Mechanisms Underlying Synergistic Killing of Polymyxin B in Combination with Cannabidiol against Acinetobacter baumannii: A Metabolomic Study
Abstract
:1. Introduction
2. Materials and Methods
2.1. Bacterial Isolates
2.2. Determination of MIC and FIC
2.3. Static Time-Kill Studies
2.4. Bacterial Culture Preparation for Metabolomics
2.5. Metabolite Extraction
2.6. Data Processing, Bioinformatics, and Statistical Analyses
3. Results
3.1. Synergy Testing of Polymyxin B–Cannabidiol Combinations (FICI Calculation)
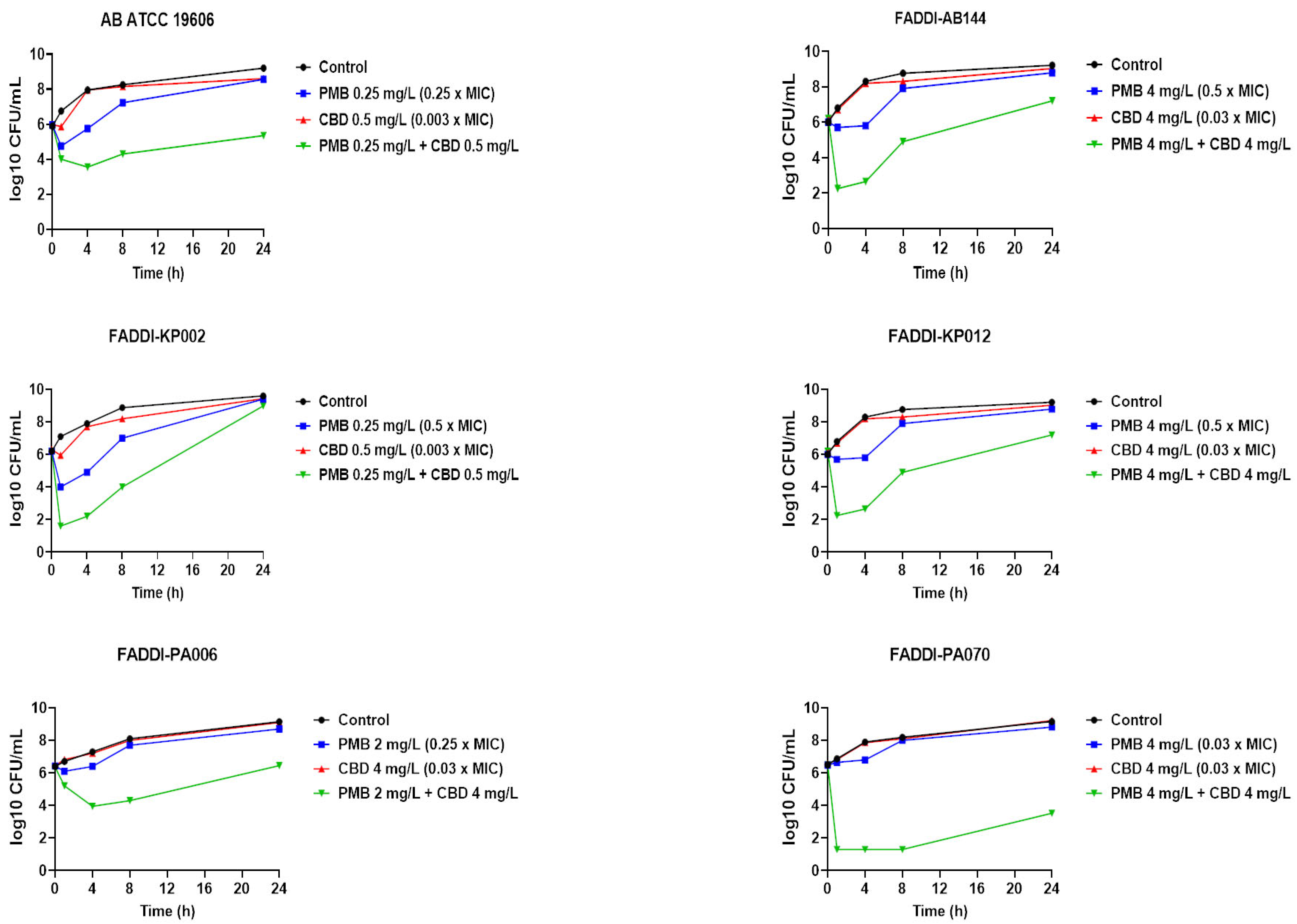
3.2. Static Time-Kill Studies
3.3. Metabolomics Analysis of A. baumannii ATCC 19606 Treated with Polymyxin B, Cannabidiol, and Their Combination
3.3.1. Analyses of Dysregulated Glycerophospholipid and Fatty Acid Metabolism
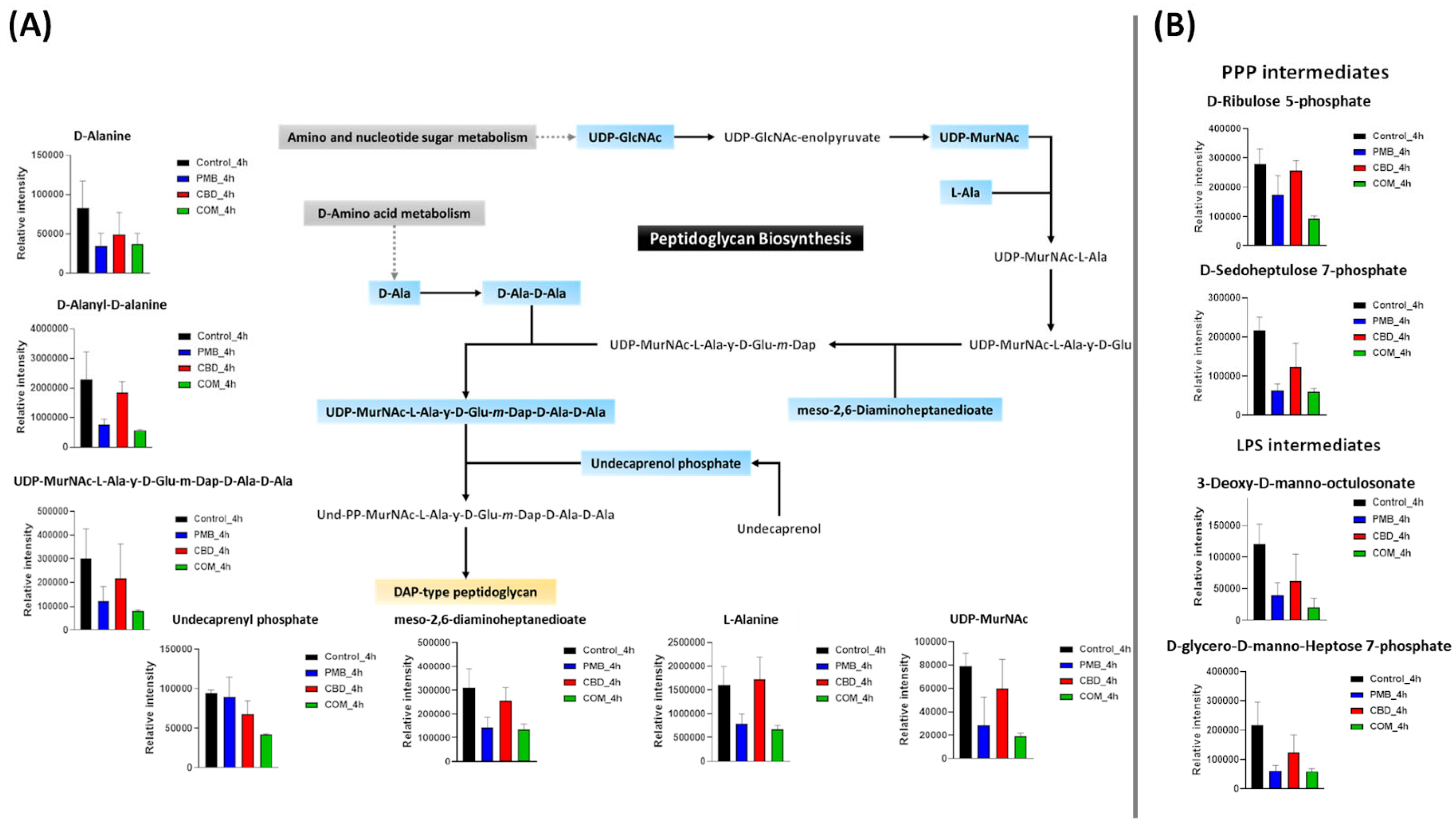
3.3.2. Analyses of Dysregulated Amino-Sugar and Nucleotide-Sugar Metabolism, the Pentose Phosphate Pathway, and Downstream Peptidoglycan and Lipopolysaccharide Biosynthesis
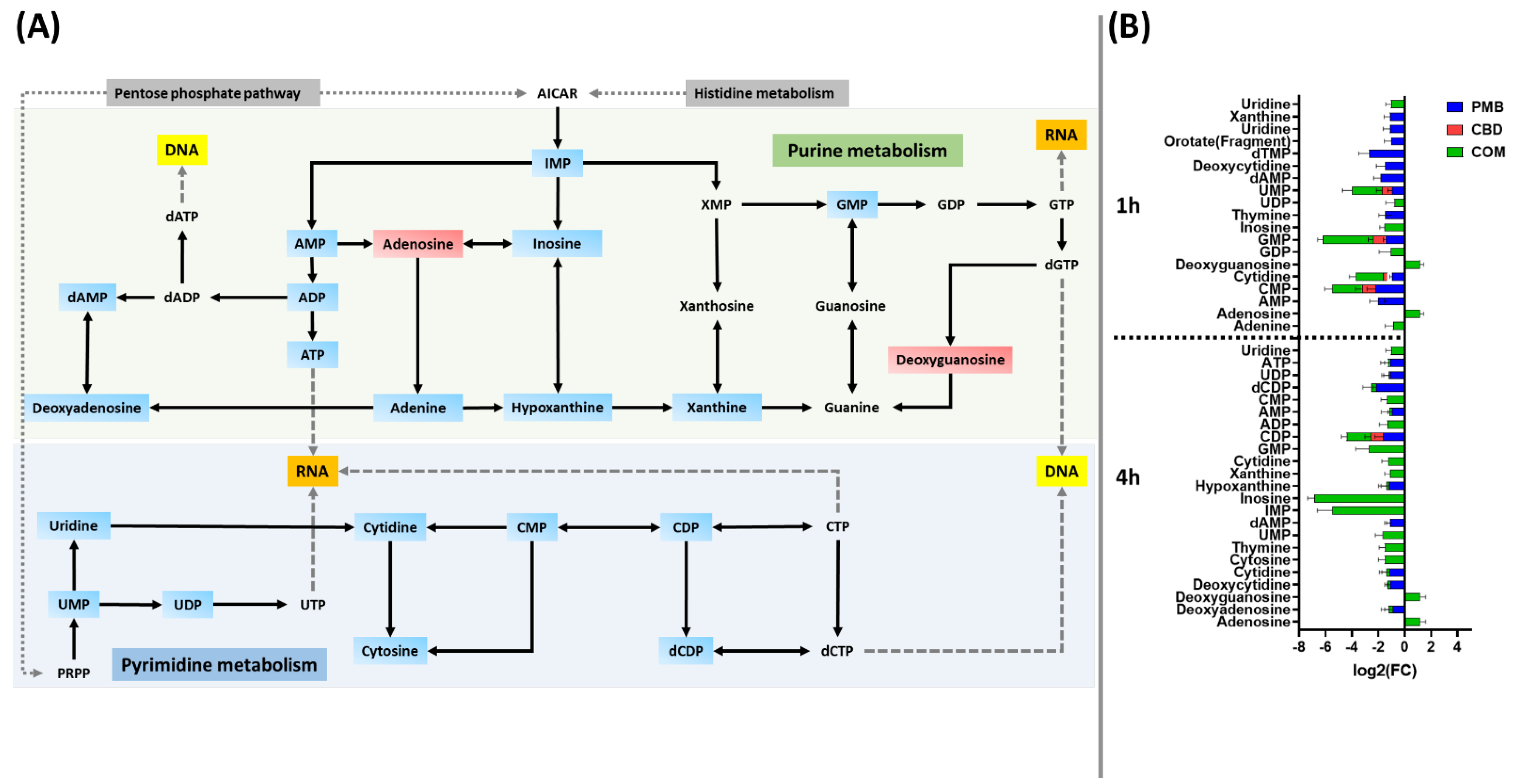
3.3.3. Analyses of Perturbations in Nucleotide and Peptide Metabolism
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Abadi, A.T.B.; Rizvanov, A.A.; Haertlé, T.; Blatt, N.L. World Health Organization report: Current crisis of antibiotic resistance. BioNanoScience 2019, 9, 778–788. [Google Scholar] [CrossRef]
- Evans, B.A.; Hamouda, A.; Amyes, S.G.B. The rise of carbapenem-resistant Acinetobacter baumannii. Curr. Pharm. Des. 2013, 19, 223–238. [Google Scholar] [CrossRef] [PubMed]
- Russo, A.; Bassetti, M.; Ceccarelli, G.; Carannante, N.; Losito, A.R.; Bartoletti, M.; Corcione, S.; Granata, G.; Santoro, A.; Giacobbe, D.R.; et al. Bloodstream infections caused by carbapenem-resistant Acinetobacter baumannii: Clinical features, therapy and outcome from a multicenter study. J. Infect. 2019, 79, 130–138. [Google Scholar] [CrossRef] [PubMed]
- Zaidan, N.; Hornak, J.P.; Reynoso, D. Extensively Drug-Resistant Acinetobacter baumannii Nosocomial Pneumonia Successfully Treated with a Novel Antibiotic Combination. Antimicrob. Agents Chemother. 2021, 65, e00924-21. [Google Scholar] [CrossRef] [PubMed]
- Fournier, P.-E.; Vallenet, D.; Barbe, V.; Audic, S.; Ogata, H.; Poirel, L.; Richet, H.; Robert, C.; Mangenot, S.; Abergel, C. Comparative genomics of multidrug resistance in Acinetobacter baumannii. PLoS Genet. 2006, 2, e7. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- World Health Organization. Global Priority List of Antibiotic-Resistant Bacteria to Guide Research, Discovery, and Development of New Antibiotics; WHO: Geneva, Switzerland, 2017.
- Asokan, G.V.; Vanitha, A. WHO global priority pathogens list on antibiotic resistance: An urgent need for action to integrate One Health data. Perspect. Public Health 2018, 138, 87–88. [Google Scholar]
- Kaye, K.S.; Pogue, J.M.; Tran, T.B.; Nation, R.L.; Li, J. Agents of last resort: Polymyxin resistance. Infect. Dis. Clin. 2016, 30, 391–414. [Google Scholar] [CrossRef]
- Velkov, T.; Thompson, P.E.; Nation, R.L.; Li, J. Structure− activity relationships of polymyxin antibiotics. J. Med. Chem. 2010, 53, 1898–1916. [Google Scholar] [CrossRef] [Green Version]
- Trimble, M.J.; Mlynárčik, P.; Kolář, M.; Hancock, R.E. Polymyxin: Alternative mechanisms of action and resistance. Cold Spring Harb. Perspect. Med. 2016, 6, a025288. [Google Scholar] [CrossRef] [Green Version]
- Li, J.; Rayner, C.R.; Nation, R.L.; Owen, R.J.; Spelman, D.; Tan, K.E.; Liolios, L. Heteroresistance to colistin in multidrug-resistant Acinetobacter baumannii. Antimicrob. Agents Chemother. 2006, 50, 2946–2950. [Google Scholar] [CrossRef] [Green Version]
- Moffatt, J.H.; Harper, M.; Harrison, P.; Hale, J.D.; Vinogradov, E.; Seemann, T.; Henry, R.; Crane, B.; St. Michael, F.; Cox, A.D. Colistin resistance in Acinetobacter baumannii is mediated by complete loss of lipopolysaccharide production. Antimicrob. Agents Chemother. 2010, 54, 4971–4977. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Olaitan, A.O.; Morand, S.; Rolain, J.-M. Mechanisms of polymyxin resistance: Acquired and intrinsic resistance in bacteria. Front. Microbiol. 2014, 5, 643. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pelletier, M.R.; Casella, L.G.; Jones, J.W.; Adams, M.D.; Zurawski, D.V.; Hazlett, K.R.O.; Doi, Y.; Robert, K. Unique structural modifications are present in the lipopolysaccharide from colistin-resistant strains of Acinetobacter baumannii. Antimicrob Agents Chemother. 2013, 5, 4831–4840. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Adams, M.D.; Nickel, G.C.; Bajaksouzian, S.; Lavender, H.; Murthy, A.R.; Jacobs, M.R.; Robert, A. Resistance to colistin in Acinetobacter baumannii associated with mutations in the PmrAB two-component system. Antimicrob. Agents Chemother. 2009, 53, 3628–3634. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ejim, L.; Farha, M.A.; Falconer, S.B.; Wildenhain, J.; Coombes, B.K.; Tyers, M.; Brown, E.D.; Wright, G.D. Combinations of antibiotics and nonantibiotic drugs enhance antimicrobial efficacy. Nat. Chem. Biol. 2011, 7, 348–350. [Google Scholar] [CrossRef]
- Hussein, M.; Han, M.-L.; Zhu, Y.; Schneider-Futschik, E.K.; Hu, X.; Zhou, Q.T.; Lin, Y.-W.; Anderson, D.; Creek, D.J.; Hoyer, D. Mechanistic insights from global metabolomics studies into synergistic bactericidal effect of a polymyxin B combination with tamoxifen against cystic fibrosis MDR Pseudomonas aeruginosa. Comput. Struct. Biotechnol. J. 2018, 16, 587–599. [Google Scholar] [CrossRef]
- Hussein, M.; Schneider-Futschik, E.K.; Paulin, O.K.; Allobawi, R.; Crawford, S.; Zhou, Q.T.; Hanif, A.; Baker, M.; Zhu, Y.; Li, J. Effective Strategy Targeting Polymyxin-Resistant Gram-Negative Pathogens: Polymyxin B in Combination with the Selective Serotonin Reuptake Inhibitor Sertraline. ACS Infect. Dis. 2020, 6, 1436–1450. [Google Scholar] [CrossRef]
- Hussein, M.; Hu, X.; Paulin, O.K.; Crawford, S.; Zhou, Q.T.; Baker, M.; Schneider-Futschik, E.K.; Zhu, Y.; Li, J.; Velkov, T. Polymyxin B combinations with FDA-approved non-antibiotic phenothiazine drugs targeting multi-drug resistance of Gram-negative pathogens. Comput. Struct. Biotechnol. J. 2020, 18, 2247–2258. [Google Scholar] [CrossRef]
- Tran, T.B.; Wang, J.; Velkov, T.; Bergen, P.J.; Li, J. Novel polymyxin combination with antineoplastic mitotane improved the bacterial killing against polymyxin-resistant multidrug-resistant gram-negative pathogens. Front. Microbiol. 2018, 9, 721. [Google Scholar] [CrossRef] [Green Version]
- Lin, Y.-W.; Abdul Rahim, N.; Zhao, J.; Han, M.-L.; Yu, H.H.; Wickremasinghe, H.; Chen, K.; Wang, J.; Paterson, D.L.; Zhu, Y. Novel polymyxin combination with the antiretroviral zidovudine exerts synergistic killing against NDM-producing multidrug-resistant Klebsiella pneumoniae. Antimicrob. Agents Chemother. 2019, 63, e02176-18. [Google Scholar] [CrossRef] [Green Version]
- Hussein, M.H.; Schneider, E.K.; Elliott, A.G.; Han, M.; Reyes-Ortega, F.; Morris, F.; Blaskovich, M.A.; Jasim, R.; Currie, B.; Mayo, M. From breast cancer to antimicrobial: Combating extremely resistant Gram-negative “superbugs” using novel combinations of polymyxin B with selective estrogen receptor modulators. Microb. Drug Resist. 2017, 23, 640–650. [Google Scholar] [CrossRef] [PubMed]
- Ng, S.; Sioson, J.; Yap, J.; Ng, F.; Ching, H.; Teo, J.; Jureen, R.; Hill, J.; Chia, C. Repurposing zidovudine in combination with tigecycline for treating carbapenem-resistant Enterobacteriaceae infections. Eur. J. Clin. Microbiol. Infect. Dis. 2018, 37, 141–148. [Google Scholar] [CrossRef]
- Vincent, I.M.; Ehmann, D.E.; Mills, S.D.; Perros, M.; Barrett, M.P. Untargeted metabolomics to ascertain antibiotic modes of action. Antimicrob. Agents Chemother. 2016, 60, 2281–2291. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kaddurah-Daouk, R.; Weinshilboum, R.M.; Network, P.R. Pharmacometabolomics: Implications for clinical pharmacology and systems pharmacology. Clin. Pharmacol. Ther. 2014, 95, 154–167. [Google Scholar] [CrossRef] [PubMed]
- Mastrangelo, A.; G Armitage, E.; García, A.; Barbas, C. Metabolomics as a tool for drug discovery and personalised medicine. A review. Curr. Top. Med. Chem. 2014, 14, 2627–2636. [Google Scholar] [CrossRef]
- Jia, J.; Zhu, F.; Ma, X.; Cao, Z.W.; Li, Y.X.; Chen, Y.Z. Mechanisms of drug combinations: Interaction and network perspectives. Nat. Rev. Drug Discov. 2009, 8, 111–128. [Google Scholar] [CrossRef]
- Naveed, M.; Khan, T.A.; Ali, I.; Hassan, A.; Ali, H.; Ud, Z.; Din, Z.H.; Tabassum, S.; Saqib, A.M.; Rehman, M.U. In vitro antibacterial activity of Cannabis sativa leaf extracts to some selective pathogenicbacterial strains. Int. J. Biosci. 2014, 4, 65–70. [Google Scholar]
- Fischedick, J.T.; Hazekamp, A.; Erkelens, T.; Choi, Y.H.; Verpoorte, R. Metabolic fingerprinting of Cannabis sativa L., cannabinoids and terpenoids for chemotaxonomic and drug standardization purposes. Phytochemistry 2010, 71, 2058–2073. [Google Scholar] [CrossRef]
- Karas, J.A.; Wong, L.J.; Paulin, O.K.; Mazeh, A.C.; Hussein, M.H.; Li, J.; Velkov, T. The antimicrobial activity of cannabinoids. Antibiotics 2020, 9, 406. [Google Scholar] [CrossRef]
- Krejci, Z. Hemp (Cannabis sativa)--antibiotic drugs. II. Method & results of bacteriological experiments & preliminary clinical experience. Pharmazie. 1958, 13, 155–166. [Google Scholar]
- AS, R.; Biu, A.; SI, Z. Isolation and investigation of antibacterial properties of preparations from wild hemp (Cannabis ruderalis) growing in the Ukraine. Mikrobiol Zh. 1959, 21, 40–48. [Google Scholar]
- Van Klingeren, B.; Ten Ham, M. Antibacterial activity of Δ9-tetrahydrocannabinol and cannabidiol. Antonie Van Leeuwenhoek. 1976, 42, 9–12. [Google Scholar] [CrossRef] [PubMed]
- Appendino, G.; Gibbons, S.; Giana, A.; Pagani, A.; Grassi, G.; Stavri, M.; Smith, E.; Rahman, M.M. Antibacterial cannabinoids from Cannabis sativa: A structure− activity study. J. Nat. Prod. 2008, 71, 1427–1430. [Google Scholar] [CrossRef] [PubMed]
- Iseppi, R.; Brighenti, V.; Licata, M.; Lambertini, A.; Sabia, C.; Messi, P.; Pellati, F.; Benvenuti, S. Chemical characterization and evaluation of the antibacterial activity of essential oils from fibre-type Cannabis sativa L.(Hemp). Molecules 2019, 24, 2302. [Google Scholar] [CrossRef] [Green Version]
- Farha, M.A.; El-Halfawy, O.M.; Gale, R.T.; MacNair, C.R.; Carfrae, L.A.; Zhang, X.; Jentsch, N.G.; Magolan, J.; Brown, E.D. Uncovering the hidden antibiotic potential of cannabis. ACS Infect. Dis. 2020, 6, 338–346. [Google Scholar] [CrossRef]
- Blaskovich, M.; Kavanagh, A.; Ramu, S.; Levy, S.; Callahan, M.; Thurn, M. Cannabidiol is a Remarkably Active Gram-Positive Antibiotic. In Proceedings of the ASM Microbe Conference, San Francisco, CA, USA; 2019. [Google Scholar]
- Blaskovich, M.A.; Kavanagh, A.M.; Elliott, A.G.; Zhang, B.; Ramu, S.; Amado, M.; Lowe, G.J.; Hinton, A.O.; Pham, D.M.T.; Zuegg, J. The antimicrobial potential of cannabidiol. Commun. Biol. 2021, 4, 1–18. [Google Scholar] [CrossRef]
- Abichabki, N.; Zacharias, L.V.; Moreira, N.C.; Bellissimo-Rodrigues, F.; Moreira, F.L.; Benzi, J.R.L.; Ogasawara, T.M.C.; Ferreira, J.C.; Pereira, L.R.L.; Braga, G.Ú.L.; et al. Cannabidiol (CBD) repurposing as antibacterial: Promising therapy of CBD plus polymyxin B against superbugs. bioRxiv 2021. [Google Scholar] [CrossRef]
- Gika, H.G.; Theodoridis, G.A.; Wingate, J.E.; Wilson, I.D. Within-day reproducibility of an HPLC-MS-based method for metabonomic analysis: Application to human urine. J. Proteome Res. 2007, 6, 3291–3303. [Google Scholar] [CrossRef]
- Hussein, M.; Karas, J.A.; Schneider-Futschik, E.K.; Chen, F.; Swarbrick, J.; Paulin, O.K.A.; Hoyer, D.; Baker, M.; Zhu, Y.; Li, J.; et al. The Killing Mechanism of Teixobactin against Methicillin-Resistant Staphylococcus aureus: An Untargeted Metabolomics Study. mSystems 2020, 5, e00077-20. [Google Scholar] [CrossRef]
- Cokol, M.; Chua, H.N.; Tasan, M.; Mutlu, B.; Weinstein, Z.B.; Suzuki, Y.; Nergiz, M.E.; Costanzo, M.; Baryshnikova, A.; Giaever, G. Systematic exploration of synergistic drug pairs. Mol. Syst. Biol. 2011, 7, 544. [Google Scholar] [CrossRef]
- Epand, R.M.; Epand, R.F. Bacterial membrane lipids in the action of antimicrobial agents. J. Pept. Sci. 2011, 17, 298–305. [Google Scholar] [CrossRef] [PubMed]
- Rowlett, V.W.; Mallampalli, V.K.; Karlstaedt, A.; Dowhan, W.; Taegtmeyer, H.; Margolin, W.; Vitrac, H. Impact of membrane phospholipid alterations in Escherichia coli on cellular function and bacterial stress adaptation. J. Bacteriol. 2017, 199, e00849-16. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Parsons, J.B.; Rock, C.O. Bacterial lipids: Metabolism and membrane homeostasis. Prog. Lipid Res. 2013, 52, 249–276. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yao, J.; Rock, C.O. Phosphatidic acid synthesis in bacteria. Biochim. Et Biophys. Acta (BBA)-Mol. Cell Biol. Lipids 2013, 1831, 495–502. [Google Scholar] [CrossRef] [Green Version]
- Whitfield, C.; Trent, M.S. Biosynthesis and export of bacterial lipopolysaccharides. Annu. Rev. Biochem. 2014, 83, 99–128. [Google Scholar] [CrossRef]
- Cho, K.Y.; Salton, M.R.J. Fatty acid composition of bacterial membrane and wall lipids. Biochim. Et Biophys. Acta (BBA)-Lipids Lipid Metab. 1966, 116, 73–79. [Google Scholar] [CrossRef]
- Silhavy, T.J.; Kahne, D.; Walker, S. The bacterial cell envelope. Cold Spring Harb. Perspect. Biol. 2010, 2, a000414. [Google Scholar] [CrossRef]
- Burke, C.R.; Lupták, A. DNA synthesis from diphosphate substrates by DNA polymerases. Proc. Natl. Acad. Sci. 2018, 115, 980–985. [Google Scholar] [CrossRef] [Green Version]
- Parker, J. RNA Polymerase. In Encyclopedia of Genetics; Brenner, S., Miller, J.H., Eds.; Academic Press: New York, NY, USA, 2001; pp. 1746–1747. [Google Scholar] [CrossRef]
- Ayoub Moubareck, C. Polymyxins and Bacterial Membranes: A Review of Antibacterial Activity and Mechanisms of Resistance. Membranes 2020, 10, 181. [Google Scholar] [CrossRef]
- Tran, T.B.; Bergen, P.J.; Creek, D.J.; Velkov, T.; Li, J. Synergistic Killing of Polymyxin B in Combination With the Antineoplastic Drug Mitotane Against Polymyxin-Susceptible and -Resistant Acinetobacter baumannii: A Metabolomic Study. Front. Pharmacol. 2018, 9, 359. [Google Scholar] [CrossRef]
- Shang, R.; Liang, J.; Yi, Y.; Liu, Y.; Wang, J. Review of platensimycin and platencin: Inhibitors of β-ketoacyl-acyl carrier protein (ACP) synthase III (FabH). Molecules 2015, 20, 16127–16141. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Di Bartolomeo, F.; Wagner, A.; Daum, G. Cell biology, physiology and enzymology of phosphatidylserine decarboxylase. Biochim. Et Biophys. Acta (BBA)-Mol. Cell Biol. Lipids 2017, 1862, 25–38. [Google Scholar] [CrossRef] [PubMed]
- Bell, R.M.; Mavis, R.D.; Osborn, M.; Vagelos, P.R. Enzymes of phospholipid metabolism: Localization in the cytoplasmic and outer membrane of the cell envelope of Escherichia coli and Salmonella typhimurium. Biochim. Et Biophys. Acta (BBA)-Biomembr. 1971, 249, 628–635. [Google Scholar] [CrossRef]
- Zhang, Y.-M.; Rock, C.O. Membrane lipid homeostasis in bacteria. Nat. Rev. Microbiol. 2008, 6, 222–233. [Google Scholar] [CrossRef]
- Plumbridge, J.; Vimr, E. Convergent pathways for utilization of the amino sugars N-acetylglucosamine, N-acetylmannosamine, and N-acetylneuraminic acid by Escherichia coli. J. Bacteriol. 1999, 181, 47–54. [Google Scholar] [CrossRef] [Green Version]
- Mengin-Lecreulx, D.; Van Heijenoort, J. Copurification of glucosamine-1-phosphate acetyltransferase and N-acetylglucosamine-1-phosphate uridyltransferase activities of Escherichia coli: Characterization of the glmU gene product as a bifunctional enzyme catalyzing two subsequent steps in the pathway for UDP-N-acetylglucosamine synthesis. J. Bacteriol. 1994, 176, 5788–5795. [Google Scholar]
- Gourlay, L.J.; Sommaruga, S.; Nardini, M.; Sperandeo, P.; Dehò, G.; Polissi, A.; Bolognesi, M. Probing the active site of the sugar isomerase domain from E. coli arabinose-5-phosphate isomerase via X-ray crystallography. Protein Sci. A Publ. Protein Soc. 2010, 19, 2430–2439. [Google Scholar] [CrossRef]
- Bouhss, A.; Trunkfield, A.E.; Bugg, T.D.; Mengin-Lecreulx, D. The biosynthesis of peptidoglycan lipid-linked intermediates. FEMS Microbiol. Rev. 2007, 32, 208–233. [Google Scholar] [CrossRef] [Green Version]
- Samuel, G.; Reeves, P. Biosynthesis of O-antigens: Genes and pathways involved in nucleotide sugar precursor synthesis and O-antigen assembly. Carbohydr. Res. 2003, 338, 2503–2519. [Google Scholar] [CrossRef]
- Kleijn, L.H.; Oppedijk, S.F.; ’t Hart, P.; van Harten, R.M.; Martin-Visscher, L.A.; Kemmink, J.; Breukink, E.; Martin, N.I. Total synthesis of Laspartomycin C and characterization of its antibacterial mechanism of action. J. Med. Chem. 2016, 59, 3569–3574. [Google Scholar] [CrossRef]
- Belete, T.M. Novel targets to develop new antibacterial agents and novel alternatives to antibacterial agents. Hum. Microbiome J. 2019, 11, 100052. [Google Scholar] [CrossRef]
- Yang, J.H.; Wright, S.N.; Hamblin, M.; McCloskey, D.; Alcantar, M.A.; Schrübbers, L.; Lopatkin, A.J.; Satish, S.; Nili, A.; Palsson, B.O.; et al. A White-Box Machine Learning Approach for Revealing Antibiotic Mechanisms of Action. Cell 2019, 177, 1649–1661.e9. [Google Scholar] [CrossRef] [PubMed]
- Lopatkin, A.J.; Yang, J.H. Digital Insights Into Nucleotide Metabolism and Antibiotic Treatment Failure. Front. Digit. Health 2021, 3, 583468. [Google Scholar] [CrossRef] [PubMed]

| MIC (mg/L) | FIC Index | |||||
|---|---|---|---|---|---|---|
| Species | Polymyxin-Susceptible Isolates | PMB | CBD | FIC PMB | FIC CBD | PMB-CBD |
| Acinetobacter baumannii | ATCC 17978 | 0.25 | >64 | 0.0625 | 0.25 | 0.25 |
| ATCC 19606 | 1 | >64 | 0.0625 | 0.5 | 0.062 | |
| FADDI-AB146 | 1 | >64 | 0.125 | 0.5 | 0.12 | |
| FADDI-AB150 | 2 | >64 | 0.0625 | 0.5 | 0.03 | |
| FADDI-AB151 | 2 | >64 | 0.0625 | 0.25 | 0.03 | |
| Polymyxin-resistant isolates | ||||||
| FADDI-AB144 | 8 | >64 | 0.5 | 1 | 0.06 | |
| FADDI-AB148 | 8 | >64 | 0.25 | 1 | 0.03 | |
| FADDI-AB143 | 16 | >64 | 0.5 | 2 | 0.04 | |
| FADDI-AB065 | 64 | 0.5 | 64 | 0.5 | 2.00 | |
| FADDI-AB060 | 128 | >64 | 4 | 2 | 0.04 | |
| Klebsiella pneumoniae | Polymyxin-susceptible isolates | |||||
| Kp BM1 | 0.25 | >64 | 0.0625 | 0.5 | 0.25 | |
| FADDI-KP002 | 0.5 | >64 | 0.0625 | 1 | 0.13 | |
| FADDI-KP069 | 0.5 | >64 | 0.0625 | 4 | 0.15 | |
| KPATCC13883 | 0.5 | >64 | 0.0625 | 2 | 0.14 | |
| ATCC 700721 | 0.5 | >64 | 0.125 | 2 | 0.26 | |
| FADDI-KP005 | 1 | >64 | 0.125 | 1 | 0.13 | |
| Polymyxin-resistant isolates | ||||||
| FADDI-KP003 | 32 | >64 | 4 | 4 | 0.15 | |
| FADDI-KP012 | 32 | >64 | 2 | 4 | 0.09 | |
| FADDI-KP070 | 64 | >64 | 2 | 4 | 0.06 | |
| KPATCC13883R | 128 | >64 | 8 | 4 | 0.09 | |
| Pseudomonas aeruginosa | Polymyxin-susceptible isolates | |||||
| FADDI-PA019 ma | 1 | >64 | 0.5 | 4 | 0.53 | |
| FADDI-PA007 n/mb | 1 | >64 | 0.25 | 1 | 0.25 | |
| FADDI-PA020 m | 2 | >64 | 0.25 | 8 | 0.18 | |
| Polymyxin-resistant isolates | ||||||
| FADDI-PA006 m | 8 | >64 | 0.5 | 0.5 | 0.06 | |
| FADDI-PA066 n/m | 32 | >64 | 2 | 4 | 0.04 | |
| FADDI-PA070 n/m | 128 | >64 | 2 | 2 | 0.02 | |
| FADDI-PA064 n/m | 128 | >64 | 8 | 8 | 0.12 | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hussein, M.; Allobawi, R.; Levou, I.; Blaskovich, M.A.T.; Rao, G.G.; Li, J.; Velkov, T. Mechanisms Underlying Synergistic Killing of Polymyxin B in Combination with Cannabidiol against Acinetobacter baumannii: A Metabolomic Study. Pharmaceutics 2022, 14, 786. https://doi.org/10.3390/pharmaceutics14040786
Hussein M, Allobawi R, Levou I, Blaskovich MAT, Rao GG, Li J, Velkov T. Mechanisms Underlying Synergistic Killing of Polymyxin B in Combination with Cannabidiol against Acinetobacter baumannii: A Metabolomic Study. Pharmaceutics. 2022; 14(4):786. https://doi.org/10.3390/pharmaceutics14040786
Chicago/Turabian StyleHussein, Maytham, Rafah Allobawi, Irini Levou, Mark A. T. Blaskovich, Gauri G. Rao, Jian Li, and Tony Velkov. 2022. "Mechanisms Underlying Synergistic Killing of Polymyxin B in Combination with Cannabidiol against Acinetobacter baumannii: A Metabolomic Study" Pharmaceutics 14, no. 4: 786. https://doi.org/10.3390/pharmaceutics14040786







