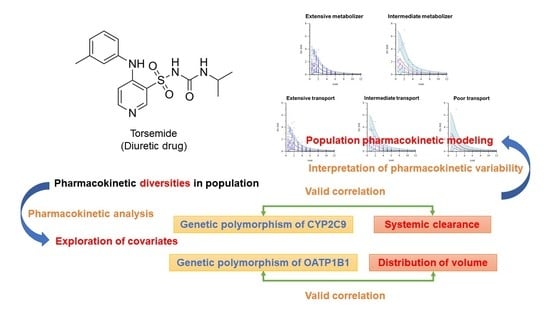
Population Pharmacokinetic (Pop-PK) Analysis of Torsemide in Healthy Korean Males Considering CYP2C9 and OATP1B1 Genetic Polymorphisms
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Subjects
2.3. Sampling
2.4. Determination of Clinical Biochemistry Parameters
2.5. Determination of Genotypes
2.5.1. Identification of CYP2C9*3 and *13 Alleles
2.5.2. Identification of SLCO1B1 388A>G and 521T>C
2.6. Determination of Serum Torsemide Concentration
2.7. PK Analysis
2.8. Model Development
2.8.1. Establishment of Structural Base Model
2.8.2. Covariate Analysis
2.9. Model Evaluation
2.10. Model Reconfirmation
3. Results and Discussion
3.1. Study Design and Demographic Analysis
3.2. Genetic Polymorphism Analysis
3.3. Determination of Torsemide Serum Concentrations
3.4. PK Results by NCA
3.5. Pop-PK Model Analysis
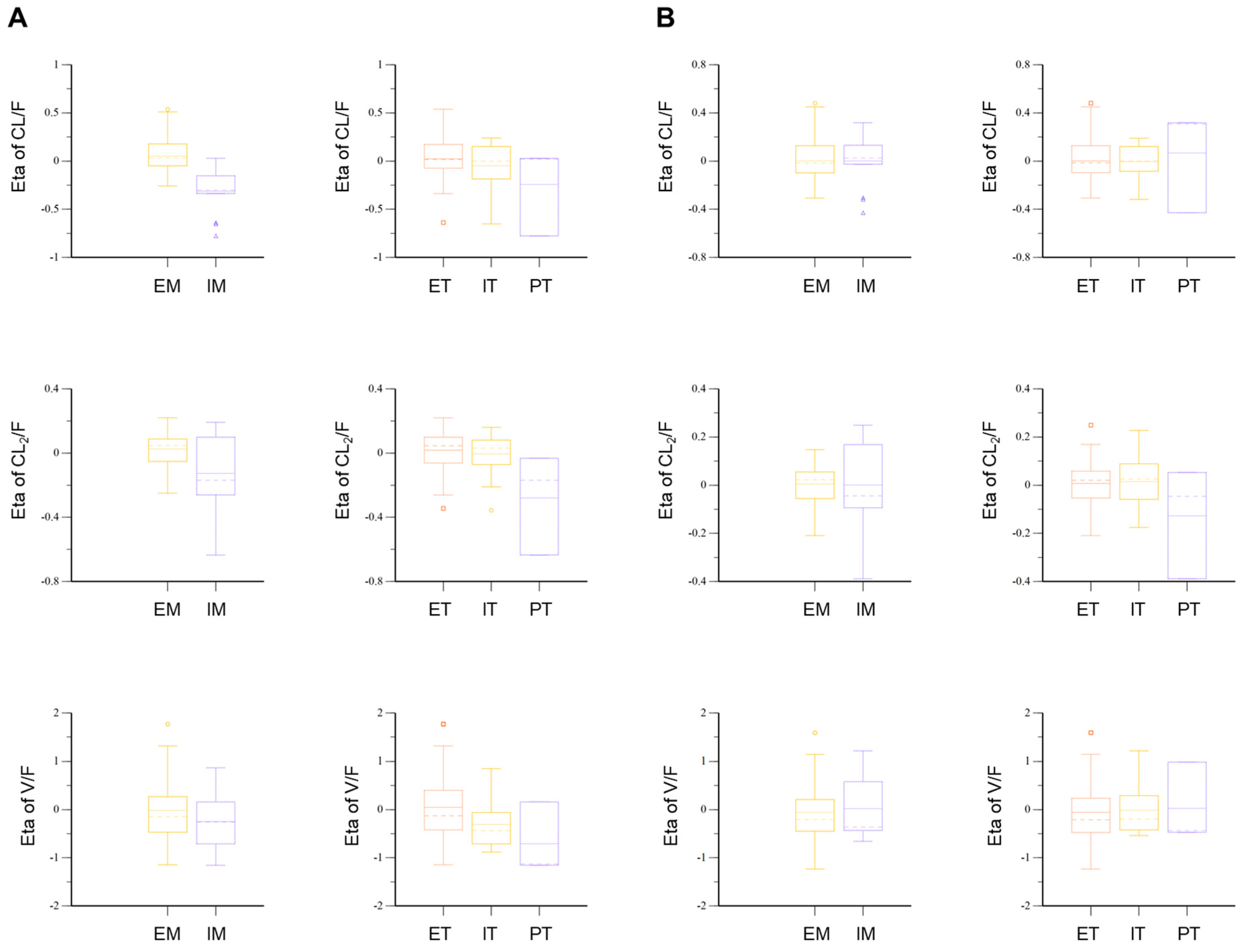
3.6. Pop-PK Model Interpretation
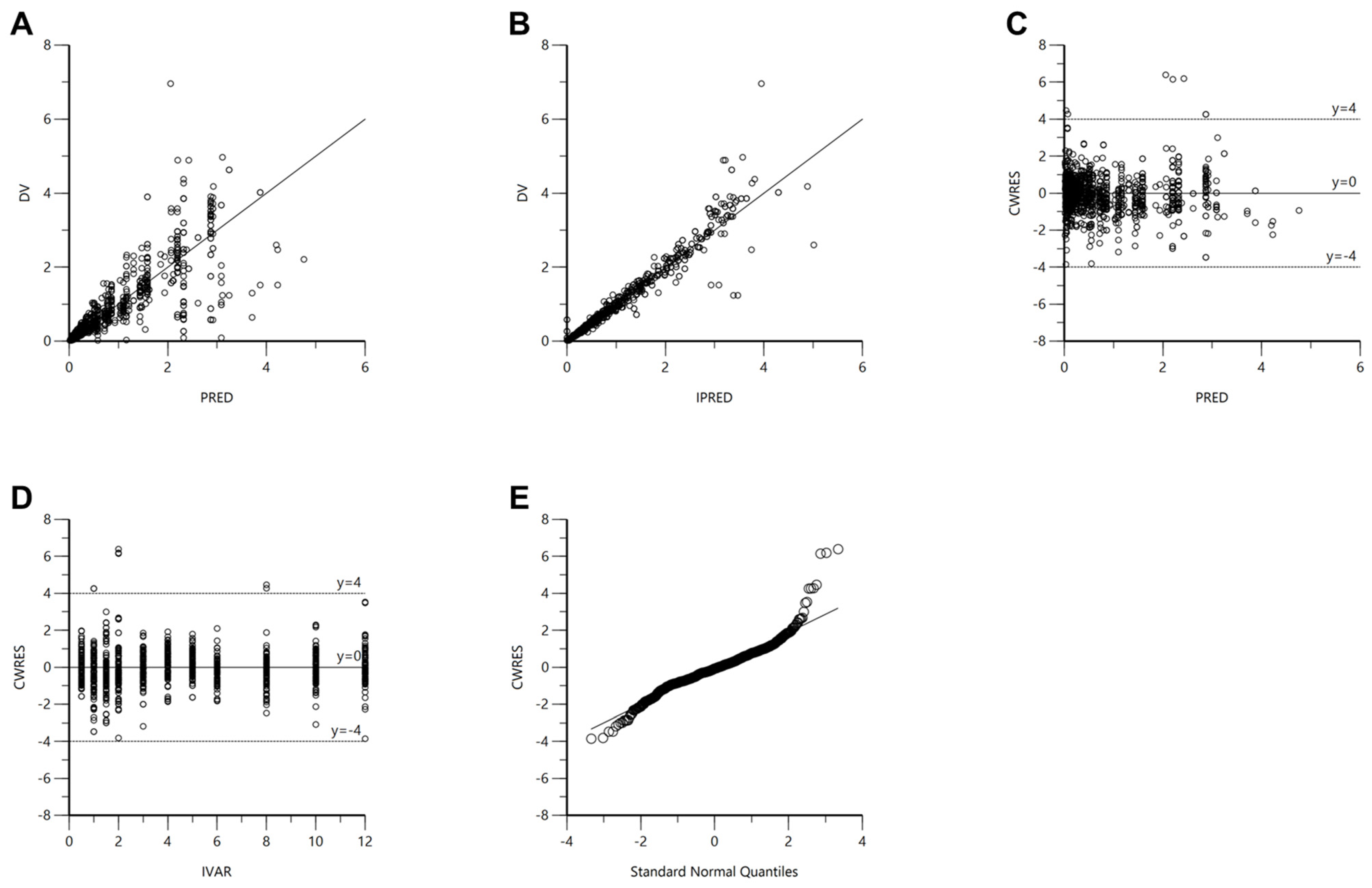
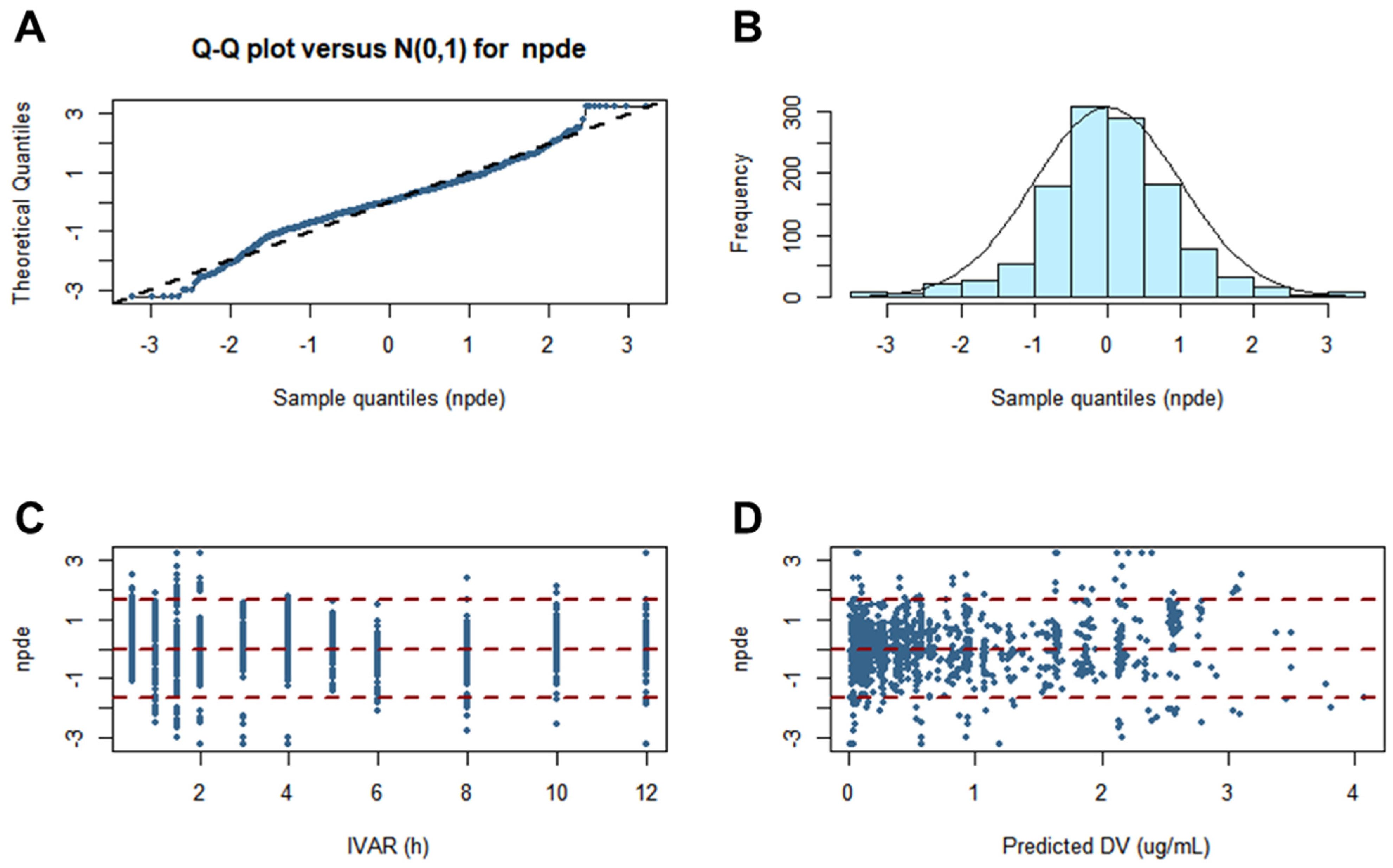
3.7. Pop-PK Model Evaluation
3.8. Reconfirmation of Torsemide Pop-PK Model
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| PK | Pharmacokinetic |
| Pop-PK | Population pharmacokinetic |
| CrCl | Creatinine clearance |
| BSA | Body surface area |
| BMI | Body mass index |
| ALP | Alkaline phosphatase |
| AST | Aspartate transaminase |
| ALT | Alanine transaminase |
| GFR | Glomerular filtration rate |
| BUN | Blood urea nitrogen |
| HPLC | High-performance liquid chromatography |
| UV | Ultraviolet |
| HPLC-UV | High-performance liquid chromatography-ultraviolet |
| PCR-RFLP | Polymerase chain reaction–restriction fragment length polymorphism |
| SD | Standard deviation |
| ANOVA | Analysis of variance |
| tv | Typical value |
| NCA | Non-compartment analysis |
| AIC | Akaike’s information criterion |
| AUC | Area under the curve |
| IIV | Inter-individual variability |
| OFV | Objective function value |
| SE | Standard error |
| RSE | Relative standard error |
| VPC | Visual predictive check |
| NPDE | Normalized prediction distribution error |
| FOCE | First-order conditional estimates |
| NLME | Nonlinear mixed-effects |
References
- Knauf, H.; Mutschler, E. Clinical pharmacokinetics and pharmacodynamics of torasemide. Clin. Pharmacokinet. 1998, 34, 1–24. [Google Scholar] [CrossRef] [PubMed]
- Brater, D.C. Clinical pharmacology of loop diuretics. Drugs 1991, 41, 14–22. [Google Scholar] [CrossRef] [PubMed]
- Qavi, A.H.; Kamal, R.; Schrier, R.W. Clinical use of diuretics in heart failure, cirrhosis, and nephrotic syndrome. Int. J. Nephrol. 2015, 2015, 1–9. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fowler, S.F.; Murray, K.M. Torsemide: A new loop diuretic. Am. J. Health Syst. Pharm. 1995, 52, 1771–1780. [Google Scholar] [CrossRef]
- Dunn, C.J.; Fitton, A.; Brogden, R.N. Torasemide. Drugs 1995, 49, 121–142. [Google Scholar] [CrossRef]
- Schwartz, S.; Brater, D.C.; Pound, D.; Green, P.K.; Kramer, W.G.; Rudy, D. Bioavailability, pharmacokinetics, and pharmacodynamics of torsemide in patients with cirrhosis. Clin. Pharmacol. Ther. 1993, 54, 90–97. [Google Scholar] [CrossRef]
- Miners, J.O.; Coulter, S.; Birkett, D.J.; Goldstein, J.A. Torsemide metabolism by CYP2C9 variants and other human CYP2C subfamily enzymes. Pharm. Genom. 2000, 10, 267–270. [Google Scholar] [CrossRef]
- Vormfelde, S.V.; Engelhardt, S.; Zirk, A.; Meineke, I.; Tuchen, F.; Kirchheiner, J.; Brockmöller, J. CYP2C9 polymorphisms and the interindividual variability in pharmacokinetics and pharmacodynamics of the loop diuretic drug torsemide. Clin. Pharm. Therap. 2004, 76, 557–566. [Google Scholar] [CrossRef]
- Vormfelde, S.; Schirmer, M.; Toliat, M.; Meineke, I.; Kirchheiner, J.; Nürnberg, P.; Brockmöller, J. Genetic variation at the CYP2C locus and its association with torsemide biotransformation. Pharm. J. 2007, 7, 200–211. [Google Scholar] [CrossRef] [Green Version]
- Vormfelde, S.V.; Schirmer, M.; Hagos, Y.; Toliat, M.R.; Engelhardt, S.; Meineke, I.; Burckhardt, G.; Nürnberg, P.; Brockmöller, J. Torsemide renal clearance and genetic variation in luminal and basolateral organic anion transporters. Br. J. Clin. Pharmacol. 2006, 62, 323–335. [Google Scholar] [CrossRef] [Green Version]
- Gehr, T.W.; Rudy, D.W.; Matzke, G.R.; Kramer, W.G.; Sica, D.A.; Brater, D.C. The pharmacokinetics of intravenous and oral torsemide in patients with chronic renal insufficiency. Clin. Pharmacol. Ther. 1994, 56, 31–38. [Google Scholar] [CrossRef] [PubMed]
- Izumi, S.; Nozaki, Y.; Maeda, K.; Komori, T.; Takenaka, O.; Kusuhara, H.; Sugiyama, Y. Investigation of the impact of substrate selection on in vitro organic anion transporting polypeptide 1B1 inhibition profiles for the prediction of drug-drug interactions. Drug Metab. Dispos. 2015, 43, 235–247. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Maeda, K. Organic anion transporting polypeptide (OATP) 1B1 and OATP1B3 as important regulators of the pharmacokinetics of substrate drugs. Biol. Pharm. Bull. 2015, 38, 155–168. [Google Scholar] [CrossRef] [Green Version]
- Werner, U.; Werner, D.; Heinbüchner, S.; Graf, B.; Ince, H.; Kische, S.; Thürmann, P.; König, J.; Fromm, M.F.; Zolk, O. Gender is an important determinant of the disposition of the loop diuretic torasemide. J. Clin. Pharmacol. 2010, 50, 160–168. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Svoboda, M.; Riha, J.; Wlcek, K.; Jaeger, W.; Thalhammer, T. Organic anion transporting polypeptides (OATPs): Regulation of expression and function. Curr. Drug Metab. 2011, 12, 139–153. [Google Scholar] [CrossRef] [PubMed]
- Jeong, S.-H.; Jang, J.-H.; Lee, Y.-B. Population pharmacokinetic analysis of lornoxicam in healthy Korean males considering creatinine clearance and CYP2C9 genetic polymorphism. J. Pharm. Investig. 2021, 52, 109–127. [Google Scholar] [CrossRef]
- Jang, J.-H.; Jeong, S.-H.; Cho, H.-Y.; Lee, Y.-B. Population Pharmacokinetics of Cis-, Trans-, and Total Cefprozil in Healthy Male Koreans. Pharmaceutics 2019, 11, 531–545. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jeong, S.-H.; Jang, J.-H.; Cho, H.-Y.; Lee, Y.-B. Population Pharmacokinetic Analysis of Tiropramide in Healthy Korean Subjects. Pharmaceutics 2020, 12, 374–393. [Google Scholar] [CrossRef]
- Jeong, S.-H.; Jang, J.-H.; Cho, H.-Y.; Lee, Y.-B. Population Pharmacokinetic Analysis of Cefaclor in Healthy Korean Subjects. Pharmaceutics 2021, 13, 754–773. [Google Scholar] [CrossRef]
- Kang, H.-A.; Yoon, H.; Lee, Y.-B. Bioequivalence of Torad tablet 5 mg to Torem tablet 5 mg (torasemide 5 mg). J. Pharm. Investig. 2013, 43, 153–159. [Google Scholar] [CrossRef]
- Cho, H.-Y.; Kang, H.-A.; Park, C.-H.; Kim, S.-M.; Kim, D.-H.; Park, S.; Kim, K.-R.; Hur, H.; Lee, Y.-B. Bioequivalence of Boryung torsemide tablet to Torem tablet (torasemide 10 mg) by high performance liquid chromatography/UV detector. J. Pharm. Investig. 2005, 35, 323–328. [Google Scholar]
- Lesne, M. Comparison of the pharmacokinetics and pharmacodynamics of torasemide and furosemide in healthy volunteers. Arzneimittelforschung 1988, 38, 160–163. [Google Scholar] [PubMed]
- Vargo, D.L.; Kramer, W.G.; Black, P.K.; Smith, W.B.; Serpas, T.; Brater, D.C. Bioavailability, pharmacokinetics, and pharmacodynamics of torsemide and furosemide in patients with congestive heart failure. Clin. Pharm. Therap. 1995, 57, 601–609. [Google Scholar] [CrossRef]
- Lameire, N.; Dodion, L. Acute and chronic effects of torasemide in healthy volunteers. Arzneimittelforschung 1988, 38, 167–171. [Google Scholar]
- Neugebauer, G.; Besenfelder, E.v.; Von Moellendorff, E. Pharmacokinetics and metabolism of torasemide in man. Arzneimittelforschung 1988, 38, 164–166. [Google Scholar]
- Barr, W.; Smith, H.; Karnes, H.; Sica, D.; Vetticaden, S.; Purich, E.; Prasad, V.; Schary, W.; Kramer, W.; Linberg, S. Comparison of bioavailability, pharmacokinetics and pharmacodynamics of torasemide in young and elderly healthy volunteers. Prog. Pharmacol. Clin. Pharmacol. 1990, 8, 15–28. [Google Scholar]
- Barr, W.; Smith, H.; Karnes, H.; Sica, D.; Vetticaden, S.; Prasad, V.; Kramer, W.; Scott, D.; Linberg, S. Torasemide dose-proportionality of pharmacokinetics and pharmacodynamics. Prog. Pharmacol. Clin. Pharmacol. 1990, 8, 29–37. [Google Scholar]
- Kramer, W. Lack of effect of cimetidine on torsemide pharmacokinetics and pharmacodynamics in healthy subjects. In Diuretics IV: Chemistry, Pharmacology and Clinical Applications; Elsevier Science Publishers: Amsterdam, The Netherlands, 1993; pp. 361–364. [Google Scholar]
- Kramer, W.G. Effect of Food on the Pharmacokinetics and Pharmacodynamics of Torsemide. Am. J. Ther. 1995, 2, 499–503. [Google Scholar] [CrossRef]
- Bae, J.W.; Kim, H.K.; Kim, J.H.; Yang, S.I.; Kim, M.J.; Jang, C.G.; Park, Y.S.; Lee, S.Y. Allele and genotype frequencies of CYP2C9 in a Korean population. Br. J. Clin. Pharmacol. 2005, 60, 418–422. [Google Scholar] [CrossRef] [Green Version]
- Wu, X.; Gong, C.; Weinstock, J.; Cheng, J.; Hu, S.; Venners, S.A.; Hsu, Y.-H.; Wu, S.; Zha, X.; Jiang, S.; et al. Associations of the SLCO1B1 polymorphisms With hepatic function, baseline lipid levels, and lipid-lowering response to simvastatin in patients With hyperlipidemia. Clin. Appl. Thromb. Hemost. 2018, 24, 240S–247S. [Google Scholar] [CrossRef] [Green Version]
- FDA. Bioanalytical Method Validation Gudiance for Industry; US Department of Health and Human Services, Food and Drug Administration, Center for Drug Evaluation and Research (CDER), Center for Veterinary Medicine (CVM): Rockville, MD, USA, 2018. [Google Scholar]
- Lim, Y.-J.; Cha, E.-Y.; Jung, H.-E.; Ghim, J.-L.; Lee, S.-J.; Kim, E.-Y.; Shin, J.-G. Genetic polymorphisms of CYP2C9, CYP2C19, CYP2D6, CYP3A4, and CYP3A5 in Vietnamese-Koreans. Transl. Clin. Pharmacol. 2014, 22, 70–77. [Google Scholar] [CrossRef] [Green Version]
- Waisayarat, J.; Dejthevaporn, C.; Srisawasdi, P.; Wongwaisayawan, S.; Sukasem, C. Genetic variations and frequencies of the two functional single nucleotide polymorphisms of SLCO1B1 in the Thai population. Front. Pharmacol. 2020, 11, 728–733. [Google Scholar]
- Ramsey, L.B.; Johnson, S.G.; Caudle, K.E.; Haidar, C.E.; Voora, D.; Wilke, R.A.; Maxwell, W.D.; McLeod, H.L.; Krauss, R.M.; Roden, D.M. The clinical pharmacogenetics implementation consortium guideline for SLCO1B1 and simvastatin-induced myopathy: 2014 update. Clin. Pharmacol. Ther. 2014, 96, 423–428. [Google Scholar] [CrossRef] [PubMed]
- Benet, L.Z. The drug transporter− metabolism alliance: Uncovering and defining the interplay. Mol. Pharm. 2009, 6, 1631–1643. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yoo, H.-D.; Cho, H.-Y.; Lee, S.-N.; Yoon, H.; Lee, Y.-B. Population pharmacokinetic analysis of risperidone and 9-hydroxyrisperidone with genetic polymorphisms of CYP2D6 and ABCB1. J. Pharmacokinet. Pharmacodyn. 2012, 39, 329–341. [Google Scholar] [CrossRef] [PubMed]
- Vormfelde, S.; Toliat, M.; Schirmer, M.; Meineke, I.; Nürnberg, P.; Brockmöller, J. The polymorphisms Asn130Asp and Val174Ala in OATP1B1 and the CYP2C9 allele* 3 independently affect torsemide pharmacokinetics and pharmacodynamics. Clin. Pharmacol. Ther. 2008, 83, 815–817. [Google Scholar] [CrossRef] [PubMed]
- Hermsen, E.D.; Maiefski, M.M.; Florescu, M.C.; Qiu, F.; Rupp, M.E. Comparison of the modification of diet in renal disease and Cockcroft‐Gault equations for dosing antimicrobials. Pharmacother. J. Hum. Pharmacol. Drug Ther. 2009, 29, 649–655. [Google Scholar] [CrossRef]
- Hotta, M.; Li, Y.; Anme, T.; Ushijima, H. Risk factors for low Kaup index among children in rural ethnic minority areas of Yunnan, China. Pediatr. Int. 2005, 47, 147–153. [Google Scholar] [CrossRef]
- El Edelbi, R.; Lindemalm, S.; Eksborg, S. Estimation of body surface area in various childhood ages–validation of the Mosteller formula. Acta Paediatr. 2012, 101, 540–544. [Google Scholar] [CrossRef]





| CYP2C9 | |||||||
|---|---|---|---|---|---|---|---|
| Genotype | *1*1 | *1*3 | *1*13 | ||||
| Subject No | 97 (86.61%) | 12 (10.71%) | 3 (2.68%) | ||||
| Phenotype | EM | IM | IM | ||||
| OATP1B1 | |||||||
| Genotype | *1a*1a | *1a*1b | *1b*1b | *1a*15 | *1b*15 | *15*15 | |
| Subject No | 11 (9.82%) | 38 (33.93%) | 37 (33.04%) | 4 (3.57%) | 19 (16.96%) | 3 (2.68%) | |
| Phenotype | ET | ET | ET | IT | IT | PT | |
| OATP1B1/CYP2C9 | |||||||
| Genotype | OATP1B1 | *1a*1a, *1a*1b, *1b*1b | *1a*1a, *1a*1b, *1b*1b | *1a*15, *1b*15 | *1a*15, *1b*15 | *15*15 | *15*15 |
| CYP2C9 | *1*1 | *1*3, *1*13 | *1*1 | *1*3, *1*13 | *1*1 | *1*3, *1*13 | |
| Subject No | 80 (71.43%) | 6 (5.36%) | 17 (15.18%) | 6 (5.36%) | 0 (0%) | 3 (2.68%) | |
| Phenotype | ET/EM | ET/IM | IT/EM | IT/IM | PT/EM | PT/IM | |
| Model | Description | nParameter | −2LL | AIC | △−2LL | △AIC | Compared with |
|---|---|---|---|---|---|---|---|
| Compartment model | |||||||
| 01 | 1-compartment | 7 | −1106.512 | −1092.512 | - | - | - |
| 02 * | 2-compartment | 11 | −2201.649 | −2179.649 | −1095.14 | −1087.14 | 01 |
| Absorption model | |||||||
| 02 | No Tlag | 11 | −2201.649 | −2179.649 | - | - | - |
| 03 * | With Tlag | 13 | −2757.066 | −2731.066 | −555.42 | −551.42 | 02 |
| Residual error model | |||||||
| 03 * | Proportional | 13 | −2757.066 | −2731.066 | - | - | - |
| 04 | Additive | 13 | 363.99 | 389.99 | 3121.06 | 3121.06 | 03 |
| 05 | Log additive | 13 | −11.59 | 14.41 | 2745.48 | 2745.48 | 03 |
| 06 | Mixed | 14 | −2529.50 | −2501.50 | 227.57 | 229.57 | 03 |
| 07 | Power | 13 | 528.70 | 554.70 | 3285.77 | 3285.77 | 03 |
| IIV model | |||||||
| 08 * | Remove IIV on Ka | 12 | −2765.86 | −2741.86 | −8.80 | −10.80 | 03 |
| 09 | Remove IIV on V/F | 12 | −2530.69 | −2506.69 | 226.37 | 224.37 | 03 |
| 10 | Remove IIV on V2/F | 12 | −2705.07 | −2643.98 | 52.00 | 87.09 | 03 |
| 11 | Remove IIV on CL/F | 12 | −1834.89 | −1810.89 | 922.18 | 920.18 | 03 |
| 12 | Remove IIV on CL2/F | 12 | −2729.3692 | −2705.3692 | 27.70 | 25.70 | 03 |
| 13 | Remove IIV on Tlag | 12 | −2477.5333 | −2453.5333 | 279.53 | 277.53 | 03 |
| Model | Description | Summary | OFV | △OFV | Compared with | nParameter |
|---|---|---|---|---|---|---|
| 1 | Base model | - | −2765.86 | - | - | 12 |
| Forward addition | ||||||
| 2 | OATP1B1 on V/F | - | −2775.354 | −9.490 | Base model | 14 |
| 3 | OATP1B1 on V2/F | - | −2766.150 | −0.286 | Base model | 14 |
| 4 | OATP1B1 on CL/F | - | −2772.458 | −6.594 | Base model | 14 |
| 5 | OATP1B1 on CL2/F | - | −2778.904 | −13.040 | Base model | 14 |
| 6 | OATP1B1 on Tlag | - | −2765.983 | −0.119 | Base model | 14 |
| 7 | CYP2C9 on V/F | - | −2767.611 | −1.747 | Base model | 13 |
| 8 | CYP2C9 on V2/F | - | −2768.067 | −2.203 | Base model | 13 |
| 9 | CYP2C9 on CL/F | - | −2813.517 | −47.652 | Base model | 13 |
| 10 | CYP2C9 on CL2/F | - | −2781.570 | −15.706 | Base model | 13 |
| 11 | CYP2C9 on Tlag | - | −2765.885 | −0.021 | Base model | 13 |
| 12 | OATP1B1 on V/F, CL/F | - | −2782.070 | −6.716 | Model 2 | 16 |
| 13 | OATP1B1 on V/F, CL/F, CL2/F | - | −2797.111 | −15.041 | Model 12 | 18 |
| 14 | OATP1B1 on V/F, CL/F, CL2/F CYP2C9 on CL/F | - | −2838.7158 | −41.605 | Model 13 | 19 |
| 15 | OATP1B1 on V/F, CL/F, CL2/F CYP2C9 on CL/F, CL2/F | Full model | −2850.9748 | −12.259 | Model 14 | 20 |
| Backward removal | ||||||
| 16 | OATP1B1 on V/F, CL/F, CL2/F CYP2C9 on CL2/F | Full model deleted CYP2C9 on CL/F | −2779.106 | 71.8688 | Model 15 | 19 |
| 17 | OATP1B1 on V/F, CL/F, CL2/F CYP2C9 on CL/F | Full model deleted CYP2C9 on CL2/F | −2837.9679 | 13.0069 | Model 15 | 19 |
| 18 | OATP1B1 on V/F, CL/F CYP2C9 on CL/F, CL2/F | Full model deleted OATP1B1 on CL2/F | −2844.7793 | 6.1955 | Model 15 | 18 |
| 19 | OATP1B1 on V/F, CL2/F CYP2C9 on CL/F, CL2/F | Full model deleted OATP1B1 on CL/F | −2850.8641 | 0.1107 | Model 15 | 18 |
| 20 | OATP1B1 on CL/F, CL2/F CYP2C9 on CL/F, CL2/F | Full model deleted OATP1B1 on V/F | −2841.5173 | 9.4575 | Model 15 | 18 |
| 21 * | OATP1B1 on V/F CYP2C9 on CL/F, CL2/F | Full model deleted OATP1B1 on CL/F, CL2/F | −2844.404 | 6.5708 | Model 15 | 16 |
| Parameter | Estimate | SE | RSE (%) | Shrinkage (%) | IIV (%) |
|---|---|---|---|---|---|
| Final model | |||||
| tvV/F (L) | 1.027 | 0.081 | 7.92 | - | - |
| tvCL/F (L/h) | 1.740 | 0.073 | 4.19 | - | - |
| tvV2/F (L) | 3.914 | 0.104 | 2.65 | - | - |
| tvCL2/F (L/h) | 0.828 | 0.056 | 6.71 | - | - |
| tvKa (1/h) | 0.861 | 0.025 | 2.93 | - | - |
| tvTlag (h) | 0.274 | 0.018 | 6.64 | - | - |
| dV/FdOATP1B1IT | −0.410 | 0.129 | 31.54 | - | - |
| dV/FdOATP1B1PT | −0.646 | 0.204 | 31.56 | - | - |
| dCL/FdCYP2C9EM | 0.510 | 0.063 | 12.37 | - | - |
| dCL2/FdCYP2C9EM | 0.365 | 0.056 | 15.23 | - | - |
| ω2v/F | 0.562 | 0.070 | 12.45 | 21.040 | 74.949 |
| ω2CL/F | 0.029 | 0.004 | 13.34 | 2.790 | 17.098 |
| ω2V2/F | 0.036 | 0.007 | 20.74 | 23.780 | 18.938 |
| ω2CL2/F | 0.022 | 0.006 | 25.28 | 38.530 | 14.899 |
| ω2Tlag | 0.341 | 0.079 | 23.16 | 23.370 | 58.367 |
| ε | 0.136 | 0.010 | 7.15 | - | - |
| Parameter | Final Model | Bootstrap (n = 1000) | ||
|---|---|---|---|---|
| Estimate | 95% CI | Median | 95% CI | |
| tvV/F (L) | 1.027 | 0.868–1.187 | 1.042 | 0.861–1.233 |
| tvCL/F (L/h) | 1.740 | 1.597–1.884 | 1.733 | 1.473–2.005 |
| tvV2/F (L) | 3.914 | 3.710–4.118 | 3.892 | 3.698–4.131 |
| tvCL2/F (L/h) | 0.828 | 0.719–0.937 | 0.826 | 0.667–1.102 |
| tvKa (1/h) | 0.861 | 0.811–0.910 | 0.859 | 0.818–0.919 |
| tvTlag (h) | 0.274 | 0.238–0.309 | 0.278 | 0.226–0.324 |
| dV/FdOATP1B1IT | −0.410 | −0.663–−0.156 | −0.409 | −0.586–−0.203 |
| dV/FdOATP1B1PT | −0.646 | −1.046–−0.246 | −0.664 | −1.000–−0.096 |
| dCL/FdCYP2C9EM | 0.510 | 0.386–0.634 | 0.505 | 0.310–0.786 |
| dCL2/FdCYP2C9EM | 0.365 | 0.256–0.473 | 0.357 | 0.093–0.659 |
| ω2v/F | 0.562 | 0.425–0.699 | 0.536 | 0.357–0.715 |
| ω2CL/F | 0.029 | 0.022–0.037 | 0.028 | 0.021–0.035 |
| ω2V2/F | 0.036 | 0.021–0.050 | 0.035 | 0.016–0.053 |
| ω2CL2/F | 0.022 | 0.011–0.033 | 0.019 | 0.002–0.036 |
| ω2Tlag | 0.341 | 0.186–0.495 | 0.356 | 0.089–0.623 |
| ε | 0.136 | 0.117–0.156 | 0.137 | 0.117–0.150 |
| Parameter | Estimate * | NONMEM−NLME Difference (%) of Estimate | RSE (%) * | NONMEM−NLME Difference of RSE | IIV (%) * | NONMEM−NLME Difference of IIV |
|---|---|---|---|---|---|---|
| Final model | ||||||
| tvV/F (L) | 1.027 | 0.00 | 8.04 | 0.12 | - | - |
| tvCL/F (L/h) | 1.740 | 0.00 | 4.32 | 0.13 | - | - |
| tvV2/F (L) | 3.914 | 0.00 | 2.68 | 0.03 | - | - |
| tvCL2/F (L/h) | 0.828 | 0.00 | 6.80 | 0.09 | - | - |
| tvKa (1/h) | 0.861 | 0.00 | 2.94 | 0.01 | - | - |
| tvTlag (h) | 0.274 | 0.00 | 6.66 | 0.02 | - | - |
| dV/FdOATP1B1IT | −0.410 | 0.00 | 31.54 | 0.00 | - | - |
| dV/FdOATP1B1PT | −0.646 | 0.00 | 31.59 | 0.03 | - | - |
| dCL/FdCYP2C9EM | 0.510 | 0.00 | 12.39 | 0.02 | - | - |
| dCL2/FdCYP2C9EM | 0.365 | 0.00 | 15.29 | 0.06 | - | - |
| ω2V/F | 0.562 | 0.00 | 12.47 | 0.02 | 74.967 | 0.02 |
| ω2CL/F | 0.029 | 0.00 | 13.38 | 0.04 | 17.029 | −0.07 |
| ω2V2/F | 0.036 | 0.00 | 20.78 | 0.04 | 18.974 | 0.04 |
| ω2CL2/F | 0.022 | 0.00 | 25.91 | 0.63 | 14.832 | −0.07 |
| ω2Tlag | 0.341 | 0.00 | 23.17 | 0.01 | 58.395 | 0.03 |
| ε | 0.136 | 0.00 | 7.15 | 0.00 | - | - |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jeong, S.-H.; Jang, J.-H.; Cho, H.-Y.; Lee, Y.-B. Population Pharmacokinetic (Pop-PK) Analysis of Torsemide in Healthy Korean Males Considering CYP2C9 and OATP1B1 Genetic Polymorphisms. Pharmaceutics 2022, 14, 771. https://doi.org/10.3390/pharmaceutics14040771
Jeong S-H, Jang J-H, Cho H-Y, Lee Y-B. Population Pharmacokinetic (Pop-PK) Analysis of Torsemide in Healthy Korean Males Considering CYP2C9 and OATP1B1 Genetic Polymorphisms. Pharmaceutics. 2022; 14(4):771. https://doi.org/10.3390/pharmaceutics14040771
Chicago/Turabian StyleJeong, Seung-Hyun, Ji-Hun Jang, Hea-Young Cho, and Yong-Bok Lee. 2022. "Population Pharmacokinetic (Pop-PK) Analysis of Torsemide in Healthy Korean Males Considering CYP2C9 and OATP1B1 Genetic Polymorphisms" Pharmaceutics 14, no. 4: 771. https://doi.org/10.3390/pharmaceutics14040771
APA StyleJeong, S.-H., Jang, J.-H., Cho, H.-Y., & Lee, Y.-B. (2022). Population Pharmacokinetic (Pop-PK) Analysis of Torsemide in Healthy Korean Males Considering CYP2C9 and OATP1B1 Genetic Polymorphisms. Pharmaceutics, 14(4), 771. https://doi.org/10.3390/pharmaceutics14040771