | Catalase | ND b | 2.5 × 10−5 ± 0.2 × 10−5 | 6.6 × 10−5 ± 0.9 × 10−5 | ND | ND | ND |
| Peroxidases | ND | ND | ND | ND | ND | 0.002 ± 0.001 | |
| Superoxide dismutase | 3.33 ± 0.01 | 1.51 ± 0.07 | 92.1 ± 2.3 | 3.40 ± 0.06 | 20.00 ± 0.01 | 76.3 ± 0.1 | |
| 2c | Catalase | ND | 5.5 × 10−5 ± 0.1 × 10−5 | 8.3 × 10−5 ± 0.1 × 10−5 | ND | ND | ND |
| Peroxidases | 0.48 ± 0.05 | 0.16 ± 0.02 | 0.03 ± 0.01 | 0.38 ± 0.03 | 1.1 ± 0.1 | 2.5 ± 0.3 | |
| Superoxide dismutase | 3.33 ± 0.01 | 4.14 ± 0.07 | 3.66 ± 0.06 | 3.40 ± 0.06 | 9.1 ± 0.1 | 20.0 ± 0.2 | |
| E. coli | P. aeruginosa | |||
|---|---|---|---|---|
| Complex | MIC | Onset Inhibitory Concentration of Biofilm | MIC | Onset Inhibitory Concentration of Biofilm |
| 2b | 0.156 | 0.039 | 0.156 | 0.020 |
| 2c | 0.313 | 0.313 | 0.313 | 0.313 |
| R |  [ImMes,R], 1 |  1′ |
| H | 0 | 9.4 |
| Me | 0 | 8.9 |
| iPr | 0 | 7.0 |
| CH2iPr | 0 | 9.0 |
| R’ |  [ImAr’,Me] |  |
| NH2 | 0 | 10.7 |
| OH | 0 | 9.4 |
| Me | 0 | 7.8 |
| H | 0 | 7.2 |
| F | 0 | 6.3 |
| Cl | 0 | 6.0 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sánchez, A.; Carrasco, C.J.; Montilla, F.; Álvarez, E.; Galindo, A.; Pérez-Aranda, M.; Pajuelo, E.; Alcudia, A. Antimicrobial Properties of Amino-Acid-Derived N-Heterocyclic Carbene Silver Complexes. Pharmaceutics 2022, 14, 748. https://doi.org/10.3390/pharmaceutics14040748
Sánchez A, Carrasco CJ, Montilla F, Álvarez E, Galindo A, Pérez-Aranda M, Pajuelo E, Alcudia A. Antimicrobial Properties of Amino-Acid-Derived N-Heterocyclic Carbene Silver Complexes. Pharmaceutics. 2022; 14(4):748. https://doi.org/10.3390/pharmaceutics14040748
Chicago/Turabian StyleSánchez, Adrián, Carlos J. Carrasco, Francisco Montilla, Eleuterio Álvarez, Agustín Galindo, María Pérez-Aranda, Eloísa Pajuelo, and Ana Alcudia. 2022. "Antimicrobial Properties of Amino-Acid-Derived N-Heterocyclic Carbene Silver Complexes" Pharmaceutics 14, no. 4: 748. https://doi.org/10.3390/pharmaceutics14040748
APA StyleSánchez, A., Carrasco, C. J., Montilla, F., Álvarez, E., Galindo, A., Pérez-Aranda, M., Pajuelo, E., & Alcudia, A. (2022). Antimicrobial Properties of Amino-Acid-Derived N-Heterocyclic Carbene Silver Complexes. Pharmaceutics, 14(4), 748. https://doi.org/10.3390/pharmaceutics14040748








