Hyaluronic Acid: Known for Almost a Century, but Still in Vogue
Abstract
:1. Introducing Hyaluronic Acid
2. A Unique Metabolism from Beginning to End
3. HA Fits Every Size
4. Old and New Partners in Action
5. New Roles in Radiology and Biology
6. That Is Not All
7. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
Abbreviations
References
- Cowman, M.K. Hyaluronan and Hyaluronan Fragments. Adv. Carbohydr. Chem. Biochem. 2017, 74, 1–59. [Google Scholar] [CrossRef]
- Laurent, T.C. Hyaluronan Research in Uppsala*. Upsala J. Med. Sci. 2007, 112, 123–142. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Couchman, J.R.; Pataki, C.A. An Introduction to Proteoglycans and Their Localization. J. Histochem. Cytochem. 2012, 60, 885–897. [Google Scholar] [CrossRef] [PubMed]
- Iozzo, R.V.; Schaefer, L. Proteoglycan Form and Function: A Comprehensive Nomenclature of Proteoglycans. Matrix Biol. 2015, 42, 11–55. [Google Scholar] [CrossRef] [PubMed]
- Kakehi, K.; Kinoshita, M.; Yasueda, S. Hyaluronic Acid: Separation and Biological Implications. J. Chromatogr. B 2003, 797, 347–355. [Google Scholar] [CrossRef]
- Necas, J.; Bartosikova, L.; Brauner, P.; Kolář, J. Hyaluronic Acid (Hyaluronan): A Review. Vet. Med. 2008, 53, 397–411. [Google Scholar] [CrossRef] [Green Version]
- Liao, Y.-H.; Jones, S.A.; Forbes, B.; Martin, G.P.; Brown, M.B. Hyaluronan: Pharmaceutical Characterization and Drug Delivery. Drug Deliv. 2005, 12, 327–342. [Google Scholar] [CrossRef]
- Meyer, K.; Palmer, J.W. The Polysaccharide of the Vitreous Humor. J. Biol. Chem. 1934, 107, 629–634. [Google Scholar] [CrossRef]
- Levene, P.A.; López-Suárez, J. Mucins and Mucoids. J. Biol. Chem. 1918, 36, 105–126. [Google Scholar] [CrossRef]
- Weissmann, B.; Meyer, K. The Structure of Hyalobiuronic Acid and of Hyaluronic Acid from Umbilical Cord1,2. J. Am. Chem. Soc. 1954, 76, 1753–1757. [Google Scholar] [CrossRef]
- Chen, W.Y.; Abatangelo, G. Functions of Hyaluronan in Wound Repair. Wound Repair Regen 1999, 7, 79–89. [Google Scholar] [CrossRef] [PubMed]
- Hargittai, I.; Hargittai, M. Molecular Structure of Hyaluronan: An Introduction. Struct. Chem. 2008, 19, 697–717. [Google Scholar] [CrossRef]
- Valcarcel, J.; Novoa-Carballal, R.; Pérez-Martín, R.I.; Reis, R.L.; Vázquez, J.A. Glycosaminoglycans from Marine Sources as Therapeutic Agents. Biotechnol. Adv. 2017, 35, 711–725. [Google Scholar] [CrossRef] [PubMed]
- Sze, J.H.; Brownlie, J.C.; Love, C.A. Biotechnological Production of Hyaluronic Acid: A Mini Review. 3 Biotech 2016, 6, 67. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Almond, A. Hyaluronan. Cell. Mol. Life Sci. 2007, 64, 1591–1596. [Google Scholar] [CrossRef]
- Dechert, T.A.; Ducale, A.E.; Ward, S.I.; Yager, D.R. Hyaluronan in Human Acute and Chronic Dermal Wounds. Wound Repair Regen. 2006, 14, 252–258. [Google Scholar] [CrossRef]
- Rah, M.J. A Review of Hyaluronan and Its Ophthalmic Applications. Optom. J. Am. Optom. Assoc. 2011, 82, 38–43. [Google Scholar] [CrossRef]
- Khunmanee, S.; Jeong, Y.; Park, H. Crosslinking Method of Hyaluronic-Based Hydrogel for Biomedical Applications. J. Tissue Eng. 2017, 8, 2041731417726464. [Google Scholar] [CrossRef] [Green Version]
- Jouon, N.; Rinaudo, M.; Milas, M.; Desbrières, J. Hydration of Hyaluronic Acid as a Function of the Counterion Type and Relative Humidity. Carbohydr. Polym. 1995, 26, 69–73. [Google Scholar] [CrossRef]
- Nakamura, M.; Hikida, M.; Nakano, T.; Ito, S.; Hamano, T.; Kinoshita, S. Characterization of Water Retentive Properties of Hyaluronan. Cornea 1993, 12, 433–436. [Google Scholar] [CrossRef]
- Huang, H.; Du, W.; Brekken, R.A. Extracellular Matrix Induction of Intracellular Reactive Oxygen Species. Antioxid. Redox Signal. 2017, 27, 774–784. [Google Scholar] [CrossRef] [PubMed]
- Waddington, R.J.; Moseley, R.; Embery, G. Periodontal Disease Mechanisms: Reactive Oxygen Species: A Potential Role in the Pathogenesis of Periodontal Diseases. Oral Dis. 2000, 6, 138–151. [Google Scholar] [CrossRef]
- Moseley, R.; Leaver, M.; Walker, M.; Waddington, R.J.; Parsons, D.; Chen, W.Y.J.; Embery, G. Comparison of the Antioxidant Properties of HYAFF®-11p75, AQUACEL® and Hyaluronan towards Reactive Oxygen Species in Vitro. Biomaterials 2002, 23, 2255–2264. [Google Scholar] [CrossRef]
- Choi, K.Y.; Saravanakumar, G.; Park, J.H.; Park, K. Hyaluronic Acid-Based Nanocarriers for Intracellular Targeting: Interfacial Interactions with Proteins in Cancer. Colloids Surf. B Biointerfac. 2012, 99, 82–94. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rodemann, H.P.; Blaese, M.A. Responses of Normal Cells to Ionizing Radiation. Semin. Radiat. Oncol. 2007, 17, 81–88. [Google Scholar] [CrossRef]
- DeAngelis, P.L. Hyaluronan Synthases: Fascinating Glycosyltransferases from Vertebrates, Bacterial Pathogens, and Algal Viruses. Cell. Mol. Life Sci. 1999, 56, 670–682. [Google Scholar] [CrossRef]
- DeAngelis, P.L.; Papaconstantinou, J.; Weigel, P.H. Molecular Cloning, Identification, and Sequence of the Hyaluronan Synthase Gene from Group A Streptococcus Pyogenes. J. Biol. Chem. 1993, 268, 19181–19184. [Google Scholar] [CrossRef]
- DeAngelis, P.L.; Papaconstantinou, J.; Weigel, P.H. Isolation of a Streptococcus Pyogenes Gene Locus That Directs Hyaluronan Biosynthesis in Acapsular Mutants and in Heterologous Bacteria. J. Biol. Chem. 1993, 268, 14568–14571. [Google Scholar] [CrossRef]
- DeAngelis, P.L.; Jing, W.; Drake, R.R.; Achyuthan, A.M. Identification and Molecular Cloning of a Unique Hyaluronan Synthase from Pasteurella Multocida. J. Biol. Chem. 1998, 273, 8454–8458. [Google Scholar] [CrossRef] [Green Version]
- Weigel, P.H. Functional Characteristics and Catalytic Mechanisms of the Bacterial Hyaluronan Synthases. IUBMB Life 2002, 54, 201–211. [Google Scholar] [CrossRef]
- Weigel, P.H.; DeAngelis, P.L. Hyaluronan Synthases: A Decade-plus of Novel Glycosyltransferases. J. Biol. Chem. 2007, 282, 36777–36781. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Weigel, P.H. Hyaluronan Synthase: The Mechanism of Initiation at the Reducing End and a Pendulum Model for Polysaccharide Translocation to the Cell Exterior. Int. J. Cell Biol. 2015, 2015, 367579. [Google Scholar] [CrossRef] [Green Version]
- Siiskonen, H.; Oikari, S.; Pasonen-Seppänen, S.; Rilla, K. Hyaluronan Synthase 1: A Mysterious Enzyme with Unexpected Functions. Front. Immunol. 2015, 6, 43. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Reitinger, S.; Müllegger, J.; Lepperdinger, G. Xenopus Kidney Hyaluronidase-1 (XKH1), a Novel Type of Membrane-Bound Hyaluronidase Solely Degrades Hyaluronan at Neutral PH. FEBS Lett. 2001, 505, 213–216. [Google Scholar] [CrossRef] [Green Version]
- Tsepilov, R.N.; Beloded, A.V. Hyaluronic Acid-an “Old” Molecule with “New” Functions: Biosynthesis and Depolymerization of Hyaluronic Acid in Bacteria and Vertebrate Tissues Including during Carcinogenesis. Biochem. Mosc. 2015, 80, 1093–1108. [Google Scholar] [CrossRef] [PubMed]
- Spicer, A.P.; McDonald, J.A. Characterization and Molecular Evolution of a Vertebrate Hyaluronan Synthase Gene Family. J. Biol. Chem. 1998, 273, 1923–1932. [Google Scholar] [CrossRef] [Green Version]
- Fallacara, A.; Baldini, E.; Manfredini, S.; Vertuani, S. Hyaluronic Acid in the Third Millennium. Polymers 2018, 10, 701. [Google Scholar] [CrossRef] [Green Version]
- Itano, N.; Kimata, K. Mammalian Hyaluronan Synthases. IUBMB Life 2002, 54, 195–199. [Google Scholar] [CrossRef]
- Lee, J.Y.; Spicer, A.P. Hyaluronan: A Multifunctional, MegaDalton, Stealth Molecule. Curr. Opin. Cell Biol. 2000, 12, 581–586. [Google Scholar] [CrossRef]
- Bai, K.-J.; Spicer, A.P.; Mascarenhas, M.M.; Yu, L.; Ochoa, C.D.; Garg, H.G.; Quinn, D.A. The Role of Hyaluronan Synthase 3 in Ventilator-Induced Lung Injury. Am. J. Respir. Crit. Care Med. 2005, 172, 92–98. [Google Scholar] [CrossRef] [Green Version]
- Itano, N.; Sawai, T.; Yoshida, M.; Lenas, P.; Yamada, Y.; Imagawa, M.; Shinomura, T.; Hamaguchi, M.; Yoshida, Y.; Ohnuki, Y.; et al. Three Isoforms of Mammalian Hyaluronan Synthases Have Distinct Enzymatic Properties. J. Biol. Chem. 1999, 274, 25085–25092. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Camenisch, T.D.; Spicer, A.P.; Brehm-Gibson, T.; Biesterfeldt, J.; Augustine, M.L.; Calabro, A.; Kubalak, S.; Klewer, S.E.; McDonald, J.A. Disruption of Hyaluronan Synthase-2 Abrogates Normal Cardiac Morphogenesis and Hyaluronan-Mediated Transformation of Epithelium to Mesenchyme. J. Clin. Investig. 2000, 106, 349–360. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tien, J.Y.L.; Spicer, A.P. Three Vertebrate Hyaluronan Synthases Are Expressed during Mouse Development in Distinct Spatial and Temporal Patterns. Dev. Dyn. 2005, 233, 130–141. [Google Scholar] [CrossRef] [PubMed]
- Laurent, T.C.; Fraser, J.R. Hyaluronan. FASEB J. 1992, 6, 2397–2404. [Google Scholar] [CrossRef] [PubMed]
- Vigetti, D.; Viola, M.; Karousou, E.; De Luca, G.; Passi, A. Metabolic Control of Hyaluronan Synthases. Matrix Biol. 2014, 35, 8–13. [Google Scholar] [CrossRef] [Green Version]
- Vigetti, D.; Karousou, E.; Viola, M.; Passi, A. Analysis of Hyaluronan Synthase Activity. In Glycosaminoglycans: Chemistry and Biology; Balagurunathan, K., Nakato, H., Desai, U.R., Eds.; Methods in Molecular Biology; Springer: New York, NY, USA, 2015; pp. 201–208. ISBN 978-1-4939-1714-3. [Google Scholar]
- Cowman, M.K.; Matsuoka, S. Experimental Approaches to Hyaluronan Structure. Carbohydr. Res. 2005, 340, 791–809. [Google Scholar] [CrossRef]
- Tammi, R.H.; Passi, A.G.; Rilla, K.; Karousou, E.; Vigetti, D.; Makkonen, K.; Tammi, M.I. Transcriptional and Post-Translational Regulation of Hyaluronan Synthesis. FEBS J. 2011, 278, 1419–1428. [Google Scholar] [CrossRef]
- Jacobson, A.; Brinck, J.; Briskin, M.J.; Spicer, A.P.; Heldin, P. Expression of Human Hyaluronan Synthases in Response to External Stimuli. Biochem. J. 2000, 348, 29–35. [Google Scholar] [CrossRef]
- Tlapak-Simmons, V.L.; Baron, C.A.; Gotschall, R.; Haque, D.; Canfield, W.M.; Weigel, P.H. Hyaluronan Biosynthesis by Class I Streptococcal Hyaluronan Synthases Occurs at the Reducing End. J. Biol. Chem. 2005, 280, 13012–13018. [Google Scholar] [CrossRef] [Green Version]
- Deen, A.J.; Rilla, K.; Oikari, S.; Kärnä, R.; Bart, G.; Häyrinen, J.; Bathina, A.R.; Ropponen, A.; Makkonen, K.; Tammi, R.H.; et al. Rab10-Mediated Endocytosis of the Hyaluronan Synthase HAS3 Regulates Hyaluronan Synthesis and Cell Adhesion to Collagen. J. Biol. Chem. 2014, 289, 8375–8389. [Google Scholar] [CrossRef] [Green Version]
- Karousou, E.; Kamiryo, M.; Skandalis, S.S.; Ruusala, A.; Asteriou, T.; Passi, A.; Yamashita, H.; Hellman, U.; Heldin, C.-H.; Heldin, P. The Activity of Hyaluronan Synthase 2 Is Regulated by Dimerization and Ubiquitination. J. Biol. Chem. 2010, 285, 23647–23654. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bart, G.; Vico, N.O.; Hassinen, A.; Pujol, F.M.; Deen, A.J.; Ruusala, A.; Tammi, R.H.; Squire, A.; Heldin, P.; Kellokumpu, S.; et al. Fluorescence Resonance Energy Transfer (FRET) and Proximity Ligation Assays Reveal Functionally Relevant Homo-and Heteromeric Complexes among Hyaluronan Synthases HAS1, HAS2, and HAS3. J. Biol. Chem. 2015, 290, 11479–11490. [Google Scholar] [CrossRef] [Green Version]
- Marcellin, E.; Steen, J.A.; Nielsen, L.K. Insight into Hyaluronic Acid Molecular Weight Control. Appl. Microbiol. Biotechnol. 2014, 98, 6947–6956. [Google Scholar] [CrossRef] [PubMed]
- Ferrer, V.P.; de Mari, T.L.; Gremski, L.H.; Trevisan Silva, D.; da Silveira, R.B.; Gremski, W.; Chaim, O.M.; Senff-Ribeiro, A.; Nader, H.B.; Veiga, S.S. A Novel Hyaluronidase from Brown Spider (Loxosceles Intermedia) Venom (Dietrich’s Hyaluronidase): From Cloning to Functional Characterization. PLoS Negl. Trop. Dis. 2013, 7, e2206. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kogan, G.; Šoltés, L.; Stern, R.; Gemeiner, P. Hyaluronic Acid: A Natural Biopolymer with a Broad Range of Biomedical and Industrial Applications. Biotechnol. Lett. 2007, 29, 17–25. [Google Scholar] [CrossRef] [PubMed]
- Takeo, S.; Fujise, M.; Akiyama, T.; Habuchi, H.; Itano, N.; Matsuo, T.; Aigaki, T.; Kimata, K.; Nakato, H. In Vivo Hyaluronan Synthesis upon Expression of the Mammalian Hyaluronan Synthase Gene in Drosophila. J. Biol. Chem. 2004, 279, 18920–18925. [Google Scholar] [CrossRef] [Green Version]
- Shiedlin, A.; Bigelow, R.; Christopher, W.; Arbabi, S.; Yang, L.; Maier, R.V.; Wainwright, N.; Childs, A.; Miller, R.J. Evaluation of Hyaluronan from Different Sources: Streptococcus Zooepidemicus, Rooster Comb, Bovine Vitreous, and Human Umbilical Cord. Biomacromolecules 2004, 5, 2122–2127. [Google Scholar] [CrossRef]
- Cowman, M.K.; Lee, H.-G.; Schwertfeger, K.L.; McCarthy, J.B.; Turley, E.A. The Content and Size of Hyaluronan in Biological Fluids and Tissues. Front. Immunol. 2015, 6, 261. [Google Scholar] [CrossRef] [Green Version]
- Papakonstantinou, E.; Karakiulakis, G. The “sweet” and “Bitter” Involvement of Glycosaminoglycans in Lung Diseases: Pharmacotherapeutic Relevance. Br. J. Pharmacol. 2009, 157, 1111–1127. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Laurent, C.; Johnson-Wells, G.; Hellström, S.; Engström-Laurent, A.; Wells, A.F. Localization of Hyaluronan in Various Muscular Tissues. Cell Tissue Res. 1991, 263, 201–205. [Google Scholar] [CrossRef]
- Armstrong, S.E.; Bell, D.R. Relationship between Lymph and Tissue Hyaluronan in Skin and Skeletal Muscle. Am. J. Physiol. Heart Circ. Physiol. 2002, 283, H2485–H2494. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- George, J.; Stern, R. Serum Hyaluronan and Hyaluronidase: Very Early Markers of Toxic Liver Injury. Clin. Chim. Acta 2004, 348, 189–197. [Google Scholar] [CrossRef]
- Toole, B.P. Hyaluronan and Its Binding Proteins, the Hyaladherins. Curr. Opin. Cell Biol. 1990, 2, 839–844. [Google Scholar] [CrossRef]
- Allison, D.D.; Grande-Allen, K.J. Review. Hyaluronan: A Powerful Tissue Engineering Tool. Tissue Eng. 2006, 12, 2131–2140. [Google Scholar] [CrossRef] [PubMed]
- Tiwari, S.; Bahadur, P. Modified Hyaluronic Acid Based Materials for Biomedical Applications. Int. J. Biol. Macromol. 2019, 121, 556–571. [Google Scholar] [CrossRef] [PubMed]
- Pomin, V.H.; Mulloy, B. Glycosaminoglycans and Proteoglycans. Pharmaceuticals 2018, 11, 27. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hardingham, T.E.; Muir, H. The Specific Interaction of Hyaluronic Acid with Cartilage Proteoglycans. Biochim. Et Biophys. Acta (BBA) Gen. Subj. 1972, 279, 401–405. [Google Scholar] [CrossRef]
- Hascall, V.C.; Heinegård, D. Aggregation of Cartilage Proteoglycans I. The Role of Hyaluronic Acid. J. Biol. Chem. 1974, 249, 4232–4241. [Google Scholar] [CrossRef]
- Laurent, U.B.G.; Reed, R.K. Turnover of Hyaluronan in the Tissues. Adv. Drug Deliv. Rev. 1991, 7, 237–256. [Google Scholar] [CrossRef]
- Triggs-Raine, B.; Natowicz, M.R. Biology of Hyaluronan: Insights from Genetic Disorders of Hyaluronan Metabolism. World J. Biol. Chem. 2015, 6, 110–120. [Google Scholar] [CrossRef]
- Buhren, B.A.; Schrumpf, H.; Hoff, N.-P.; Bölke, E.; Hilton, S.; Gerber, P.A. Hyaluronidase: From Clinical Applications to Molecular and Cellular Mechanisms. Eur. J. Med. Res. 2016, 21, 5. [Google Scholar] [CrossRef] [Green Version]
- Stern, R.; Jedrzejas, M.J. The Hyaluronidases: Their Genomics, Structures, and Mechanisms of Action. Chem. Rev. 2006, 106, 818–839. [Google Scholar] [CrossRef] [Green Version]
- Rigden, D.J.; Jedrzejas, M.J. Structures of Streptococcus Pneumoniae Hyaluronate Lyase in Complex with Chondroitin and Chondroitin Sulfate Disaccharides. Insights into Specificity and Mechanism of Action. J. Biol. Chem. 2003, 278, 50596–50606. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Meyer, K. 11 Hyaluronidases. In The Enzymes; Boyer, P.D., Ed.; Academic Press: Cambridge, MA, USA, 1971; Volume 5, pp. 307–320. [Google Scholar]
- Jedrzejas, M.J. Structural and Functional Comparison of Polysaccharide-Degrading Enzymes. Crit. Rev. Biochem. Mol. Biol. 2000, 35, 221–251. [Google Scholar] [CrossRef] [PubMed]
- Khan, N.; Niazi, Z.R.; Rehman, F.U.; Akhtar, A.; Khan, M.M.; Khan, S.; Baloch, N.; Khan, S. Hyaluronidases: A Therapeutic Enzyme. Available online: https://www.ingentaconnect.com/content/ben/ppl/2018/00000025/00000007/art00010 (accessed on 2 April 2019).
- Stern, R. Devising a Pathway for Hyaluronan Catabolism: Are We There Yet? Glycobiology 2003, 13, 105R–115R. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kreil, G. Hyaluronidases-A Group of Neglected Enzymes. Protein Sci. 1995, 4, 1666–1669. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Csóka, A.B.; Scherer, S.W.; Stern, R. Expression Analysis of Six Paralogous Human Hyaluronidase Genes Clustered on Chromosomes 3p21 and 7q31. Genomics 1999, 60, 356–361. [Google Scholar] [CrossRef] [PubMed]
- Csóka, A.B.; Frost, G.I.; Wong, T.; Stern, R.; Csóka, T.B. Purification and Microsequencing of Hyaluronidase Isozymes from Human Urine. FEBS Lett. 1997, 417, 307–310. [Google Scholar] [CrossRef] [Green Version]
- Frost, G.I.; Csóka, A.B.; Wong, T.; Stern, R.; Csóka, T.B. Purification, Cloning, and Expression of Human Plasma Hyaluronidase. Biochem. Biophys. Res. Commun. 1997, 236, 10–15. [Google Scholar] [CrossRef]
- Frost, G.I.; Mohapatra, G.; Wong, T.M.; Csóka, A.B.; Gray, J.W.; Stern, R. HYAL1LUCA-1, a Candidate Tumor Suppressor Gene on Chromosome 3p21.3, Is Inactivated in Head and Neck Squamous Cell Carcinomas by Aberrant Splicing of Pre-MRNA. Oncogene 2000, 19, 870–877. [Google Scholar] [CrossRef] [Green Version]
- Lepperdinger, G.; Müllegger, J.; Kreil, G. Hyal2-Less Active, but More Versatile? Matrix Biol. 2001, 20, 509–514. [Google Scholar] [CrossRef]
- Rodén, L.; Campbell, P.; Fraser, J.R.; Laurent, T.C.; Pertoft, H.; Thompson, J.N. Enzymic Pathways of Hyaluronan Catabolism. Ciba Found. Symp. 1989, 143, 60–76; discussion 76–86, 281–285. [Google Scholar] [PubMed]
- Lepperdinger, G.; Strobl, B.; Kreil, G. HYAL2, a Human Gene Expressed in Many Cells, Encodes a Lysosomal Hyaluronidase with a Novel Type of Specificity. J. Biol. Chem. 1998, 273, 22466–22470. [Google Scholar] [CrossRef] [Green Version]
- Vigetti, D.; Karousou, E.; Viola, M.; Deleonibus, S.; De Luca, G.; Passi, A. Hyaluronan: Biosynthesis and Signaling. Biochim. Et Biophys. Acta (BBA) Gen. Subj. 2014, 1840, 2452–2459. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stern, R. Hyaluronidases in Cancer Biology. Semin. Cancer Biol. 2008, 18, 275–280. [Google Scholar] [CrossRef] [PubMed]
- Stern, R. Hyaluronan Catabolism: A New Metabolic Pathway. Eur. J. Cell Biol. 2004, 83, 317–325. [Google Scholar] [CrossRef]
- Erickson, M.; Stern, R. Chain Gangs: New Aspects of Hyaluronan Metabolism. Biochem. Res. Int. 2011, 2012, e893947. [Google Scholar] [CrossRef] [Green Version]
- Lin, Y.; Mahan, K.; Lathrop, W.F.; Myles, D.G.; Primakoff, P. A Hyaluronidase Activity of the Sperm Plasma Membrane Protein PH-20 Enables Sperm to Penetrate the Cumulus Cell Layer Surrounding the Egg. J. Cell Biol. 1994, 125, 1157–1163. [Google Scholar] [CrossRef]
- Maciej-Hulme, M.L. New Insights into Human Hyaluronidase 4/Chondroitin Sulphate Hydrolase. Front. Cell Dev. Biol. 2021, 9, 1–7. [Google Scholar] [CrossRef]
- Yamamoto, H.; Tobisawa, Y.; Inubushi, T.; Irie, F.; Ohyama, C.; Yamaguchi, Y. A Mammalian Homolog of the Zebrafish Transmembrane Protein 2 (TMEM2) Is the Long-Sought-after Cell-Surface Hyaluronidase. J. Biol. Chem. 2017, 292, 7304–7313. [Google Scholar] [CrossRef] [Green Version]
- Stern, R.; Maibach, H.I. Hyaluronan in Skin: Aspects of Aging and Its Pharmacologic Modulation. Clin. Dermatol. 2008, 26, 106–122. [Google Scholar] [CrossRef] [PubMed]
- Stern, R.; Kogan, G.; Jedrzejas, M.J.; Soltés, L. The Many Ways to Cleave Hyaluronan. Biotechnol. Adv. 2007, 25, 537–557. [Google Scholar] [CrossRef] [PubMed]
- Monzon, M.E.; Fregien, N.; Schmid, N.; Falcon, N.S.; Campos, M.; Casalino-Matsuda, S.M.; Forteza, R.M. Reactive Oxygen Species and Hyaluronidase 2 Regulate Airway Epithelial Hyaluronan Fragmentation. J. Biol. Chem. 2010, 285, 26126–26134. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Salwowska, N.M.; Bebenek, K.A.; Żądło, D.A.; Wcisło-Dziadecka, D.L. Physiochemical Properties and Application of Hyaluronic Acid: A Systematic Review. J. Cosmet. Derm. 2016, 15, 520–526. [Google Scholar] [CrossRef]
- Heitzmann, E.; Thumm, D.; Baudouin, C. A Review of the Efficacy, Safety and Tolerability of Lacrycon® Eye Drops for the Treatment of Dry Eye Syndrome. J. Français D’Ophtalmologie 2019, 42, 642–654. [Google Scholar] [CrossRef]
- López-Ruiz, E.; Jiménez, G.; Álvarez de Cienfuegos, L.; Antic, C.; Sabata, R.; Marchal, J.A.; Gálvez-Martín, P. Advances of Hyaluronic Acid in Stem Cell Therapy and Tissue Engineering, Including Current Clinical Trials. Eur. Cell. Mater. 2019, 37, 186–213. [Google Scholar] [CrossRef]
- Huang, G.; Huang, H. Hyaluronic Acid-Based Biopharmaceutical Delivery and Tumor-Targeted Drug Delivery System. J. Control. Release 2018, 278, 122–126. [Google Scholar] [CrossRef]
- Kim, J.H.; Moon, M.J.; Kim, D.Y.; Heo, S.H.; Jeong, Y.Y. Hyaluronic Acid-Based Nanomaterials for Cancer Therapy. Polymers 2018, 10, 1133. [Google Scholar] [CrossRef] [Green Version]
- Toole, B.P. Hyaluronan: From Extracellular Glue to Pericellular Cue. Nat. Rev. Cancer 2004, 4, 528–539. [Google Scholar] [CrossRef]
- Šafránková, B.; Hermannová, M.; Nešporová, K.; Velebný, V.; Kubala, L. Absence of Differences among Low, Middle, and High Molecular Weight Hyaluronan in Activating Murine Immune Cells in Vitro. Int. J. Biol. Macromol. 2018, 107, 1–8. [Google Scholar] [CrossRef]
- Yuan, H.; Amin, R.; Ye, X.; De La Motte, C.A.; Cowman, M.K. Determination of Hyaluronan Molecular Mass Distribution in Human Breast Milk. Anal. Biochem. 2015, 474, 78–88. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Balazs, E.A. Viscoelastic Properties of Hyaluronic Acid and Biological Lubrication. Univ. Mich. Med. Cent. J. 1968, 255–259. Available online: https://pubmed.ncbi.nlm.nih.gov/5728249/ (accessed on 20 February 2022).
- Lokeshwar, V.B.; Selzer, M.G. Differences in Hyaluronic Acid-Mediated Functions and Signaling in Arterial, Microvessel, and Vein-Derived Human Endothelial Cells. J. Biol. Chem. 2000, 275, 27641–27649. [Google Scholar] [CrossRef] [Green Version]
- West, D.C.; Kumar, S. The Effect of Hyaluronate and Its Oligosaccharides on Endothelial Cell Proliferation and Monolayer Integrity. Exp. Cell Res. 1989, 183, 179–196. [Google Scholar] [CrossRef]
- Lesley, J.; Hascall, V.C.; Tammi, M.; Hyman, R. Hyaluronan Binding by Cell Surface CD44. J. Biol. Chem. 2000, 275, 26967–26975. [Google Scholar] [CrossRef]
- Day, A.J.; de la Motte, C.A. Hyaluronan Cross-Linking: A Protective Mechanism in Inflammation? Trends Immunol. 2005, 26, 637–643. [Google Scholar] [CrossRef] [PubMed]
- Bano, F.; Banerji, S.; Howarth, M.; Jackson, D.G.; Richter, R.P. A Single Molecule Assay to Probe Monovalent and Multivalent Bonds between Hyaluronan and Its Key Leukocyte Receptor CD44 under Force. Sci. Rep. 2016, 6, 34176. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Suzuki, T.; Suzuki, M.; Ogino, S.; Umemoto, R.; Nishida, N.; Shimada, I. Mechanical Force Effect on the Two-State Equilibrium of the Hyaluronan-Binding Domain of CD44 in Cell Rolling. Proc. Natl. Acad. Sci. USA 2015, 112, 6991–6996. [Google Scholar] [CrossRef] [Green Version]
- Neuman, M.G.; Nanau, R.M.; Oruña-Sanchez, L.; Coto, G. Hyaluronic Acid and Wound Healing. J. Pharm. Pharm. Sci. 2015, 18, 53–60. [Google Scholar] [CrossRef] [Green Version]
- Delmage, J.M.; Powars, D.R.; Jaynes, P.K.; Allerton, S.E. The Selective Suppression of Immunogenicity by Hyaluronic Acid. Ann. Clin. Lab. Sci. 1986, 16, 303–310. [Google Scholar]
- Nakamura, K.; Yokohama, S.; Yoneda, M.; Okamoto, S.; Tamaki, Y.; Ito, T.; Okada, M.; Aso, K.; Makino, I. High, but Not Low, Molecular Weight Hyaluronan Prevents T-Cell-Mediated Liver Injury by Reducing Proinflammatory Cytokines in Mice. J. Gastroenterol. 2004, 39, 346–354. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.-Y.; Lee, C.-H.; Dedaj, R.; Zhao, H.; Mrabat, H.; Sheidlin, A.; Syrkina, O.; Huang, P.-M.; Garg, H.G.; Hales, C.A.; et al. High-Molecular-Weight Hyaluronan-a Possible New Treatment for Sepsis-Induced Lung Injury: A Preclinical Study in Mechanically Ventilated Rats. Crit. Care 2008, 12, R102. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jiang, D.; Liang, J.; Fan, J.; Yu, S.; Chen, S.; Luo, Y.; Prestwich, G.D.; Mascarenhas, M.M.; Garg, H.G.; Quinn, D.A.; et al. Regulation of Lung Injury and Repair by Toll-like Receptors and Hyaluronan. Nat. Med. 2005, 11, 1173–1179. [Google Scholar] [CrossRef] [PubMed]
- Miki, Y.; Teramura, T.; Tomiyama, T.; Onodera, Y.; Matsuoka, T.; Fukuda, K.; Hamanishi, C. Hyaluronan Reversed Proteoglycan Synthesis Inhibited by Mechanical Stress: Possible Involvement of Antioxidant Effect. Inflamm. Res. 2010, 59, 471–477. [Google Scholar] [CrossRef]
- Cooper, C.A.; Brown, K.K.; Meletis, C.D.; Zabriskie, N. Inflammation and Hyaluronic Acid. Altern. Complement. Ther. 2008, 14, 78–84. [Google Scholar] [CrossRef] [Green Version]
- Stern, R. Hyaluronan Metabolism: A Major Paradox in Cancer Biology. Pathol. Biol. 2005, 53, 372–382. [Google Scholar] [CrossRef]
- Auvinen, P.; Rilla, K.; Tumelius, R.; Tammi, M.; Sironen, R.; Soini, Y.; Kosma, V.-M.; Mannermaa, A.; Viikari, J.; Tammi, R. Hyaluronan Synthases (HAS1–3) in Stromal and Malignant Cells Correlate with Breast Cancer Grade and Predict Patient Survival. Breast Cancer Res. Treat. 2014, 143, 277–286. [Google Scholar] [CrossRef]
- Paiva, P.; Van Damme, M.-P.; Tellbach, M.; Jones, R.L.; Jobling, T.; Salamonsen, L.A. Expression Patterns of Hyaluronan, Hyaluronan Synthases and Hyaluronidases Indicate a Role for Hyaluronan in the Progression of Endometrial Cancer. Gynecol. Oncol. 2005, 98, 193–202. [Google Scholar] [CrossRef]
- Udabage, L.; Brownlee, G.R.; Nilsson, S.K.; Brown, T.J. The Over-Expression of HAS2, Hyal-2 and CD44 Is Implicated in the Invasiveness of Breast Cancer. Exp. Cell Res. 2005, 310, 205–217. [Google Scholar] [CrossRef]
- Cyphert, J.M.; Trempus, C.S.; Garantziotis, S. Size Matters: Molecular Weight Specificity of Hyaluronan Effects in Cell Biology. Int. J. Cell Biol. 2015, 2015, 563818. [Google Scholar] [CrossRef] [Green Version]
- Monslow, J.; Govindaraju, P.; Puré, E. Hyaluronan-A Functional and Structural Sweet Spot in the Tissue Microenvironment. Front. Immunol 2015, 6, 231. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rayahin, J.E.; Buhrman, J.S.; Zhang, Y.; Koh, T.J.; Gemeinhart, R.A. High and Low Molecular Weight Hyaluronic Acid Differentially Influence Macrophage Activation. ACS Biomater. Sci. Eng. 2015, 1, 481–493. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pardue, E.L.; Ibrahim, S.; Ramamurthi, A. Role of Hyaluronan in Angiogenesis and Its Utility to Angiogenic Tissue Engineering. Organogenesis 2008, 4, 203–214. [Google Scholar] [CrossRef] [Green Version]
- Petrey, A.C.; de la Motte, C.A. Hyaluronan, a Crucial Regulator of Inflammation. Front. Immunol. 2014, 5, 101. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ruppert, S.M.; Hawn, T.R.; Arrigoni, A.; Wight, T.N.; Bollyky, P.L. Tissue Integrity Signals Communicated by High-Molecular Weight Hyaluronan and the Resolution of Inflammation. Immunol. Res. 2014, 58, 186–192. [Google Scholar] [CrossRef] [Green Version]
- Scheibner, K.A.; Lutz, M.A.; Boodoo, S.; Fenton, M.J.; Powell, J.D.; Horton, M.R. Hyaluronan Fragments Act as an Endogenous Danger Signal by Engaging TLR2. J. Immunol. 2006, 177, 1272–1281. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Termeer, C.; Benedix, F.; Sleeman, J.; Fieber, C.; Voith, U.; Ahrens, T.; Miyake, K.; Freudenberg, M.; Galanos, C.; Simon, J.C. Oligosaccharides of Hyaluronan Activate Dendritic Cells via Toll-like Receptor 4. J. Exp. Med. 2002, 195, 99–111. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Campo, G.M.; Avenoso, A.; Campo, S.; D’Ascola, A.; Nastasi, G.; Calatroni, A. Molecular Size Hyaluronan Differently Modulates Toll-like Receptor-4 in LPS-Induced Inflammation in Mouse Chondrocytes. Biochimie 2010, 92, 204–215. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, Y.; Li, L.; Kamiryo, M.; Asteriou, T.; Moustakas, A.; Yamashita, H.; Heldin, P. Hyaluronan Fragments Induce Endothelial Cell Differentiation in a CD44-and CXCL1/GRO1-Dependent Manner. J. Biol. Chem. 2005, 280, 24195–24204. [Google Scholar] [CrossRef] [Green Version]
- Ohno, S.; Im, H.-J.; Knudson, C.B.; Knudson, W. Hyaluronan Oligosaccharide-Induced Activation of Transcription Factors in Bovine Articular Chondrocytes. Arthritis Rheum. 2005, 52, 800–809. [Google Scholar] [CrossRef] [Green Version]
- Ohno, S.; Im, H.-J.; Knudson, C.B.; Knudson, W. Hyaluronan Oligosaccharides Induce Matrix Metalloproteinase 13 via Transcriptional Activation of NFκB and P38 MAP Kinase in Articular Chondrocytes. J. Biol. Chem. 2006, 281, 17952–17960. [Google Scholar] [CrossRef] [Green Version]
- Xu, H.; Ito, T.; Tawada, A.; Maeda, H.; Yamanokuchi, H.; Isahara, K.; Yoshida, K.; Uchiyama, Y.; Asari, A. Effect of Hyaluronan Oligosaccharides on the Expression of Heat Shock Protein 72. J. Biol. Chem. 2002, 277, 17308–17314. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Suzuki, Y.; Yamaguchi, T. Effects of Hyaluronic Acid on Macrophage Phagocytosis and Active Oxygen Release. Agents Actions 1993, 38, 32–37. [Google Scholar] [CrossRef] [PubMed]
- McKee, C.M.; Penno, M.B.; Cowman, M.; Burdick, M.D.; Strieter, R.M.; Bao, C.; Noble, P.W. Hyaluronan (HA) Fragments Induce Chemokine Gene Expression in Alveolar Macrophages. The Role of HA Size and CD44. J. Clin. Investig. 1996, 98, 2403–2413. [Google Scholar] [CrossRef] [Green Version]
- Horton, M.R.; Burdick, M.D.; Strieter, R.M.; Bao, C.; Noble, P.W. Regulation of Hyaluronan-Induced Chemokine Gene Expression by IL-10 and IFN-Gamma in Mouse Macrophages. J. Immunol. 1998, 160, 3023–3030. [Google Scholar] [PubMed]
- Hodge-Dufour, J.; Noble, P.W.; Horton, M.R.; Bao, C.; Wysoka, M.; Burdick, M.D.; Strieter, R.M.; Trinchieri, G.; Puré, E. Induction of IL-12 and Chemokines by Hyaluronan Requires Adhesion-Dependent Priming of Resident but Not Elicited Macrophages. J. Immunol. 1997, 159, 2492–2500. [Google Scholar] [PubMed]
- Lyle, D.B.; Breger, J.C.; Baeva, L.F.; Shallcross, J.C.; Durfor, C.N.; Wang, N.S.; Langone, J.J. Low Molecular Weight Hyaluronic Acid Effects on Murine Macrophage Nitric Oxide Production. J. Biomed. Mater. Res. A 2010, 94, 893–904. [Google Scholar] [CrossRef]
- McKee, C.M.; Lowenstein, C.J.; Horton, M.R.; Wu, J.; Bao, C.; Chin, B.Y.; Choi, A.M.; Noble, P.W. Hyaluronan Fragments Induce Nitric-Oxide Synthase in Murine Macrophages through a Nuclear Factor KappaB-Dependent Mechanism. J. Biol. Chem. 1997, 272, 8013–8018. [Google Scholar] [CrossRef] [Green Version]
- Termeer, C.C.; Hennies, J.; Voith, U.; Ahrens, T.; Weiss, J.M.; Prehm, P.; Simon, J.C. Oligosaccharides of Hyaluronan Are Potent Activators of Dendritic Cells. J. Immunol. 2000, 165, 1863–1870. [Google Scholar] [CrossRef] [Green Version]
- Jiang, D.; Liang, J.; Noble, P.W. Hyaluronan in Tissue Injury and Repair. Annu. Rev. Cell Dev. Biol. 2007, 23, 435–461. [Google Scholar] [CrossRef] [Green Version]
- Stern, R.; Asari, A.A.; Sugahara, K.N. Hyaluronan Fragments: An Information-Rich System. Eur. J. Cell Biol. 2006, 85, 699–715. [Google Scholar] [CrossRef]
- Powell, J.D.; Horton, M.R. Threat Matrix: Low-Molecular-Weight Hyaluronan (HA) as a Danger Signal. Immunol. Res. 2005, 31, 207–218. [Google Scholar] [CrossRef]
- Misra, S.; Hascall, V.C.; Markwald, R.R.; Ghatak, S. Interactions between Hyaluronan and Its Receptors (CD44, RHAMM) Regulate the Activities of Inflammation and Cancer. Front. Immunol 2015, 6, 201. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bohaumilitzky, L.; Huber, A.-K.; Stork, E.M.; Wengert, S.; Woelfl, F.; Boehm, H. A Trickster in Disguise: Hyaluronan’s Ambivalent Roles in the Matrix. Front. Oncol. 2017, 7, 242. [Google Scholar] [CrossRef] [PubMed]
- Lagunas-Rangel, F.A.; Chávez-Valencia, V. Learning of Nature: The Curious Case of the Naked Mole Rat. Mech. Ageing Dev. 2017, 164, 76–81. [Google Scholar] [CrossRef]
- Piersigilli, A.; Meyerholz, D.K. The “Naked Truth”: Naked Mole-Rats Do Get Cancer. Vet. Pathol. 2016, 53, 519–520. [Google Scholar] [CrossRef] [Green Version]
- Tian, X.; Azpurua, J.; Hine, C.; Vaidya, A.; Myakishev-Rempel, M.; Ablaeva, J.; Mao, Z.; Nevo, E.; Gorbunova, V.; Seluanov, A. High Molecular Weight Hyaluronan Mediates the Cancer Resistance of the Naked Mole-Rat. Nature 2013, 499, 346. [Google Scholar] [CrossRef] [Green Version]
- Garg, H.G.; Hales, C.A. Chemistry and Biology of Hyaluronan; Elsevier: Amsterdam, The Netherlands, 2004; ISBN 978-0-08-047222-5. [Google Scholar]
- Knudson, C.B.; Knudson, W. Hyaluronan-Binding Proteins in Development, Tissue Homeostasis, and Disease. FASEB J. 1993, 7, 1233–1241. [Google Scholar] [CrossRef]
- Girish, K.S.; Kemparaju, K. The Magic Glue Hyaluronan and Its Eraser Hyaluronidase: A Biological Overview. Life Sci. 2007, 80, 1921–1943. [Google Scholar] [CrossRef]
- Perkins, S.J.; Nealis, A.S.; Dudhia, J.; Hardingham, T.E. Immunoglobulin Fold and Tandem Repeat Structures in Proteoglycan N-Terminal Domains and Link Protein. J. Mol. Biol. 1989, 206, 737–748. [Google Scholar] [CrossRef]
- Kohda, D.; Morton, C.J.; Parkar, A.A.; Hatanaka, H.; Inagaki, F.M.; Campbell, I.D.; Day, A.J. Solution Structure of the Link Module: A Hyaluronan-Binding Domain Involved in Extracellular Matrix Stability and Cell Migration. Cell 1996, 86, 767–775. [Google Scholar] [CrossRef]
- Watanabe, H.; Cheung, S.C.; Itano, N.; Kimata, K.; Yamada, Y. Identification of Hyaluronan-Binding Domains of Aggrecan. J. Biol. Chem. 1997, 272, 28057–28065. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hasegawa, K.; Yoneda, M.; Kuwabara, H.; Miyaishi, O.; Itano, N.; Ohno, A.; Zako, M.; Isogai, Z. Versican, a Major Hyaluronan-Binding Component in the Dermis, Loses Its Hyaluronan-Binding Ability in Solar Elastosis. J. Investig. Dermatol. 2007, 127, 1657–1663. [Google Scholar] [CrossRef] [Green Version]
- Rauch, U.; Clement, A.; Retzler, C.; Fröhlich, L.; Fässler, R.; Göhring, W.; Faissner, A. Mapping of a Defined Neurocan Binding Site to Distinct Domains of Tenascin-C. J. Biol. Chem. 1997, 272, 26905–26912. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yamada, H.; Watanabe, K.; Shimonaka, M.; Yamaguchi, Y. Molecular Cloning of Brevican, a Novel Brain Proteoglycan of the Aggrecan/Versican Family. J. Biol. Chem. 1994, 269, 10119–10126. [Google Scholar] [CrossRef]
- Hascall, V.C. Interaction of Cartilage Proteoglycans with Hyaluronic Acid. J. Supramol. Struct. 1977, 7, 101–120. [Google Scholar] [CrossRef] [PubMed]
- Yamaguchi, Y. Lecticans: Organizers of the Brain Extracellular Matrix. Cell. Mol. Life Sci. 2000, 57, 276–289. [Google Scholar] [CrossRef]
- Teriete, P.; Banerji, S.; Noble, M.; Blundell, C.D.; Wright, A.J.; Pickford, A.R.; Lowe, E.; Mahoney, D.J.; Tammi, M.I.; Kahmann, J.D.; et al. Structure of the Regulatory Hyaluronan Binding Domain in the Inflammatory Leukocyte Homing Receptor CD44. Mol. Cell 2004, 13, 483–496. [Google Scholar] [CrossRef] [Green Version]
- Banerji, S.; Day, A.J.; Kahmann, J.D.; Jackson, D.G. Characterization of a Functional Hyaluronan-Binding Domain from the Human CD44 Molecule Expressed in Escherichia Coli. Protein Expr. Purif. 1998, 14, 371–381. [Google Scholar] [CrossRef]
- Zhou, B.; Weigel, J.A.; Fauss, L.; Weigel, P.H. Identification of the Hyaluronan Receptor for Endocytosis (HARE). J. Biol. Chem. 2000, 275, 37733–37741. [Google Scholar] [CrossRef] [Green Version]
- Wisniewski, H.-G.; Snitkin, E.S.; Mindrescu, C.; Sweet, M.H.; Vilcek, J. TSG-6 Protein Binding to Glycosaminoglycans: Formation of Stable Complexes with Hyaluronan and Binding to Chondroitin Sulfates. J. Biol. Chem. 2005, 280, 14476–14484. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, B.; Yang, B.L.; Savani, R.C.; Turley, E.A. Identification of a Common Hyaluronan Binding Motif in the Hyaluronan Binding Proteins RHAMM, CD44 and Link Protein. EMBO J. 1994, 13, 286–296. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Entwistle, J.; Hou, G.; Li, Q.; Turley, E.A. The Characterization of a Human RHAMM CDNA: Conservation of the Hyaluronan-Binding Domains. Gene 1996, 174, 299–306. [Google Scholar] [CrossRef]
- Grammatikakis, N.; Grammatikakis, A.; Yoneda, M.; Yu, Q.; Banerjee, S.D.; Toole, B.P. A Novel Glycosaminoglycan-Binding Protein Is the Vertebrate Homologue of the Cell Cycle Control Protein, Cdc37. J. Biol. Chem. 1995, 270, 16198–16205. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bost, F.; Diarra-Mehrpour, M.; Martin, J.-P. Inter-α-Trypsin Inhibitor Proteoglycan Family. Eur. J. Biochem. 1998, 252, 339–346. [Google Scholar] [CrossRef] [PubMed]
- Nishina, H.; Inageda, K.; Takahashi, K.; Hoshino, S.; Ikeda, K.; Katada, T. Cell Surface Antigen CD38 Identified as Ecto-Enzyme of NAD Glycohydrolase Has Hyaluronate-Binding Activity. Biochem. Biophys. Res. Commun. 1994, 203, 1318–1323. [Google Scholar] [CrossRef]
- Becerra, S.P.; Perez-Mediavilla, L.A.; Weldon, J.E.; Locatelli-Hoops, S.; Senanayake, P.; Notari, L.; Notario, V.; Hollyfield, J.G. Pigment Epithelium-Derived Factor Binds to Hyaluronan. J. Biol. Chem. 2008, 283, 33310–33320. [Google Scholar] [CrossRef] [Green Version]
- Amemiya, K.; Nakatani, T.; Saito, A.; Suzuki, A.; Munakata, H. Hyaluronan-Binding Motif Identified by Panning a Random Peptide Display Library. Biochim. Et Biophys. Acta (BBA) Gen. Subj. 2005, 1724, 94–99. [Google Scholar] [CrossRef]
- Dicker, K.T.; Gurski, L.A.; Pradhan-Bhatt, S.; Witt, R.L.; Farach-Carson, M.C.; Jia, X. Hyaluronan: A Simple Polysaccharide with Diverse Biological Functions. Acta Biomater. 2014, 10, 1558–1570. [Google Scholar] [CrossRef] [Green Version]
- Day, A.J.; Prestwich, G.D. Hyaluronan-Binding Proteins: Tying Up the Giant. J. Biol. Chem. 2002, 277, 4585–4588. [Google Scholar] [CrossRef] [Green Version]
- Garantziotis, S.; Savani, R.C. Hyaluronan Biology: A Complex Balancing Act of Structure, Function, Location and Context. Matrix Biol. 2019, 78–79, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Ponta, H.; Sherman, L.; Herrlich, P.A. CD44: From Adhesion Molecules to Signalling Regulators. Nat. Rev. Mol. Cell Biol. 2003, 4, 33–45. [Google Scholar] [CrossRef]
- Jackson, D.G. Hyaluronan in the Lymphatics: The Key Role of the Hyaluronan Receptor LYVE-1 in Leucocyte Trafficking. Matrix Biol. 2019, 78–79, 219–235. [Google Scholar] [CrossRef]
- Harris, E.N.; Baker, E. Role of the Hyaluronan Receptor, Stabilin-2/HARE, in Health and Disease. Int. J. Mol. Sci. 2020, 21, 3504. [Google Scholar] [CrossRef] [PubMed]
- Lesley, J.; Gál, I.; Mahoney, D.J.; Cordell, M.R.; Rugg, M.S.; Hyman, R.; Day, A.J.; Mikecz, K. TSG-6 Modulates the Interaction between Hyaluronan and Cell Surface CD44*. J. Biol. Chem. 2004, 279, 25745–25754. [Google Scholar] [CrossRef] [Green Version]
- Miller, C.M.; Donner, A.J.; Blank, E.E.; Egger, A.W.; Kellar, B.M.; Østergaard, M.E.; Seth, P.P.; Harris, E.N. Stabilin-1 and Stabilin-2 Are Specific Receptors for the Cellular Internalization of Phosphorothioate-Modified Antisense Oligonucleotides (ASOs) in the Liver. Nucleic Acids Res. 2016, 44, 2782–2794. [Google Scholar] [CrossRef] [PubMed]
- Spicer, A.P.; Joo, A.; Bowling, R.A. A Hyaluronan Binding Link Protein Gene Family Whose Members Are Physically Linked Adjacent to Chondroitin Sulfate Proteoglycan Core Protein Genes: The Missing Links. J. Biol. Chem. 2003, 278, 21083–21091. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hirakawa, S.; Oohashi, T.; Su, W.-D.; Yoshioka, H.; Murakami, T.; Arata, J.; Ninomiya, Y. The Brain Link Protein-1 (BRAL1): CDNA Cloning, Genomic Structure, and Characterization as a Novel Link Protein Expressed in Adult Brain. Biochem. Biophys. Res. Commun. 2000, 276, 982–989. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Qadri, M.; Almadani, S.; Jay, G.D.; Elsaid, K.A. Role of CD44 in Regulating Toll-like Receptor 2 (TLR2) Activation of Human Macrophages and Downstream Expression of Proinflammatory Cytokines. J. Immunol. 2018, 200, 758–767. [Google Scholar] [CrossRef]
- Oertli, B.; Beck-Schimmer, B.; Fan, X.; Wüthrich, R.P. Mechanisms of Hyaluronan-Induced up-Regulation of ICAM-1 and VCAM-1 Expression by Murine Kidney Tubular Epithelial Cells: Hyaluronan Triggers Cell Adhesion Molecule Expression through a Mechanism Involving Activation of Nuclear Factor-Kappa B and Activating Protein-1. J. Immunol. 1998, 161, 3431–3437. [Google Scholar]
- Bono, P.; Rubin, K.; Higgins, J.M.; Hynes, R.O. Layilin, A Novel Integral Membrane Protein, Is a Hyaluronan Receptor. Mol. Biol. Cell 2001, 12, 891–900. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Harris, E.; Weigel, P. Hyaluronan-Binding Proteoglycans. Biochemistry-Faculty Publications: 2009. Available online: https://digitalcommons.unl.edu/biochemfacpub/66 (accessed on 20 February 2022).
- LeBoeuf, R.D.; Raja, R.H.; Fuller, G.M.; Weigel, P.H. Human Fibrinogen Specifically Binds Hyaluronic Acid. J. Biol. Chem. 1986, 261, 12586–12592. [Google Scholar] [CrossRef]
- Fries, E.; Kaczmarczyk, A. Inter-Alpha-Inhibitor, Hyaluronan and Inflammation. Acta Biochim. Pol. 2003, 50, 735–742. [Google Scholar] [CrossRef]
- Yoshino, Y.; Ishisaka, M.; Tsuruma, K.; Shimazawa, M.; Yoshida, H.; Inoue, S.; Shimoda, M.; Okada, Y.; Hara, H. Distribution and Function of Hyaluronan Binding Protein Involved in Hyaluronan Depolymerization (HYBID, KIAA1199) in the Mouse Central Nervous System. Neuroscience 2017, 347, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Acharya, S.; Rodriguez, I.R.; Moreira, E.F.; Midura, R.J.; Misono, K.; Todres, E.; Hollyfield, J.G. SPACR, A Novel Interphotoreceptor Matrix Glycoprotein in Human Retina That Interacts with Hyaluronan *. J. Biol. Chem. 1998, 273, 31599–31606. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Acharya, S.; Foletta, V.C.; Lee, J.W.; Rayborn, M.E.; Rodriguez, I.R.; Young, W.S.; Hollyfield, J.G. SPACRCAN, a Novel Human Interphotoreceptor Matrix Hyaluronan-Binding Proteoglycan Synthesized by Photoreceptors and Pinealocytes. J. Biol. Chem. 2000, 275, 6945–6955. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Assmann, V.; Jenkinson, D.; Marshall, J.F.; Hart, I.R. The Intracellular Hyaluronan Receptor RHAMM/IHABP Interacts with Microtubules and Actin Filaments. J. Cell. Sci. 1999, 112, 3943–3954. [Google Scholar] [CrossRef] [PubMed]
- Ramakrishna, S.; Suresh, B.; Bae, S.-M.; Ahn, W.-S.; Lim, K.-H.; Baek, K.-H. Hyaluronan Binding Motifs of USP17 and SDS3 Exhibit Anti-Tumor Activity. PLoS ONE 2012, 7, e37772. [Google Scholar] [CrossRef] [Green Version]
- Huang, L.; Grammatikakis, N.; Yoneda, M.; Banerjee, S.D.; Toole, B.P. Molecular Characterization of a Novel Intracellular Hyaluronan-Binding Protein. J. Biol. Chem. 2000, 275, 29829–29839. [Google Scholar] [CrossRef] [Green Version]
- Dalchau, R.; Kirkley, J.; Fabre, J.W. Monoclonal Antibody to a Human Leukocyte-Specific Membrane Glycoprotein Probably Homologous to the Leukocyte-Common (L-C) Antigen of the Rat. Eur. J. Immunol. 1980, 10, 737–744. [Google Scholar] [CrossRef]
- Underhill, C.B.; Green, S.J.; Comoglio, P.M.; Tarone, G. The Hyaluronate Receptor Is Identical to a Glycoprotein of Mr 85,000 (Gp85) as Shown by a Monoclonal Antibody That Interferes with Binding Activity. J. Biol. Chem. 1987, 262, 13142–13146. [Google Scholar] [CrossRef]
- Naor, D.; Sionov, R.V.; Ish-Shalom, D. CD44: Structure, Function and Association with the Malignant Process. In Advances in Cancer Research; Vande Woude, G.F., Klein, G., Eds.; Academic Press: Cambridge, MA, USA, 1997; Volume 71, pp. 241–319. [Google Scholar]
- Senbanjo, L.T.; Chellaiah, M.A. CD44: A Multifunctional Cell Surface Adhesion Receptor Is a Regulator of Progression and Metastasis of Cancer Cells. Front. Cell Dev. Biol. 2017, 5, 18. [Google Scholar] [CrossRef] [Green Version]
- Naor, D. CD44. In Encyclopedia of Immunology, 2nd ed.; Delves, P.J., Ed.; Elsevier: Oxford, UK, 1998; pp. 488–491. ISBN 978-0-12-226765-9. [Google Scholar]
- Zeilstra, J.; Joosten, S.P.J.; van Andel, H.; Tolg, C.; Berns, A.; Snoek, M.; van de Wetering, M.; Spaargaren, M.; Clevers, H.; Pals, S.T. Stem Cell CD44v Isoforms Promote Intestinal Cancer Formation in Apc (Min) Mice Downstream of Wnt Signaling. Oncogene 2014, 33, 665–670. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sneath, R.J.; Mangham, D.C. The Normal Structure and Function of CD44 and Its Role in Neoplasia. Mol. Pathol. 1998, 51, 191–200. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Naor, D.; Wallach-Dayan, S.B.; Zahalka, M.A.; Sionov, R.V. Involvement of CD44, a Molecule with a Thousand Faces, in Cancer Dissemination. Semin. Cancer Biol. 2008, 18, 260–267. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Zuo, X.; Xie, K.; Wei, D. The Role of CD44 and Cancer Stem Cells. Methods Mol. Biol. 2018, 1692, 31–42. [Google Scholar] [CrossRef]
- Iida, N.; Bourguignon, L.Y.W. New CD44 Splice Variants Associated with Human Breast Cancers. J. Cell. Physiol. 1995, 162, 127–133. [Google Scholar] [CrossRef]
- Bourguignon, L.Y.; Gunja-Smith, Z.; Iida, N.; Zhu, H.B.; Young, L.J.; Muller, W.J.; Cardiff, R.D. CD44v(3,8-10) Is Involved in Cytoskeleton-Mediated Tumor Cell Migration and Matrix Metalloproteinase (MMP-9) Association in Metastatic Breast Cancer Cells. J. Cell. Physiol. 1998, 176, 206–215. [Google Scholar] [CrossRef]
- Franzmann, E.J.; Weed, D.T.; Civantos, F.J.; Goodwin, W.J.; Bourguignon, L.Y. A Novel CD44 v3 Isoform Is Involved in Head and Neck Squamous Cell Carcinoma Progression. Otolaryngol. Head Neck Surg. 2001, 124, 426–432. [Google Scholar] [CrossRef]
- Wang, S.J.; Wreesmann, V.B.; Bourguignon, L.Y.W. Association of CD44 V3-Containing Isoforms with Tumor Cell Growth, Migration, Matrix Metalloproteinase Expression, and Lymph Node Metastasis in Head and Neck Cancer. Head Neck 2007, 29, 550–558. [Google Scholar] [CrossRef]
- Ni, J.; Cozzi, P.J.; Hao, J.L.; Beretov, J.; Chang, L.; Duan, W.; Shigdar, S.; Delprado, W.J.; Graham, P.H.; Bucci, J.; et al. CD44 Variant 6 Is Associated with Prostate Cancer Metastasis and Chemo-/Radioresistance. Prostate 2014, 74, 602–617. [Google Scholar] [CrossRef]
- Piselli, P.; Vendetti, S.; Vismara, D.; Cicconi, R.; Poccia, F.; Colizzi, V.; Delpino, A. Different Expression of CD44, ICAM-1, and HSP60 on Primary Tumor and Metastases of a Human Pancreatic Carcinoma Growing in Scid Mice. Anticancer. Res. 2000, 20, 825–831. [Google Scholar] [PubMed]
- Zhou, G.; Chiu, D.; Qin, D.; Niu, L.; Cai, J.; He, L.; Huang, W.; Xu, K. Detection and Clinical Significance of CD44v6 and Integrin-Β1 in Pancreatic Cancer Patients Using a Triplex Real-Time RT-PCR Assay. Appl. Biochem. Biotechnol. 2012, 167, 2257–2268. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Chen, K.; Jiang, P.; Zhang, X.; Li, X.; Li, Z. CD44v/CD44s Expression Patterns Are Associated with the Survival of Pancreatic Carcinoma Patients. Diagn. Pathol. 2014, 9, 79. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Todaro, M.; Gaggianesi, M.; Catalano, V.; Benfante, A.; Iovino, F.; Biffoni, M.; Apuzzo, T.; Sperduti, I.; Volpe, S.; Cocorullo, G.; et al. CD44v6 Is a Marker of Constitutive and Reprogrammed Cancer Stem Cells Driving Colon Cancer Metastasis. Cell Stem Cell 2014, 14, 342–356. [Google Scholar] [CrossRef] [Green Version]
- Mulder, J.W.; Kruyt, P.M.; Sewnath, M.; Oosting, J.; Seldenrijk, C.A.; Weidema, W.F.; Offerhaus, G.J.; Pals, S.T. Colorectal Cancer Prognosis and Expression of Exon-v6-Containing CD44 Proteins. Lancet 1994, 344, 1470–1472. [Google Scholar] [CrossRef]
- Yan, Y.; Zuo, X.; Wei, D. Concise Review: Emerging Role of CD44 in Cancer Stem Cells: A Promising Biomarker and Therapeutic Target. Stem Cells Transl. Med. 2015, 4, 1033–1043. [Google Scholar] [CrossRef]
- Weber, G.F.; Ashkar, S.; Cantor, H. Interaction between CD44 and Osteopontin as a Potential Basis for Metastasis Formation. Proc. Assoc. Am. Physicians 1997, 109, 1–9. [Google Scholar]
- Gupta, A.; Cao, W.; Sadashivaiah, K.; Chen, W.; Schneider, A.; Chellaiah, M.A. Promising Noninvasive Cellular Phenotype in Prostate Cancer Cells Knockdown of Matrix Metalloproteinase 9. Sci. World J. 2013, 2013, 493689. [Google Scholar] [CrossRef]
- Konstantopoulos, K.; Thomas, S.N. Cancer Cells in Transit: The Vascular Interactions of Tumor Cells. Annu. Rev. Biomed. Eng. 2009, 11, 177–202. [Google Scholar] [CrossRef]
- Bourguignon, L.Y.W.; Shiina, M.; Li, J.-J. Hyaluronan–CD44 Interaction Promotes Oncogenic Signaling, MicroRNA Functions, Chemoresistance, and Radiation Resistance in Cancer Stem Cells Leading to Tumor Progression. In Advances in Cancer Research; Simpson, M.A., Heldin, P., Eds.; Hyaluronan Signaling and Turnover; Academic Press: Cambridge, MA, USA, 2014; Volume 123, pp. 255–275. [Google Scholar]
- Chen, C.; Zhao, S.; Karnad, A.; Freeman, J.W. The Biology and Role of CD44 in Cancer Progression: Therapeutic Implications. J. Hematol. Oncol. 2018, 11, 64. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lesley, J.; Hyman, R.; Kincade, P.W. CD44 and Its Interaction with Extracellular Matrix. Adv. Immunol. 1993, 54, 271–335. [Google Scholar] [PubMed]
- Hardwick, C.; Hoare, K.; Owens, R.; Hohn, H.P.; Hook, M.; Moore, D.; Cripps, V.; Austen, L.; Nance, D.M.; Turley, E.A. Molecular Cloning of a Novel Hyaluronan Receptor That Mediates Tumor Cell Motility. J. Cell Biol. 1992, 117, 1343–1350. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Turley, E.A.; Moore, D.; Hayden, L.J. Characterization of Hyaluronate Binding Proteins Isolated from 3T3 and Murine Sarcoma Virus Transformed 3T3 Cells. Biochemistry 1987, 26, 2997–3005. [Google Scholar] [CrossRef]
- Spicer, A.P.; Roller, M.L.; Camper, S.A.; McPherson, J.D.; Wasmuth, J.J.; Hakim, S.; Wang, C.; Turley, E.A.; McDonald, J.A. The Human and Mouse Receptors for Hyaluronan-Mediated Motility, RHAMM, Genes (HMMR) Map to Human Chromosome 5q33.2-Qter and Mouse Chromosome 11. Genomics 1995, 30, 115–117. [Google Scholar] [CrossRef]
- Turley, E.A.; Noble, P.W.; Bourguignon, L.Y.W. Signaling Properties of Hyaluronan Receptors. J. Biol. Chem. 2002, 277, 4589–4592. [Google Scholar] [CrossRef] [Green Version]
- Turley, E.A. Hyaluronan and Cell Locomotion. Cancer Metastasis Rev. 1992, 11, 21–30. [Google Scholar] [CrossRef]
- Lynn, B.D.; Turley, E.A.; Nagy, J.I. Subcellular Distribution, Calmodulin Interaction, and Mitochondrial Association of the Hyaluronan-Binding Protein RHAMM in Rat Brain. J. Neurosci. Res. 2001, 65, 6–16. [Google Scholar] [CrossRef]
- Tolg, C.; McCarthy, J.B.; Yazdani, A.; Turley, E.A. Hyaluronan and RHAMM in Wound Repair and the “Cancerization” of Stromal Tissues. Available online: https://www.hindawi.com/journals/bmri/2014/103923/ (accessed on 9 June 2019).
- Nikitovic, D.; Kouvidi, K.; Kavasi, R.-M.; Berdiaki, A.; Tzanakakis, G.N. Hyaluronan/Hyaladherins-A Promising Axis for Targeted Drug Delivery in Cancer. Curr. Drug Deliv. 2016, 13, 500–511. [Google Scholar] [CrossRef]
- Zhang, S.; Chang, M.C.; Zylka, D.; Turley, S.; Harrison, R.; Turley, E.A. The Hyaluronan Receptor RHAMM Regulates Extracellular-Regulated Kinase. J. Biol. Chem. 1998, 273, 11342–11348. [Google Scholar] [CrossRef] [Green Version]
- Hall, C.L.; Wang, C.; Lange, L.A.; Turley, E.A. Hyaluronan and the Hyaluronan Receptor RHAMM Promote Focal Adhesion Turnover and Transient Tyrosine Kinase Activity. J. Cell Biol. 1994, 126, 575–588. [Google Scholar] [CrossRef] [PubMed]
- Hall, C.L.; Turley, E.A. Hyaluronan: RHAMM Mediated Cell Locomotion and Signaling in Tumorigenesis. J. Neurooncol. 1995, 26, 221–229. [Google Scholar] [CrossRef] [PubMed]
- Hamilton, S.R.; Fard, S.F.; Paiwand, F.F.; Tolg, C.; Veiseh, M.; Wang, C.; McCarthy, J.B.; Bissell, M.J.; Koropatnick, J.; Turley, E.A. The Hyaluronan Receptors CD44 and Rhamm (CD168) Form Complexes with ERK1,2 That Sustain High Basal Motility in Breast Cancer Cells. J. Biol. Chem. 2007, 282, 16667–16680. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Maxwell, C.A.; McCarthy, J.; Turley, E. Cell-Surface and Mitotic-Spindle RHAMM: Moonlighting or Dual Oncogenic Functions? J. Cell Sci. 2008, 121, 925–932. [Google Scholar] [CrossRef] [Green Version]
- Nedvetzki, S.; Gonen, E.; Assayag, N.; Reich, R.; Williams, R.O.; Thurmond, R.L.; Huang, J.-F.; Neudecker, B.A.; Wang, F.-S.; Turley, E.A.; et al. RHAMM, a Receptor for Hyaluronan-Mediated Motility, Compensates for CD44 in Inflamed CD44-Knockout Mice: A Different Interpretation of Redundancy. Proc. Natl. Acad. Sci. USA 2004, 101, 18081–18086. [Google Scholar] [CrossRef] [Green Version]
- Assmann, V.; Marshall, J.F.; Fieber, C.; Hofmann, M.; Hart, I.R. The Human Hyaluronan Receptor RHAMM Is Expressed as an Intracellular Protein in Breast Cancer Cells. J. Cell. Sci. 1998, 111, 1685–1694. [Google Scholar] [CrossRef]
- Tolg, C.; Hamilton, S.R.; Morningstar, L.; Zhang, J.; Zhang, S.; Esguerra, K.V.; Telmer, P.G.; Luyt, L.G.; Harrison, R.; McCarthy, J.B.; et al. RHAMM Promotes Interphase Microtubule Instability and Mitotic Spindle Integrity through MEK1/ERK1/2 Activity. J. Biol. Chem. 2010, 285, 26461–26474. [Google Scholar] [CrossRef] [Green Version]
- Evanko, S.P.; Wight, T.N. Intracellular Localization of Hyaluronan in Proliferating Cells. J. Histochem. Cytochem. 1999, 47, 1331–1341. [Google Scholar] [CrossRef]
- Evanko, S.P.; Parks, W.T.; Wight, T.N. Intracellular Hyaluronan in Arterial Smooth Muscle Cells: Association with Microtubules, RHAMM, and the Mitotic Spindle. J. Histochem. Cytochem. 2004, 52, 1525–1535. [Google Scholar] [CrossRef] [Green Version]
- Telmer, P.G.; Tolg, C.; McCarthy, J.B.; Turley, E.A. How Does a Protein with Dual Mitotic Spindle and Extracellular Matrix Receptor Functions Affect Tumor Susceptibility and Progression? Commun. Integr. Biol. 2011, 4, 182–185. [Google Scholar] [CrossRef]
- Choi, S.; Wang, D.; Chen, X.; Tang, L.H.; Verma, A.; Chen, Z.; Kim, B.J.; Selesner, L.; Robzyk, K.; Zhang, G.; et al. Function and Clinical Relevance of RHAMM Isoforms in Pancreatic Tumor Progression. Mol. Cancer 2019, 18, 92. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Maxwell, C.A.; Rasmussen, E.; Zhan, F.; Keats, J.J.; Adamia, S.; Strachan, E.; Crainie, M.; Walker, R.; Belch, A.R.; Pilarski, L.M.; et al. RHAMM Expression and Isoform Balance Predict Aggressive Disease and Poor Survival in Multiple Myeloma. Blood 2004, 104, 1151–1158. [Google Scholar] [CrossRef]
- Kouvidi, K.; Berdiaki, A.; Nikitovic, D.; Katonis, P.; Afratis, N.; Hascall, V.C.; Karamanos, N.K.; Tzanakakis, G.N. Role of Receptor for Hyaluronic Acid-Mediated Motility (RHAMM) in Low Molecular Weight Hyaluronan (LMWHA)-Mediated Fibrosarcoma Cell Adhesion. J. Biol. Chem. 2011, 286, 38509–38520. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nikitovic, D.; Kouvidi, K.; Karamanos, N.K.; Tzanakakis, G.N. The Roles of Hyaluronan/RHAMM/CD44 and Their Respective Interactions along the Insidious Pathways of Fibrosarcoma Progression. Biomed. Res. Int. 2013, 2013, 929531. [Google Scholar] [CrossRef] [PubMed]
- Mele, V.; Sokol, L.; Kölzer, V.H.; Pfaff, D.; Muraro, M.G.; Keller, I.; Stefan, Z.; Centeno, I.; Terracciano, L.M.; Dawson, H.; et al. The Hyaluronan-Mediated Motility Receptor RHAMM Promotes Growth, Invasiveness and Dissemination of Colorectal Cancer. Oncotarget 2017, 8, 70617–70629. [Google Scholar] [CrossRef] [Green Version]
- Jackson, D.G. The Lymphatics Revisited: New Perspectives from the Hyaluronan Receptor LYVE-1. Trends Cardiovasc. Med. 2003, 13, 1–7. [Google Scholar] [CrossRef]
- Jackson, D.G. Immunological Functions of Hyaluronan and Its Receptors in the Lymphatics. Immunol. Rev. 2009, 230, 216–231. [Google Scholar] [CrossRef]
- Lee, L.K.; Ghorbanian, Y.; Wang, W.; Wang, Y.; Kim, Y.J.; Weissman, I.L.; Inlay, M.A.; Mikkola, H.K.A. LYVE1 Marks the Divergence of Yolk Sac Definitive Hemogenic Endothelium from the Primitive Erythroid Lineage. Cell Rep. 2016, 17, 2286–2298. [Google Scholar] [CrossRef] [Green Version]
- DeLeve, L.D.; Maretti-Mira, A.C. Liver Sinusoidal Endothelial Cell: An Update. Semin. Liver Dis. 2017, 37, 377–387. [Google Scholar] [CrossRef]
- Zheng, M.; Kimura, S.; Nio-Kobayashi, J.; Iwanaga, T. The Selective Distribution of LYVE-1-Expressing Endothelial Cells and Reticular Cells in the Reticulo-Endothelial System (RES). Biomed. Res. 2016, 37, 187–198. [Google Scholar] [CrossRef] [Green Version]
- Lim, H.Y.; Lim, S.Y.; Tan, C.K.; Thiam, C.H.; Goh, C.C.; Carbajo, D.; Chew, S.H.S.; See, P.; Chakarov, S.; Wang, X.N.; et al. Hyaluronan Receptor LYVE-1-Expressing Macrophages Maintain Arterial Tone through Hyaluronan-Mediated Regulation of Smooth Muscle Cell Collagen. Immunity 2018, 49, 326–341.e7. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schledzewski, K.; Falkowski, M.; Moldenhauer, G.; Metharom, P.; Kzhyshkowska, J.; Ganss, R.; Demory, A.; Falkowska-Hansen, B.; Kurzen, H.; Ugurel, S.; et al. Lymphatic Endothelium-Specific Hyaluronan Receptor LYVE-1 Is Expressed by Stabilin-1+, F4/80+, CD11b+ Macrophages in Malignant Tumours and Wound Healing Tissue in Vivo and in Bone Marrow Cultures in Vitro: Implications for the Assessment of Lymphangiogenesis. J. Pathol. 2006, 209, 67–77. [Google Scholar] [CrossRef]
- Jackson, D.G.; Prevo, R.; Clasper, S.; Banerji, S. LYVE-1, the Lymphatic System and Tumor Lymphangiogenesis. Trends Immunol. 2001, 22, 317–321. [Google Scholar] [CrossRef]
- Gale, N.W.; Prevo, R.; Espinosa, J.; Ferguson, D.J.; Dominguez, M.G.; Yancopoulos, G.D.; Thurston, G.; Jackson, D.G. Normal Lymphatic Development and Function in Mice Deficient for the Lymphatic Hyaluronan Receptor LYVE-1. Mol. Cell. Biol. 2007, 27, 595–604. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tammela, T.; Alitalo, K. Lymphangiogenesis: Molecular Mechanisms and Future Promise. Cell 2010, 140, 460–476. [Google Scholar] [CrossRef] [Green Version]
- Banerji, S.; Hide, B.R.S.; James, J.R.; Noble, M.E.M.; Jackson, D.G. Distinctive Properties of the Hyaluronan-Binding Domain in the Lymphatic Endothelial Receptor Lyve-1 and Their Implications for Receptor Function. J. Biol. Chem. 2010, 285, 10724–10735. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Banerji, S.; Ni, J.; Wang, S.X.; Clasper, S.; Su, J.; Tammi, R.; Jones, M.; Jackson, D.G. LYVE-1, a New Homologue of the CD44 Glycoprotein, Is a Lymph-Specific Receptor for Hyaluronan. J. Cell Biol. 1999, 144, 789–801. [Google Scholar] [CrossRef]
- Harris, E.N.; Parry, S.; Sutton-Smith, M.; Pandey, M.S.; Panico, M.; Morris, H.R.; Haslam, S.M.; Dell, A.; Weigel, P.H. N-Glycans on the Link Domain of Human HARE/Stabilin-2 Are Needed for Hyaluronan Binding to Purified Ecto-Domain, but Not for Cellular Endocytosis of Hyaluronan. Glycobiology 2010, 20, 991–1001. [Google Scholar] [CrossRef] [Green Version]
- Harris, E.N.; Weigel, P.H. The Ligand-Binding Profile of HARE: Hyaluronan and Chondroitin Sulfates A, C, and D Bind to Overlapping Sites Distinct from the Sites for Heparin, Acetylated Low-Density Lipoprotein, Dermatan Sulfate, and CS-E. Glycobiology 2008, 18, 638–648. [Google Scholar] [CrossRef] [Green Version]
- Weigel, P.H.; Baggenstoss, B.A. What Is Special about 200 KDa Hyaluronan That Activates Hyaluronan Receptor Signaling? Glycobiology 2017, 27, 868–877. [Google Scholar] [CrossRef] [Green Version]
- Pandey, M.S.; Baggenstoss, B.A.; Washburn, J.; Harris, E.N.; Weigel, P.H. The Hyaluronan Receptor for Endocytosis (HARE) Activates NF-ΚB-Mediated Gene Expression in Response to 40-400-KDa, but Not Smaller or Larger, Hyaluronans. J. Biol. Chem. 2013, 288, 14068–14079. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kyosseva, S.V.; Harris, E.N.; Weigel, P.H. The Hyaluronan Receptor for Endocytosis Mediates Hyaluronan-Dependent Signal Transduction via Extracellular Signal-Regulated Kinases. J. Biol. Chem. 2008, 283, 15047–15055. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kawai, T.; Akira, S. The Role of Pattern-Recognition Receptors in Innate Immunity: Update on Toll-like Receptors. Nat. Immunol. 2010, 11, 373. [Google Scholar] [CrossRef] [PubMed]
- Jin, M.S.; Lee, J.-O. Structures of the Toll-like Receptor Family and Its Ligand Complexes. Immunity 2008, 29, 182–191. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bauer, S.; Müller, T.; Hamm, S. Pattern Recognition by Toll-like Receptors. In Target Pattern Recognition in Innate Immunity; Kishore, U., Ed.; Springer: New York, NY, USA, 2009; pp. 15–34. ISBN 978-1-4419-0901-5. [Google Scholar]
- Hamblin, M.J.; Eberlein, M.; Black, K.; Hallowell, R.; Collins, S.; Chan-Li, Y.; Horton, M.R. Lovastatin Inhibits Low Molecular Weight Hyaluronan Induced Chemokine Expression via LFA-1 and Decreases Bleomycin-Induced Pulmonary Fibrosis. Int. J. Biomed. Sci. 2014, 10, 146–157. [Google Scholar] [PubMed]
- Campo, G.M.; Avenoso, A.; D’Ascola, A.; Prestipino, V.; Scuruchi, M.; Nastasi, G.; Calatroni, A.; Campo, S. Hyaluronan Differently Modulates TLR-4 and the Inflammatory Response in Mouse Chondrocytes. BioFactors 2012, 38, 69–76. [Google Scholar] [CrossRef]
- Ebid, R.; Lichtnekert, J.; Anders, H.-J. Hyaluronan Is Not a Ligand but a Regulator of Toll-Like Receptor Signaling in Mesangial Cells: Role of Extracellular Matrix in Innate Immunity. ISRN Nephrol. 2014, 2014, 714081. [Google Scholar] [CrossRef] [Green Version]
- Bourguignon, L.Y.W.; Wong, G.; Earle, C.A.; Xia, W. Interaction of Low Molecular Weight Hyaluronan with CD44 and Toll-like Receptors Promotes the Actin Filament-Associated Protein 110-Actin Binding and MyD88-NFκB Signaling Leading to Proinflammatory Cytokine/Chemokine Production and Breast Tumor Invasion. Cytoskeleton 2011, 68, 671–693. [Google Scholar] [CrossRef] [Green Version]
- Yoshida, H.; Nagaoka, A.; Kusaka-Kikushima, A.; Tobiishi, M.; Kawabata, K.; Sayo, T.; Sakai, S.; Sugiyama, Y.; Enomoto, H.; Okada, Y.; et al. KIAA1199, a Deafness Gene of Unknown Function, is a New Hyaluronan Binding Protein Involved in Hyaluronan Depolymerization. Proc. Natl. Acad. Sci. USA 2013, 110, 5612–5617. [Google Scholar] [CrossRef] [Green Version]
- Deroyer, C.; Charlier, E.; Neuville, S.; Malaise, O.; Gillet, P.; Kurth, W.; Chariot, A.; Malaise, M.; de Seny, D. CEMIP (KIAA1199) Induces a Fibrosis-like Process in Osteoarthritic Chondrocytes. Cell Death Dis. 2019, 10, 103. [Google Scholar] [CrossRef] [Green Version]
- Kanse, S.M.; Parahuleva, M.; Muhl, L.; Kemkes-Matthes, B.; Sedding, D.; Preissner, K.T. Factor VII-Activating Protease (FSAP): Vascular Functions and Role in Atherosclerosis. Thromb. Haemost. 2008, 99, 286–289. [Google Scholar] [CrossRef] [PubMed]
- Mambetsariev, N.; Mirzapoiazova, T.; Mambetsariev, B.; Sammani, S.; Lennon, F.E.; Garcia, J.G.N.; Singleton, P.A. HABP2 Is a Novel Regulator of Vascular Integrity. Arter. Thromb. Vasc. Biol. 2010, 30, 483–490. [Google Scholar] [CrossRef] [PubMed]
- Wygrecka, M.; Markart, P.; Fink, L.; Guenther, A.; Preissner, K.T. Raised Protein Levels and Altered Cellular Expression of Factor VII Activating Protease (FSAP) in the Lungs of Patients with Acute Respiratory Distress Syndrome (ARDS). Thorax 2007, 62, 880–888. [Google Scholar] [CrossRef] [Green Version]
- Mirzapoiazova, T.; Mambetsariev, N.; Lennon, F.E.; Mambetsariev, B.; Berlind, J.E.; Salgia, R.; Singleton, P.A. HABP2 Is a Novel Regulator of Hyaluronan-Mediated Human Lung Cancer Progression. Front. Oncol. 2015, 5, 164. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jaffray, D.A.; Gospodarowicz, M.K. Radiation Therapy for Cancer. In Cancer: Disease Control Priorities, 3rd ed.; Gelband, H., Jha, P., Sankaranarayanan, R., Horton, S., Eds.; The International Bank for Reconstruction and Development/The World Bank: Washington, DC, USA, 2015; Volume 3, ISBN 978-1-4648-0349-9. [Google Scholar]
- Lukoff, J.; Olmos, J. Minimizing Medical Radiation Exposure by Incorporating a New Radiation “Vital Sign” into the Electronic Medical Record: Quality of Care and Patient Safety. Perm. J. 2017, 21, 1–8. [Google Scholar] [CrossRef] [Green Version]
- Santivasi, W.L.; Xia, F. Ionizing Radiation-Induced DNA Damage, Response, and Repair. Antioxid. Redox Signal. 2013, 21, 251–259. [Google Scholar] [CrossRef]
- Le Caër, S. Water Radiolysis: Influence of Oxide Surfaces on H2 Production under Ionizing Radiation. Water 2011, 3, 235–253. [Google Scholar] [CrossRef] [Green Version]
- Rosen, E.M.; Day, R.; Singh, V.K. New Approaches to Radiation Protection. Front. Oncol. 2015, 4, 381. [Google Scholar] [CrossRef] [Green Version]
- Wirsdörfer, F.; Jendrossek, V. Modeling DNA Damage-Induced Pneumopathy in Mice: Insight from Danger Signaling Cascades. Radiat. Oncol. 2017, 12, 142. [Google Scholar] [CrossRef]
- Kirsch, D.G.; Diehn, M.; Kesarwala, A.H.; Maity, A.; Morgan, M.A.; Schwarz, J.K.; Bristow, R.; Demaria, S.; Eke, I.; Griffin, R.J.; et al. The Future of Radiobiology. J. Natl. Cancer Inst. 2018, 110, 329–340. [Google Scholar] [CrossRef]
- Mothersill, C. Are Epigenetic Mechanisms Involved in Radiation-Induced Bystander Effects? Front. Genet. 2012, 3, 74. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mukherjee, S.; Chakraborty, A. Radiation-Induced Bystander Phenomenon: Insight and Implications in Radiotherapy. Int. J. Radiat. Biol. 2019, 95, 243–263. [Google Scholar] [CrossRef]
- Šoltés, L.; Mendichi, R.; Kogan, G.; Schiller, J.; Stankovská, M.; Arnhold, J. Degradative Action of Reactive Oxygen Species on Hyaluronan. Biomacromolecules 2006, 7, 659–668. [Google Scholar] [CrossRef] [PubMed]
- Parsons, B.J. Free Radical Studies of Components of the Extracellular Matrix: Contributions to Protection of Biomolecules and Biomaterials from Sterilising Doses of Ionising Radiation. Cell Tissue Bank 2017, 19, 201–213. [Google Scholar] [CrossRef] [Green Version]
- Schoenberg, M.D.; Brooks, R.E.; Hall, J.J.; Schneiderman, H. Effect of X-Irradiation on the Hyaluronïdasehyaluronic Acid System. Arch. Biochem. 1951, 30, 333–340. [Google Scholar] [PubMed]
- Caputo, A. Depolymerization of Hyaluronic Acid by X-Rays. Nature 1957, 179, 1133–1134. [Google Scholar] [CrossRef]
- Daar, E.; King, L.; Nisbet, A.; Thorpe, R.B.; Bradley, D.A. Viscosity Changes in Hyaluronic Acid: Irradiation and Rheological Studies. Appl. Radiat. Isot. 2010, 68, 746–750. [Google Scholar] [CrossRef]
- Al-Assaf, S.; Navaratnam, S.; Parsons, B.J.; Phillips, G.O. Chain Scission of Hyaluronan by Carbonate and Dichloride Radical Anions: Potential Reactive Oxidative Species in Inflammation? Free. Radic. Biol. Med. 2006, 40, 2018–2027. [Google Scholar] [CrossRef]
- Deeble, D.J.; Phillips, G.O.; Bothe, E.; Schuchmann, H.-P.; von Sonntag, C. The Radiation-Induced Degradation of Hyaluronic Acid. International Journal of Radiation Applications and Instrumentation. Part C. Radiat. Phys. Chem. 1991, 37, 115–118. [Google Scholar] [CrossRef]
- Al-Assaf, S.; Meadows, J.; Phillips, G.O.; Williams, P.A.; Parsons, B.J. The Effect of Hydroxyl Radicals on the Rheological Performance of Hylan and Hyaluronan. Int. J. Biol. Macromol. 2000, 27, 337–348. [Google Scholar] [CrossRef]
- Ahmad, A.F.; Mohd, H.M.K.; bin Ayob, M.T.M.; Rosli, N.R.A.M.; Mohamed, F.; Radiman, S.; Rahman, I.A. Effect of Gamma Irradiation on Hyaluronic Acid and Dipalmitoylphosphatidylcholine (DPPC). Interaction 2014, 1614, 69–73. [Google Scholar] [CrossRef]
- Huang, Y.-C.; Huang, K.-Y.; Lew, W.-Z.; Fan, K.-H.; Chang, W.-J.; Huang, H.-M. Gamma-Irradiation-Prepared Low Molecular Weight Hyaluronic Acid Promotes Skin Wound Healing. Polymers 2019, 11, 1214. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bardaweel, S.K.; Gul, M.; Alzweiri, M.; Ishaqat, A.; ALSalamat, H.A.; Bashatwah, R.M. Reactive Oxygen Species: The Dual Role in Physiological and Pathological Conditions of the Human Body. Eurasian J. Med. 2018, 50, 193–201. [Google Scholar] [CrossRef] [PubMed]
- Volpi, N.; Schiller, J.; Stern, R.; Soltés, L. Role, Metabolism, Chemical Modifications and Applications of Hyaluronan. Curr. Med. Chem. 2009, 16, 1718–1745. [Google Scholar] [CrossRef] [PubMed]
- Kumari, S.; Badana, A.K.; Mohan, G.M.; Shailender, G.; Malla, R. Reactive Oxygen Species: A Key Constituent in Cancer Survival. Biomark. Insights 2018, 13, 1177271918755391. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Azzam, E.I. What Does Radiation Biology Tell Us about Potential Health Effects at Low Dose and Low Dose Rates. J. Radiol. Prot. 2019, 39, 4. [Google Scholar] [CrossRef]
- Lapčík, L.; Schurz, J. Photochemical Degradation of Hyaluronic Acid by Singlet Oxygen. Colloid Polym. Sci. 1991, 269, 633–635. [Google Scholar] [CrossRef]
- Caspersen, M.B.; Roubroeks, J.P.; Qun, L.; Shan, H.; Fogh, J.; RuiDong, Z.; Tømmeraas, K. Thermal Degradation and Stability of Sodium Hyaluronate in Solid State. Carbohydr. Polym. 2014, 107, 25–30. [Google Scholar] [CrossRef]
- Šoltés, L.; Valachová, K.; Mendichi, R.; Kogan, G.; Arnhold, J.; Gemeiner, P. Solution Properties of High-Molar-Mass Hyaluronans: The Biopolymer Degradation by Ascorbate. Carbohydr. Res. 2007, 342, 1071–1077. [Google Scholar] [CrossRef]
- Chen, H.; Qin, J.; Hu, Y. Efficient Degradation of High-Molecular-Weight Hyaluronic Acid by a Combination of Ultrasound, Hydrogen Peroxide, and Copper Ion. Molecules 2019, 24, 617. [Google Scholar] [CrossRef] [Green Version]
- Duan, J.; Kasper, D.L. Oxidative Depolymerization of Polysaccharides by Reactive Oxygen/Nitrogen Species. Glycobiology 2011, 21, 401–409. [Google Scholar] [CrossRef]
- Kennett, E.C.; Davies, M.J. Degradation of Matrix Glycosaminoglycans by Peroxynitrite/Peroxynitrous Acid: Evidence for a Hydroxyl-Radical-like Mechanism. Free Radic. Biol. Med. 2007, 42, 1278–1289. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Rosenfeld, L.; Vilar, R.E.; Cowman, M.K. Degradation of Hyaluronan by Peroxynitrite. Arch. Biochem. Biophys. 1997, 341, 245–250. [Google Scholar] [CrossRef] [PubMed]
- Riehl, T.E.; Foster, L.; Stenson, W.F. Hyaluronic Acid Is Radioprotective in the Intestine through a TLR4 and COX-2-Mediated Mechanism. Am. J. Physiol. Gastrointest. Liver Physiol. 2012, 302, G309–G316. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ratikan, J.A.; Micewicz, E.D.; Xie, M.W.; Schaue, D. Radiation Takes Its Toll. Cancer Lett. 2015, 368, 238–245. [Google Scholar] [CrossRef] [Green Version]
- Hanson, W.R.; Houseman, K.A.; Collins, P.W. Radiation Protection in Vivo by Prostaglandins and Related Compounds of the Arachidonic Acid Cascade. Pharmacol. Ther. 1988, 39, 347–356. [Google Scholar] [CrossRef]
- Macià i Garau, M.; Lucas Calduch, A.; López, E.C. Radiobiology of the Acute Radiation Syndrome. Rep. Pract. Oncol. Radiother. 2011, 16, 123–130. [Google Scholar] [CrossRef] [Green Version]
- Kiang, J.G.; Olabisi, A.O. Radiation: A Poly-Traumatic Hit Leading to Multi-Organ Injury. Cell Biosci. 2019, 9, 25. [Google Scholar] [CrossRef] [Green Version]
- Johnson, P.; Arif, A.A.; Lee-Sayer, S.S.M.; Dong, Y. Hyaluronan and Its Interactions with Immune Cells in the Healthy and Inflamed Lung. Front. Immunol. 2018, 9, 2787. [Google Scholar] [CrossRef]
- Lennon, F.E.; Singleton, P.A. Role of Hyaluronan and Hyaluronan-Binding Proteins in Lung Pathobiology. Am. J. Physiol. Lung Cell Mol. Physiol. 2011, 301, L137–L147. [Google Scholar] [CrossRef] [Green Version]
- Garantziotis, S.; Brezina, M.; Castelnuovo, P.; Drago, L. The Role of Hyaluronan in the Pathobiology and Treatment of Respiratory Disease. Am. J. Physiol. Lung Cell Mol. Physiol. 2016, 310, L785–L795. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lierova, A.; Jelicova, M.; Nemcova, M.; Proksova, M.; Pejchal, J.; Zarybnicka, L.; Sinkorova, Z. Cytokines and Radiation-Induced Pulmonary Injuries. J. Radiat. Res. 2018, 59, 709–753. [Google Scholar] [CrossRef] [PubMed]
- Kliment, C.R.; Oury, T.D. Oxidative Stress, Extracellular Matrix Targets, and Idiopathic Pulmonary Fibrosis. Free. Radic. Biol. Med. 2010, 49, 707–717. [Google Scholar] [CrossRef] [PubMed]
- Gao, F.; Koenitzer, J.R.; Tobolewski, J.M.; Jiang, D.; Liang, J.; Noble, P.W.; Oury, T.D. Extracellular Superoxide Dismutase Inhibits Inflammation by Preventing Oxidative Fragmentation of Hyaluronan. J. Biol. Chem. 2008, 283, 6058–6066. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zelko, I.N.; Folz, R.J. Extracellular Superoxide Dismutase Attenuates Release of Pulmonary Hyaluronan from the Extracellular Matrix Following Bleomycin Exposure. FEBS Lett. 2010, 584, 2947–2952. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kang, S.K.; Rabbani, Z.N.; Folz, R.J.; Golson, M.L.; Huang, H.; Yu, D.; Samulski, T.S.; Dewhirst, M.W.; Anscher, M.S.; Vujaskovic, Z. Overexpression of Extracellular Superoxide Dismutase Protects Mice from Radiation-Induced Lung Injury. Int. J. Radiat. Oncol. Biol. Phys. 2003, 57, 1056–1066. [Google Scholar] [CrossRef]
- Wei, L.; Zhang, J.; Yang, Z.-L.; You, H. Extracellular Superoxide Dismutase Increased the Therapeutic Potential of Human Mesenchymal Stromal Cells in Radiation Pulmonary Fibrosis. Cytotherapy 2017, 19, 586–602. [Google Scholar] [CrossRef]
- Rabbani, Z.N.; Anscher, M.S.; Folz, R.J.; Archer, E.; Huang, H.; Chen, L.; Golson, M.L.; Samulski, T.S.; Dewhirst, M.W.; Vujaskovic, Z. Overexpression of Extracellular Superoxide Dismutase Reduces Acute Radiation Induced Lung Toxicity. BMC Cancer 2005, 5, 59. [Google Scholar] [CrossRef] [Green Version]
- Rosenbaum, D.; Peric, S.; Holecek, M.; Ward, H.E. Hyaluronan in Radiation-Induced Lung Disease in the Rat. Radiat. Res. 1997, 147, 585–591. [Google Scholar] [CrossRef]
- Nilsson, K.; Henriksson, R.; Hellström, S.; Tengblad, A.; Bjermer, L. Hyaluronan Reflects the Pre-Fibrotic Inflammation in Irradiated Rat Lung: Concomitant Analysis of Parenchymal Tissues and Bronchoalveolar Lavage. Int. J. Radiat. Biol. 1990, 58, 519–530. [Google Scholar] [CrossRef]
- Li, Y.; Rahmanian, M.; Widström, C.; Lepperdinger, G.; Frost, G.I.; Heldin, P. Irradiation-Induced Expression of Hyaluronan (HA) Synthase 2 and Hyaluronidase 2 Genes in Rat Lung Tissue Accompanies Active Turnover of HA and Induction of Types I and III Collagen Gene Expression. Am. J. Respir. Cell Mol. Biol. 2000, 23, 411–418. [Google Scholar] [CrossRef] [PubMed]
- Shen, Y.N.; Shin, H.-J.; Joo, H.-Y.; Park, E.-R.; Kim, S.-H.; Hwang, S.-G.; Park, S.J.; Kim, C.-H.; Lee, K.-H. Inhibition of HAS2 Induction Enhances the Radiosensitivity of Cancer Cells via Persistent DNA Damage. Biochem. Biophys. Res. Commun. 2014, 443, 796–801. [Google Scholar] [CrossRef]
- Kim, J.H.; Jenrow, K.A.; Brown, S.L. Mechanisms of Radiation-Induced Normal Tissue Toxicity and Implications for Future Clinical Trials. Radiat. Oncol. J. 2014, 32, 103–115. [Google Scholar] [CrossRef] [Green Version]
- Li, Y.; Jiang, D.; Liang, J.; Meltzer, E.B.; Gray, A.; Miura, R.; Wogensen, L.; Yamaguchi, Y.; Noble, P.W. Severe Lung Fibrosis Requires an Invasive Fibroblast Phenotype Regulated by Hyaluronan and CD44. J. Exp. Med. 2011, 208, 1459–1471. [Google Scholar] [CrossRef] [Green Version]
- Colgan, S.P.; Eltzschig, H.K.; Eckle, T.; Thompson, L.F. Physiological Roles for Ecto-5′-Nucleotidase (CD73). Purinergic Signal. 2006, 2, 351. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wirsdörfer, F.; de Leve, S.; Cappuccini, F.; Eldh, T.; Meyer, A.V.; Gau, E.; Thompson, L.F.; Chen, N.-Y.; Karmouty-Quintana, H.; Fischer, U.; et al. Extracellular Adenosine Production by Ecto-5′-Nucleotidase (CD73). Enhanc. Radiat. Induc. Lung Fibrosis. Cancer Res. 2016, 76, 3045–3056. [Google Scholar] [CrossRef] [Green Version]
- De Leve, S.; Wirsdörfer, F.; Jendrossek, V. Targeting the Immunomodulatory CD73/Adenosine System to Improve the Therapeutic Gain of Radiotherapy. Front. Immunol. 2019, 10, 698. [Google Scholar] [CrossRef] [Green Version]
- De Leve, S.; Wirsdörfer, F.; Cappuccini, F.; Schütze, A.; Meyer, A.V.; Röck, K.; Thompson, L.F.; Fischer, J.W.; Stuschke, M.; Jendrossek, V. Loss of CD73 Prevents Accumulation of Alternatively Activated Macrophages and the Formation of Prefibrotic Macrophage Clusters in Irradiated Lungs. FASEB J. 2017, 31, 2869–2880. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sohr, S.; Engeland, K. RHAMM Is Differentially Expressed in the Cell Cycle and Downregulated by the Tumor Suppressor P53. Cell Cycle 2008, 7, 3448–3460. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fei, P.; El-Deiry, W.S. P53 and Radiation Responses. Oncogene 2003, 22, 5774. [Google Scholar] [CrossRef] [Green Version]
- Lee, C.-L.; Blum, J.M.; Kirsch, D.G. Role of P53 in Regulating Tissue Response to Radiation by Mechanisms Independent of Apoptosis. Transl. Cancer Res. 2013, 2, 412–421. [Google Scholar] [PubMed]
- Uddin, M.A.; Barabutis, N. P53 in the Impaired Lungs. DNA Repair 2020, 95, 102952. [Google Scholar] [CrossRef] [PubMed]
- Wirostko, B.; Mann, B.K.; Williams, D.L.; Prestwich, G.D. Ophthalmic Uses of a Thiol-Modified Hyaluronan-Based Hydrogel. Adv. Wound Care 2014, 3, 708–716. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Widjaja, L.K.; Bora, M.; Chan, P.N.P.H.; Lipik, V.; Wong, T.T.L.; Venkatraman, S.S. Hyaluronic Acid-Based Nanocomposite Hydrogels for Ocular Drug Delivery Applications. J. Biomed. Mater. Res. A 2014, 102, 3056–3065. [Google Scholar] [CrossRef] [PubMed]
- Lee, D.; Lu, Q.; Sommerfeld, S.D.; Chan, A.; Menon, N.G.; Schmidt, T.A.; Elisseeff, J.H.; Singh, A. Targeted Delivery of Hyaluronic Acid to the Ocular Surface by a Polymer-Peptide Conjugate System for Dry Eye Disease. Acta Biomater. 2017, 55, 163–171. [Google Scholar] [CrossRef] [PubMed]
- Bowman, S.; Awad, M.E.; Hamrick, M.W.; Hunter, M.; Fulzele, S. Recent Advances in Hyaluronic Acid Based Therapy for Osteoarthritis. Clin. Transl. Med. 2018, 7, 6. [Google Scholar] [CrossRef] [Green Version]
- Tashiro, T.; Seino, S.; Sato, T.; Matsuoka, R.; Masuda, Y.; Fukui, N. Oral Administration of Polymer Hyaluronic Acid Alleviates Symptoms of Knee Osteoarthritis: A Double-Blind, Placebo-Controlled Study over a 12-Month Period. Sci. World J. 2012, 2012, 167928. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kaya, S.; Schmidl, D.; Schmetterer, L.; Witkowska, K.J.; Unterhuber, A.; Aranha Dos Santos, V.; Baar, C.; Garhöfer, G.; Werkmeister, R.M. Effect of Hyaluronic Acid on Tear Film Thickness as Assessed with Ultra-High Resolution Optical Coherence Tomography. Acta Ophthalmol. 2015, 93, 439–443. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sionkowska, A.; Kaczmarek, B.; Michalska, M.; Lewandowska, K.; Grabska, S. Preparation and Characterization of Collagen/Chitosan/Hyaluronic Acid Thin Films for Application in Hair Care Cosmetics. Pure Appl. Chem. 2017, 89, 1829–1839. [Google Scholar] [CrossRef]
- Kašparová, J.; Arnoldová, K.; Korecká, L.; Česlová, L. Determination of Hyaluronic Acid in Pharmaceutical Products by Spectrophotometry and HPLC Coupled to Fluorescence or Mass Spectrometric Detection; Faculty of Chemical Technology: Pardubice, Czech Republic, 2018. [Google Scholar]
- Cartier, H.; Hedén, P.; Delmar, H.; Bergentz, P.; Skoglund, C.; Edwartz, C.; Norberg, M.; Kestemont, P. Repeated Full-Face Aesthetic Combination Treatment with AbobotulinumtoxinA, Hyaluronic Acid Filler, and Skin-Boosting Hyaluronic Acid After Monotherapy with AbobotulinumtoxinA or Hyaluronic Acid Filler. Derm. Surg. 2020, 46, 475–482. [Google Scholar] [CrossRef]
- Al-Sibani, M.; Al-Harrasi, A.; Neubert, R.H.H. Effect of Hyaluronic Acid Initial Concentration on Cross-Linking Efficiency of Hyaluronic Acid-Based Hydrogels Used in Biomedical and Cosmetic Applications. Pharmazie 2017, 72, 81–86. [Google Scholar] [CrossRef] [PubMed]
- Rohrich, R.J.; Ghavami, A.; Crosby, M.A. The Role of Hyaluronic Acid Fillers (Restylane) in Facial Cosmetic Surgery: Review and Technical Considerations. Plast. Reconstr. Surg. 2007, 120, 41S–54S. [Google Scholar] [CrossRef] [PubMed]
- Urdiales-Gálvez, F.; Martín-Sánchez, S.; Maíz-Jiménez, M.; Castellano-Miralla, A.; Lionetti-Leone, L. Concomitant Use of Hyaluronic Acid and Laser in Facial Rejuvenation. Aesthetic Plast. Surg. 2019, 43, 1061–1070. [Google Scholar] [CrossRef] [Green Version]
- Rzany, B.; Cartier, H.; Kestemont, P.; Trevidic, P.; Sattler, G.; Kerrouche, N.; Dhuin, J.-C.; Ma, Y.M. Full-Face Rejuvenation Using a Range of Hyaluronic Acid Fillers: Efficacy, Safety, and Patient Satisfaction over 6 Months. Derm. Surg. 2012, 38, 1153–1161. [Google Scholar] [CrossRef] [PubMed]
- Benzaquen, M.; Fongue, J.; Pauly, V.; Collet-Villette, A.-M. Laser-Assisted Hyaluronic Acid Delivery by Fractional Carbon Dioxide Laser in Facial Skin Remodeling: A Prospective Randomized Split-Face Study in France. Lasers Surg. Med. 2021, 53, 1166–1172. [Google Scholar] [CrossRef]
- Wu, M.; Cao, M.; He, Y.; Liu, Y.; Yang, C.; Du, Y.; Wang, W.; Gao, F. A Novel Role of Low Molecular Weight Hyaluronan in Breast Cancer Metastasis. FASEB J. 2015, 29, 1290–1298. [Google Scholar] [CrossRef]
- Ossipov, D.A. Nanostructured Hyaluronic Acid-Based Materials for Active Delivery to Cancer. Expert Opin. Drug Deliv. 2010, 7, 681–703. [Google Scholar] [CrossRef]
- Hemshekhar, M.; Thushara, R.M.; Chandranayaka, S.; Sherman, L.S.; Kemparaju, K.; Girish, K.S. Emerging Roles of Hyaluronic Acid Bioscaffolds in Tissue Engineering and Regenerative Medicine. Int. J. Biol. Macromol. 2016, 86, 917–928. [Google Scholar] [CrossRef]
- Hussain, Z.; Thu, H.E.; Katas, H.; Bukhari, S.N.A. Hyaluronic Acid-Based Biomaterials: A Versatile and Smart Approach to Tissue Regeneration and Treating Traumatic, Surgical, and Chronic Wounds. Polym. Rev. 2017, 57, 594–630. [Google Scholar] [CrossRef]
- Margolis, R.K.; Crockett, C.P.; Kiang, W.-L.; Margolis, R.U. Glycosaminoglycans and Glycoproteins Associated with Rat Brain Nuclei. Biochim. Et Biophys. Acta (BBA) Gen. Subj. 1976, 451, 465–469. [Google Scholar] [CrossRef]
- Stein, G.S.; Roberts, R.M.; Davis, J.L.; Head, W.J.; Stein, J.L.; Thrall, C.L.; Veen, J.V.; Welch, D.W. Are Glycoproteins and Glycosaminoglycans Components of the Eukaryotic Genome? Nature 1975, 258, 639. [Google Scholar] [CrossRef] [PubMed]
- Skandalis, S.S.; Karalis, T.; Heldin, P. Intracellular Hyaluronan: Importance for Cellular Functions. Semin. Cancer Biol. 2019, 62, 20–30. [Google Scholar] [CrossRef]
- Hascall, V.C.; Majors, A.K.; De La Motte, C.A.; Evanko, S.P.; Wang, A.; Drazba, J.A.; Strong, S.A.; Wight, T.N. Intracellular Hyaluronan: A New Frontier for Inflammation? Biochim. Biophys. Acta 2004, 1673, 3–12. [Google Scholar] [CrossRef] [PubMed]
- Wang, A.; Hascall, V.C. Hyaluronan Structures Synthesized by Rat Mesangial Cells in Response to Hyperglycemia Induce Monocyte Adhesion. J. Biol. Chem. 2004, 279, 10279–10285. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nagy, N.; Sunkari, V.G.; Kaber, G.; Hasbun, S.; Lam, D.N.; Speake, C.; Sanda, S.; McLaughlin, T.L.; Wight, T.N.; Long, S.R.; et al. Hyaluronan Levels Are Increased Systemically in Human Type 2 but Not Type 1 Diabetes Independently of Glycemic Control. Matrix Biol. 2019, 80, 46–58. [Google Scholar] [CrossRef] [PubMed]
- Pu, Y.; Cai, F.; Wang, D.; Wang, J.-X.; Chen, J.-F. Colloidal Synthesis of Semiconductor Quantum Dots toward Large-Scale Production: A Review. Ind. Eng. Chem. Res. 2018, 57, 1790–1802. [Google Scholar] [CrossRef]
- Hardman, R. A Toxicologic Review of Quantum Dots: Toxicity Depends on Physicochemical and Environmental Factors. Env. Health Perspect. 2006, 114, 165–172. [Google Scholar] [CrossRef] [PubMed]
- D’Amico, M.; Fiorica, C.; Palumbo, F.S.; Militello, V.; Leone, M.; Dubertret, B.; Pitarresi, G.; Giammona, G. Uptake of Silica Covered Quantum Dots into Living Cells: Long Term Vitality and Morphology Study on Hyaluronic Acid Biomaterials. Mater Sci. Eng. C Mater Biol. Appl. 2016, 67, 231–236. [Google Scholar] [CrossRef]
- Wang, H.; Sun, H.; Wei, H.; Xi, P.; Nie, S.; Ren, Q. Biocompatible Hyaluronic Acid Polymer-Coated Quantum Dots for CD44+ Cancer Cell-Targeted Imaging. J. Nanopart. Res. 2014, 16, 2621. [Google Scholar] [CrossRef]
- Choi, K.Y.; Min, K.H.; Yoon, H.Y.; Kim, K.; Park, J.H.; Kwon, I.C.; Choi, K.; Jeong, S.Y. PEGylation of Hyaluronic Acid Nanoparticles Improves Tumor Targetability in Vivo. Biomaterials 2011, 32, 1880–1889. [Google Scholar] [CrossRef]
- Lee, H.; Lee, K.; Kim, I.K.; Park, T.G. Synthesis, Characterization, and in Vivo Diagnostic Applications of Hyaluronic Acid Immobilized Gold Nanoprobes. Biomaterials 2008, 29, 4709–4718. [Google Scholar] [CrossRef] [PubMed]
- Russo, M.; Bevilacqua, P.; Netti, P.A.; Torino, E. A Microfluidic Platform to Design Crosslinked Hyaluronic Acid Nanoparticles (CHANPs) for Enhanced MRI. Sci. Rep. 2016, 6, 37906. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.; Lee, H.; Kim, Y.B.; Kim, J.; Hyeon, T.; Park, H.; Messersmith, P.B.; Park, T.G. Bioinspired Surface Immobilization of Hyaluronic Acid on Monodisperse Magnetite Nanocrystals for Targeted Cancer Imaging. Adv. Mater. 2008, 20, 4154–4157. [Google Scholar] [CrossRef]
- Rao, N.V.; Rho, J.G.; Um, W.; Ek, P.K.; Nguyen, V.Q.; Oh, B.H.; Kim, W.; Park, J.H. Hyaluronic Acid Nanoparticles as Nanomedicine for Treatment of Inflammatory Diseases. Pharmaceutics 2020, 12, 931. [Google Scholar] [CrossRef] [PubMed]
- Holubova, L.; Korecka, L.; Podzimek, S.; Moravcova, V.; Rotkova, J.; Ehlova, T.; Velebny, V.; Bilkova, Z. Enhanced Multiparametric Hyaluronan Degradation for Production of Molar-Mass-Defined Fragments. Carbohydr. Polym. 2014, 112, 271–276. [Google Scholar] [CrossRef]
- Lierova, A.; Kasparova, J.; Pejchal, J.; Kubelkova, K.; Jelicova, M.; Palarcik, J.; Korecka, L.; Bilkova, Z.; Sinkorova, Z. Attenuation of Radiation-Induced Lung Injury by Hyaluronic Acid Nanoparticles. Front. Pharm. 2020, 11, 1199. [Google Scholar] [CrossRef]
- Malfanti, A.; Catania, G.; Degros, Q.; Wang, M.; Bausart, M.; Préat, V. Design of Bio-Responsive Hyaluronic Acid–Doxorubicin Conjugates for the Local Treatment of Glioblastoma. Pharmaceutics 2022, 14, 124. [Google Scholar] [CrossRef]
- Kim, H.; Shin, M.; Han, S.; Kwon, W.; Hahn, S.K. Hyaluronic Acid Derivatives for Translational Medicines. Biomacromolecules 2019, 20, 2889–2903. [Google Scholar] [CrossRef]
- Choi, K.Y.; Han, H.S.; Lee, E.S.; Shin, J.M.; Almquist, B.D.; Lee, D.S.; Park, J.H. Hyaluronic Acid–Based Activatable Nanomaterials for Stimuli-Responsive Imaging and Therapeutics: Beyond CD44-Mediated Drug Delivery. Adv. Mater. 2019, 31, 1803549. [Google Scholar] [CrossRef]

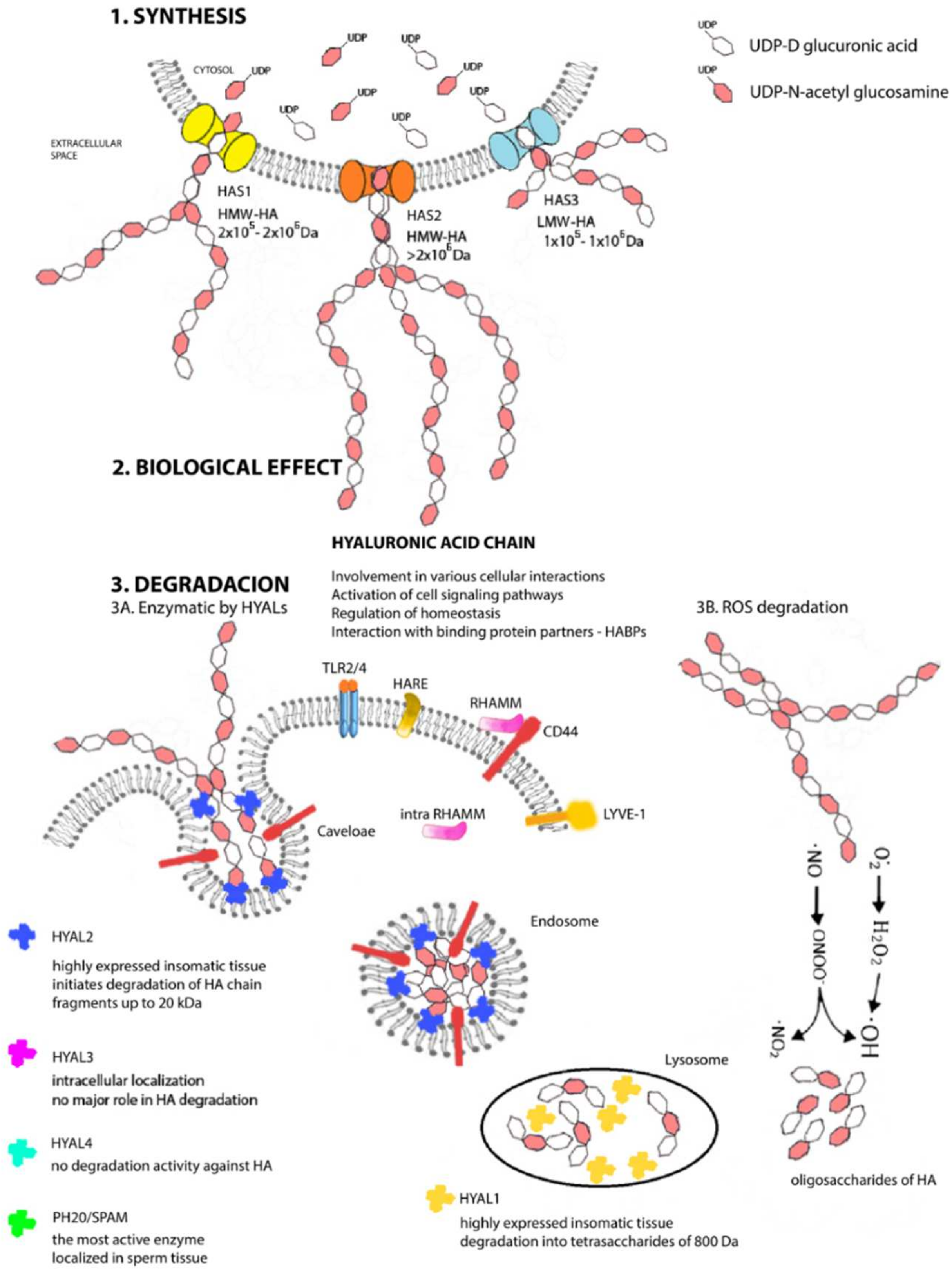
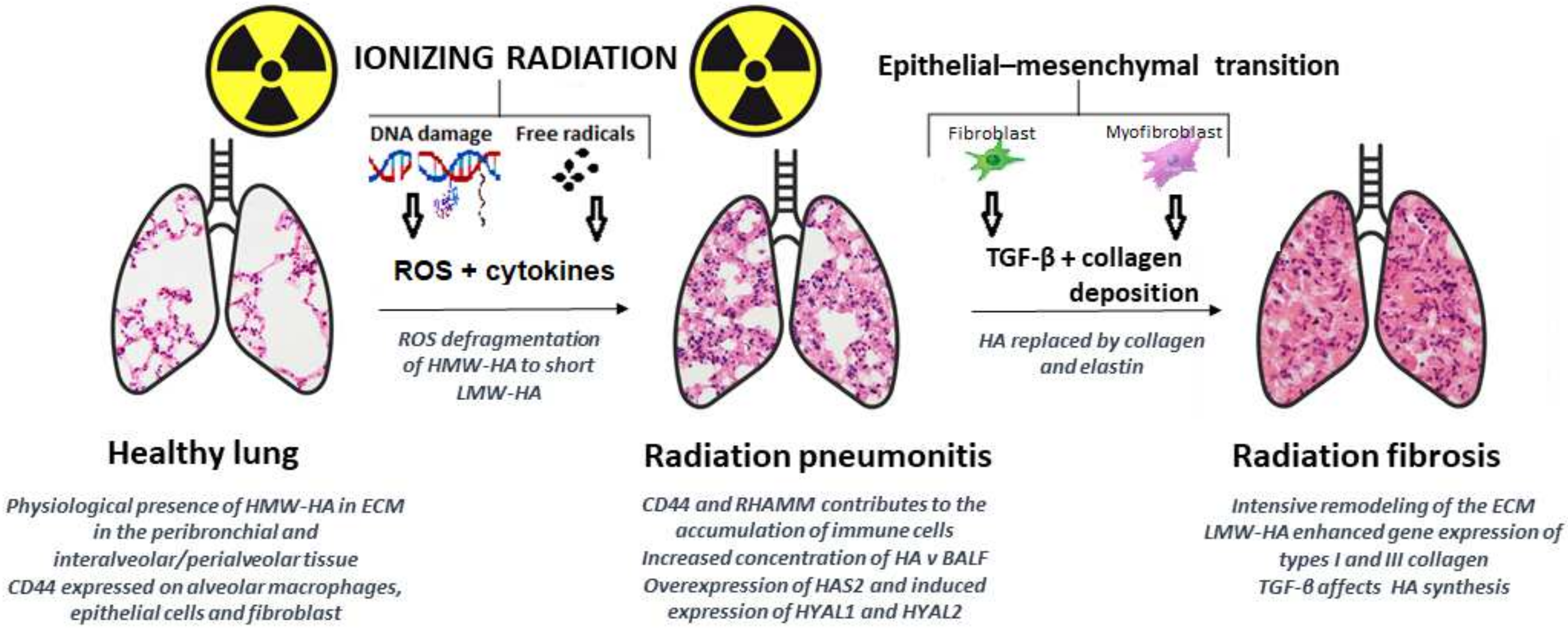
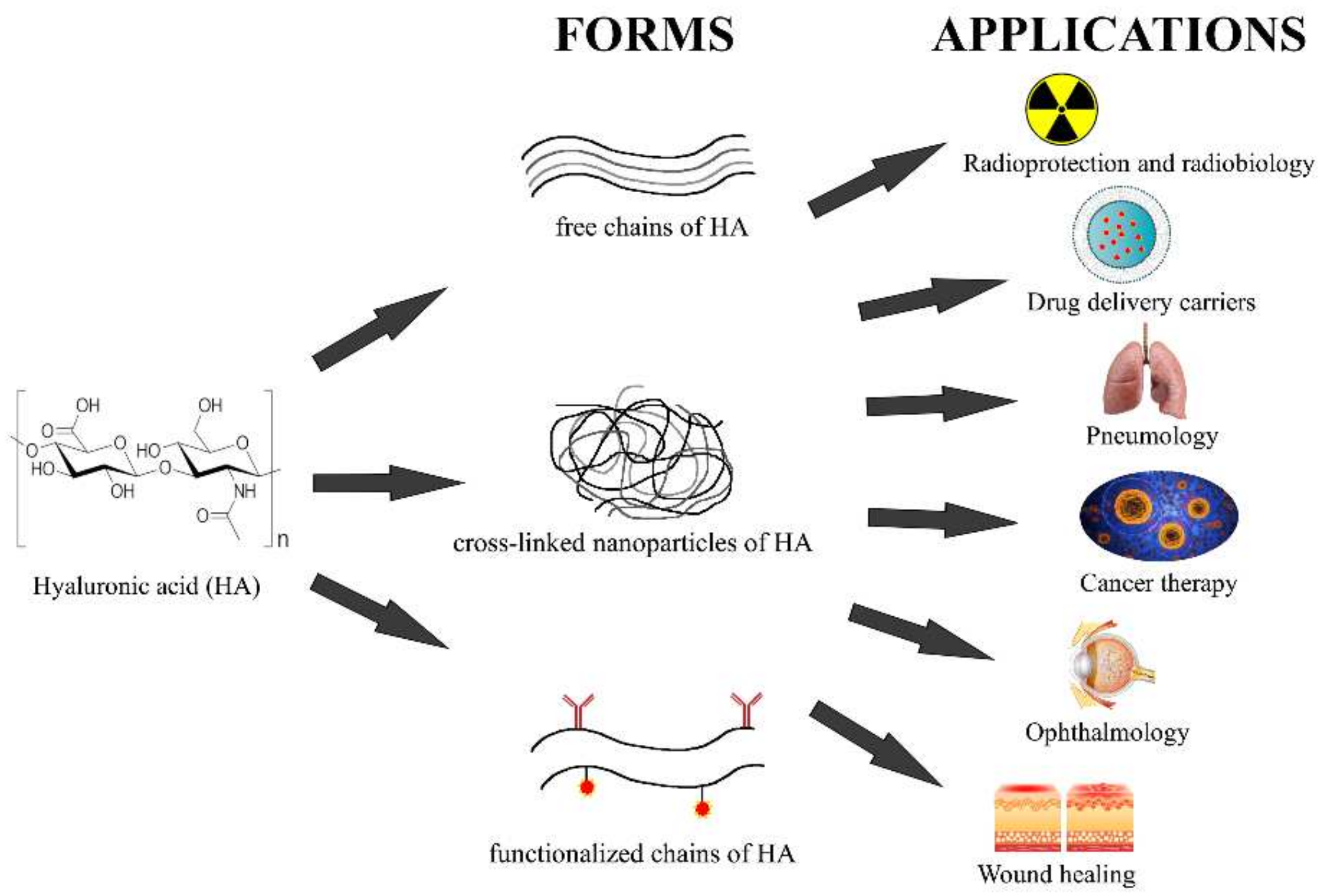
| Localization | Domain | Binding Protein | Function | Reference |
|---|---|---|---|---|
| Cell-surface | Link module domain | CD44 | regulation of cell–cell interaction and cell–matrix interface; mediation of cell adhesion; intracellular signaling pathways (Ras, MAPK, and PI3K) | [176] |
| LYVE-1 | lymphatic trafficking; hyaluronan degradation; intracellular signaling pathways for endothelial junctional retraction; regulation of lymphatic endothelial proliferation; lymphatic endothelium marker | [177] | ||
| HARE/Stabilin-2 | regulation of ligand binding and endocytic activity; mediation of hyaluronan clearance | [178] | ||
| TSG-6 | modulation of hyaluronan-CD44 interaction | [179] | ||
| Stabilin-1 | regulation of endocytic activity | [180] | ||
| HAPLN 1–4 | regulation of HA binding; HAPLN2 and HAPLN4 are specific for brain/CNS tissue | [181] | ||
| Bral1 | formation of the hyaluronan-associated matrix in the CNS | [182] | ||
| B(X7)B motif | RHAMM/CD168 | critical component of the inflammatory response | [146] | |
| Other | TLR2, TLR4 | macrophage activation and proinflammatory response; stimulation of endothelial recognition | [183] | |
| ICAM-1 | regulation of cell adhesion | [184] | ||
| Layilin | mediation of cell adhesion | [185] | ||
| Extracellular | Link module domain | Hyalectins: versican, aggrecan, neurocan, brevican (BEHAB) | regulation of HA binding; forming aggregates with HA in ECM | [186] |
| fibrinogen | regulation of HA binding in ECM | [187] | ||
| B(X7)B motif | Trypsin inhibitor (IαI) | mediation of HA-TSG-6 binding | [188] | |
| Other | CEMIP (KIAA1199/HYBID) | included in cell-migration; hyaluronan depolymerization | [189] | |
| SPACR, SPACRCAN | protein in interphotoreceptor matrix in subretinal space; organization and support to photoreceptor function | [190,191] | ||
| Intracellular | B(X7)B motif | iRHAMM | cell division; binding to the mitotic spindle; interacting with microtubules and microfilaments | [192] |
| USP17 (mouse SDS3) | regulator of cell proliferation and survival; essential for chemotaxis and chemokinesis | [193] | ||
| Other | IHABP4 | involved in cell interaction | [194] | |
| CDC37 | cell division; essential cell cycle regulatory factor | [168] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lierova, A.; Kasparova, J.; Filipova, A.; Cizkova, J.; Pekarova, L.; Korecka, L.; Mannova, N.; Bilkova, Z.; Sinkorova, Z. Hyaluronic Acid: Known for Almost a Century, but Still in Vogue. Pharmaceutics 2022, 14, 838. https://doi.org/10.3390/pharmaceutics14040838
Lierova A, Kasparova J, Filipova A, Cizkova J, Pekarova L, Korecka L, Mannova N, Bilkova Z, Sinkorova Z. Hyaluronic Acid: Known for Almost a Century, but Still in Vogue. Pharmaceutics. 2022; 14(4):838. https://doi.org/10.3390/pharmaceutics14040838
Chicago/Turabian StyleLierova, Anna, Jitka Kasparova, Alzbeta Filipova, Jana Cizkova, Lenka Pekarova, Lucie Korecka, Nikola Mannova, Zuzana Bilkova, and Zuzana Sinkorova. 2022. "Hyaluronic Acid: Known for Almost a Century, but Still in Vogue" Pharmaceutics 14, no. 4: 838. https://doi.org/10.3390/pharmaceutics14040838
APA StyleLierova, A., Kasparova, J., Filipova, A., Cizkova, J., Pekarova, L., Korecka, L., Mannova, N., Bilkova, Z., & Sinkorova, Z. (2022). Hyaluronic Acid: Known for Almost a Century, but Still in Vogue. Pharmaceutics, 14(4), 838. https://doi.org/10.3390/pharmaceutics14040838







