Combination of Six Individual Derivatives of the Pom-1 Antibiofilm Peptide Doubles Their Efficacy against Invasive and Multi-Resistant Clinical Isolates of the Pathogenic Yeast Candida albicans
Abstract
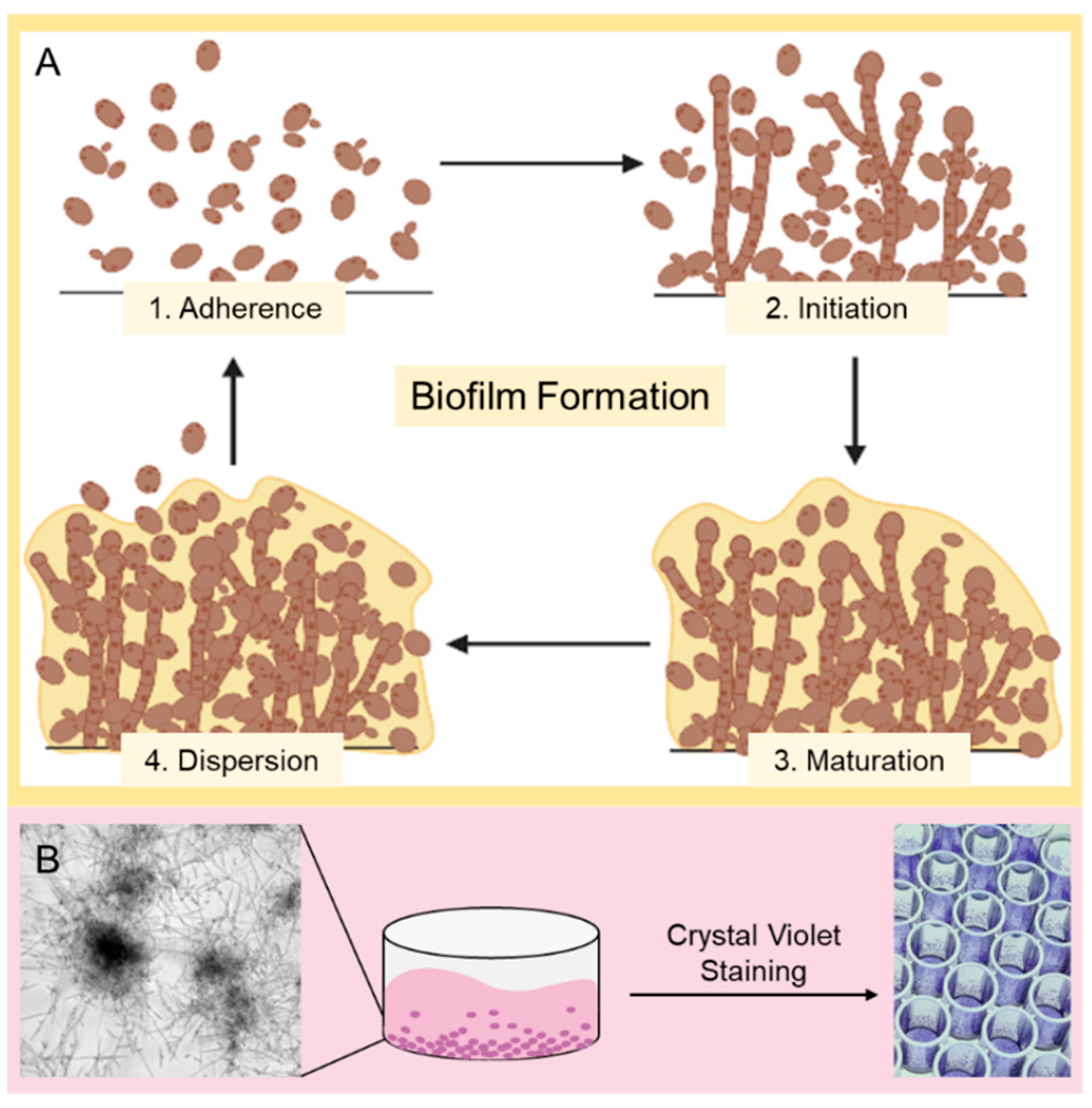
:1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Methods
2.2.1. Cultivation of Candida spp.
2.2.2. Peptide Optimization
2.2.3. Biofilm Formation and Crystal Violet Assay/Biomass Quantification
2.2.4. Cell Culture
2.2.5. Passaging Adherent Cell Cultures
2.2.6. Viability Assays for Cell Cultures
2.2.7. ExPASy ProtParam
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Pappas, P.G.; Lionakis, M.S.; Arendrup, M.C.; Ostrosky-Zeichner, L.; Kullberg, B.J. Invasive Candidiasis. Nat. Rev. Dis. Primers 2018, 4, 18026. [Google Scholar] [CrossRef] [PubMed]
- Lockhart, S.R. Current Epidemiology of Candida Infection. Clin. Microbiol. Newsl. 2014, 36, 131–136. [Google Scholar] [CrossRef]
- Kullberg, B.J.; Arendrup, M.C. Invasive Candidiasis. N. Engl. J. Med. 2015, 373, 1445–1456. [Google Scholar] [CrossRef] [Green Version]
- Yang, Y.L.; Lo, H.J. Mechanisms of Antifungal Agent Resistance. J. Microbiol. Immunol. Infect. 2001, 34, 79–86. [Google Scholar]
- Fakhim, H.; Vaezi, A.; Dannaoui, E.; Chowdhary, A.; Nasiry, D.; Faeli, L.; Meis, J.F.; Badali, H. Comparative Virulence of Candida auris with Candida haemulonii, Candida glabrata and Candida albicans in a Murine Model. Mycoses 2018, 61, 377–382. [Google Scholar] [CrossRef] [Green Version]
- Berkow, E.L.; Lockhart, S.R. Fluconazole Resistance in Candida Species: A Current Perspective. Infect. Drug Resist. 2017, 10, 237–245. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Grela, E.; Zdybicka-Barabas, A.; Pawlikowska-Pawlega, B.; Cytrynska, M.; Wlodarczyk, M.; Grudzinski, W.; Luchowski, R.; Gruszecki, W.I. Modes of the Antibiotic Activity of Amphotericin B against Candida albicans. Sci. Rep. 2019, 9, 17029. [Google Scholar] [CrossRef] [Green Version]
- Nett, J.E.; Andes, D.R. Antifungal Agents. Infect. Dis. Clin. N. Am. 2016, 30, 51–83. [Google Scholar] [CrossRef]
- Berkow, E.L.; Lockhart, S.R.; Ostrosky-Zeichner, L. Antifungal Susceptibility Testing: Current Approaches. Clin. Microbiol. Rev. 2020, 33, e00069-19. [Google Scholar] [CrossRef]
- Dannaoui, E.; Desnos-Ollivier, M.; Garcia-Hermoso, D.; Grenouillet, F.; Cassaing, S.; Baixench, M.-T.; Bretagne, S.; Dromer, F.; Lortholary, O. Candida Spp. with Acquired Echinocandin Resistance, France, 2004–2010. Emerg. Infect. Dis. 2012, 18, 86–90. [Google Scholar] [CrossRef]
- Perlin, D.S. Resistance to Echinocandin-Class Antifungal Drugs. Drug Resist. Updates 2007, 10, 121–130. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tanwar, J.; Das, S.; Fatima, Z.; Hameed, S. Multidrug Resistance: An Emerging Crisis. Interdiscip. Perspect. Infect. Dis. 2014, 2014, 541340. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wiederhold, N.P. Antifungal Resistance: Current Trends and Future Strategies to Combat. Infect. Drug Resist. 2017, 10, 249–259. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ellis, D. Amphotericin B: Spectrum and Resistance. J. Antimicrob. Chemother. 2002, 49, 7–10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McCarty, T.P.; Pappas, P.G. Invasive Candidiasis. Infect. Dis. Clin. N. Am. 2016, 30, 103–124. [Google Scholar] [CrossRef]
- Sobel, J.D.; Faro, S.; Force, R.W.; Foxman, B.; Ledger, W.J.; Nyirjesy, P.R.; Reed, B.D.; Summers, P.R. Vulvovaginal Candidiasis: Epidemiologic, Diagnostic, and Therapeutic Considerations. Am. J. Obstet. Gynecol. 1998, 178, 203–211. [Google Scholar] [CrossRef]
- Cleveland, A.A.; Harrison, L.H.; Farley, M.M.; Hollick, R.; Stein, B.; Chiller, T.M.; Lockhart, S.R.; Park, B.J. Declining Incidence of Candidemia and the Shifting Epidemiology of Candida Resistance in Two US Metropolitan Areas, 2008–2013: Results from Population-Based Surveillance. PLoS ONE 2015, 10, e0120452. [Google Scholar] [CrossRef] [Green Version]
- Sheppard, D.C.; Filler, S.G. Host Cell Invasion by Medically Important Fungi. Cold Spring Harb. Perspect. Med. 2015, 5, a019687. [Google Scholar] [CrossRef] [Green Version]
- Berman, J.; Sudbery, P.E. Candida albicans: A Molecular Revolution Built on Lessons from Budding Yeast. Nat. Rev. Genet. 2002, 3, 918–931. [Google Scholar] [CrossRef]
- Murad, A.M.A. NRG1 Represses Yeast-Hypha Morphogenesis and Hypha-Specific Gene Expression in Candida albicans. EMBO J. 2001, 20, 4742–4752. [Google Scholar] [CrossRef] [Green Version]
- Ramage, G.; Martínez, J.P.; López-Ribot, J.L. Candida Biofilms on Implanted Biomaterials: A Clinically Significant Problem. FEMS Yeast Res. 2006, 6, 979–986. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Desai, J.V.; Mitchell, A.P.; Andes, D.R. Fungal Biofilms, Drug Resistance, and Recurrent Infection. Cold Spring Harb. Perspect. Med. 2014, 4, a019729. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- LaFleur, M.D.; Kumamoto, C.A.; Lewis, K. Candida albicans Biofilms Produce Antifungal-Tolerant Persister Cells. Antimicrob. Agents Chemother. 2006, 50, 3839–3846. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Raber, H.F.; Sejfijaj, J.; Kissmann, A.K.; Wittgens, A.; Gonzalez-Garcia, M.; Alba, A.; Vázquez, A.A.; Vicente, F.E.M.; Erviti, J.P.; Kubiczek, D.; et al. Antimicrobial Peptides Pom-1 and Pom-2 from Pomacea poeyana Are Active against Candida auris, C. parapsilosis and C. albicans Biofilms. Pathogens 2021, 10, 496. [Google Scholar] [CrossRef] [PubMed]
- Taff, H.T.; Mitchell, K.F.; Edward, J.A.; Andes, D.R. Mechanisms of Candida Biofilm Drug Resistance. Future Microbiol. 2013, 8, 1325–1337. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pierce, C.; Vila, T.; Romo, J.; Montelongo-Jauregui, D.; Wall, G.; Ramasubramanian, A.; Lopez-Ribot, J. The Candida albicans Biofilm Matrix: Composition, Structure and Function. J. Fungi 2017, 3, 14. [Google Scholar] [CrossRef] [Green Version]
- Sherry, L.; Ramage, G.; Kean, R.; Borman, A.; Johnson, E.M.; Richardson, M.D.; Rautemaa-Richardson, R. Biofilm-Forming Capability of Highly Virulent, Multidrug-Resistant Candida auris. Emerg. Infect. Dis. 2017, 23, 328–331. [Google Scholar] [CrossRef] [Green Version]
- Darouiche, R.O. Treatment of Infections Associated with Surgical Implants. N. Engl. J. Med. 2004, 350, 1422–1429. [Google Scholar] [CrossRef]
- Hancock, R.E.W.; Haney, E.F.; Gill, E.E. The Immunology of Host Defence Peptides: Beyond Antimicrobial Activity. Nat. Rev. Immunol. 2016, 16, 321–334. [Google Scholar] [CrossRef]
- Jenssen, H.; Hamill, P.; Hancock, R.E.W. Peptide Antimicrobial Agents. Clin. Microbiol. Rev. 2006, 19, 491–511. [Google Scholar] [CrossRef] [Green Version]
- Powers, J.-P.S.; Hancock, R.E.W. The Relationship between Peptide Structure and Antibacterial Activity. Peptides 2003, 24, 1681–1691. [Google Scholar] [CrossRef] [PubMed]
- Van der Weerden, N.L.; Hancock, R.E.W.; Anderson, M.A. Permeabilization of Fungal Hyphae by the Plant Defensin NaD1 Occurs through a Cell Wall-Dependent Process. J. Biol. Chem. 2010, 285, 37513–37520. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jean-François, F.; Elezgaray, J.; Berson, P.; Vacher, P.; Dufourc, E.J. Pore Formation Induced by an Antimicrobial Peptide: Electrostatic Effects. Biophys. J. 2008, 95, 5748–5756. [Google Scholar] [CrossRef] [Green Version]
- Peters, B.M.; Shirtliff, M.E.; Jabra-Rizk, M.A. Antimicrobial Peptides: Primeval Molecules or Future Drugs? PLoS Pathog. 2010, 6, 4–7. [Google Scholar] [CrossRef] [PubMed]
- Nicolas, P. Multifunctional Host Defense Peptides: Intracellular-Targeting Antimicrobial Peptides. FEBS J. 2009, 276, 6483–6496. [Google Scholar] [CrossRef] [PubMed]
- Moerman, L.; Bosteels, S.; Noppe, W.; Willems, J.; Clynen, E.; Schoofs, L.; Thevissen, K.; Tytgat, J.; van Eldere, J.; van der Walt, J.; et al. Antibacterial and Antifungal Properties of α-Helical, Cationic Peptides in the Venom of Scorpions from Southern Africa. Eur. J. Biochem. 2002, 269, 4799–4810. [Google Scholar] [CrossRef] [PubMed]
- Browne, K.; Chakraborty, S.; Chen, R.; Willcox, M.D.; Black, D.S.; Walsh, W.R.; Kumar, N. A New Era of Antibiotics: The Clinical Potential of Antimicrobial Peptides. Int. J. Mol. Sci. 2020, 21, 7047. [Google Scholar] [CrossRef]
- Dennison, S.; Wallace, J.; Harris, F.; Phoenix, D. Amphiphilic α-Helical Antimicrobial Peptides and Their Structure/Function Relationships. Protein Pept. Lett. 2005, 12, 31–39. [Google Scholar] [CrossRef] [Green Version]
- Tossi, A.; Sandri, L.; Giangaspero, A. Amphipathic, α-Helical Antimicrobial Peptides. Pept. Sci. 2004, 55, 4–30. [Google Scholar] [CrossRef]
- Jiang, Z.; Vasil, A.I.; Hale, J.; Hancock, R.E.W.; Vasil, M.L.; Hodges, R.S. Effects of Net Charge and the Number of Positively Charged Residues on the Biological Activity of Amphipathic α-Helical Cationic Antimicrobial Peptides. In Peptides for Youth; Springer: Berlin/Heidelberg, Germany, 2009; pp. 561–562. [Google Scholar]
- Huang, Y.; Huang, J.; Chen, Y. Alpha-Helical Cationic Antimicrobial Peptides: Relationships of Structure and Function. Protein Cell 2010, 1, 143–152. [Google Scholar] [CrossRef]
- Kustanovich, I.; Shalev, D.E.; Mikhlin, M.; Gaidukov, L.; Mor, A. Structural Requirements for Potent Versus Selective Cytotoxicity for Antimicrobial Dermaseptin S4 Derivatives. J. Biol. Chem. 2002, 277, 16941–16951. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zelezetsky, I.; Pacor, S.; Pag, U.; Papo, N.; Shai, Y.; Sahl, H.-G.; Tossi, A. Controlled Alteration of the Shape and Conformational Stability of α-Helical Cell-Lytic Peptides: Effect on Mode of Action and Cell Specificity. Biochem. J. 2005, 390, 177–188. [Google Scholar] [CrossRef] [PubMed]
- Fernández-Vidal, M.; Jayasinghe, S.; Ladokhin, A.S.; White, S.H. Folding Amphipathic Helices into Membranes: Amphiphilicity Trumps Hydrophobicity. J. Mol. Biol. 2007, 370, 459–470. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, Y.; Mant, C.T.; Farmer, S.W.; Hancock, R.E.W.; Vasil, M.L.; Hodges, R.S. Rational Design of α-Helical Antimicrobial Peptides with Enhanced Activities and Specificity/Therapeutic Index. J. Biol. Chem. 2005, 280, 12316–12329. [Google Scholar] [CrossRef] [Green Version]
- Di Somma, A.; Moretta, A.; Canè, C.; Cirillo, A.; Duilio, A. Antimicrobial and Antibiofilm Peptides. Biomolecules 2020, 10, 652. [Google Scholar] [CrossRef] [Green Version]
- Zasloff, M. Antimicrobial Peptides of Multicellular Organisms. Nature 2002, 415, 389–395. [Google Scholar] [CrossRef]
- Kang, H.K.; Kim, C.; Seo, C.H.; Park, Y. The therapeutic applications of antimicrobial peptides (AMPs): A patent review. J. Microbiol. 2017, 55, 1–12. [Google Scholar] [CrossRef]
- Radek, K.; Gallo, R. Antimicrobial Peptides: Natural Effectors of the Innate Immune System. Semin. Immunopathol. 2007, 29, 27–43. [Google Scholar] [CrossRef]
- Conlon, J.M.; Sonnevend, A. Antimicrobial Peptides in Frog Skin Secretions. In Antimicrobial Peptides; Springer: Berlin/Heidelberg, Germany, 2010; pp. 3–14. [Google Scholar]
- De Zoysa, G.H.; Cameron, A.J.; Hegde, V.V.; Raghothama, S.; Sarojini, V. Antimicrobial Peptides with Potential for Biofilm Eradication: Synthesis and Structure Activity Relationship Studies of Battacin Peptides. J. Med. Chem. 2015, 58, 625–639. [Google Scholar] [CrossRef]
- Dutta, D.; Kumar, N.; Willcox, M.D.P. Antimicrobial Activity of Four Cationic Peptides Immobilised to Poly-Hydroxyethylmethacrylate. Biofouling 2016, 32, 429–438. [Google Scholar] [CrossRef]
- Zharkova, M.S.; Orlov, D.S.; Golubeva, O.Y.; Chakchir, O.B.; Eliseev, I.E.; Grinchuk, T.M.; Shamova, O.V. Application of Antimicrobial Peptides of the Innate Immune System in Combination with Conventional Antibiotics—A Novel Way to Combat Antibiotic Resistance? Front. Cell. Infect. Microbiol. 2019, 9, 128. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rajchakit, U.; Sarojini, V. Recent Developments in Antimicrobial-Peptide-Conjugated Gold Nanoparticles. Bioconjug. Chem. 2017, 28, 2673–2686. [Google Scholar] [CrossRef] [PubMed]
- Munir, M.U.; Ahmad, M.M. Nanomaterials Aiming to Tackle Antibiotic-Resistant Bacteria. Pharmaceutics 2022, 14, 582. [Google Scholar] [CrossRef]
- Munir, M.U.; Ahmed, A.; Usman, M.; Salman, S. Recent Advances in Nanotechnology-Aided Materials in Combating Microbial Resistance and Functioning as Antibiotics Substitutes. Int. J. Nanomed. 2020, 15, 7329–7358. [Google Scholar] [CrossRef] [PubMed]
- Naghmouchi, K.; le Lay, C.; Baah, J.; Drider, D. Antibiotic and Antimicrobial Peptide Combinations: Synergistic Inhibition of Pseudomonas fluorescens and Antibiotic-Resistant Variants. Res. Microbiol. 2012, 163, 101–108. [Google Scholar] [CrossRef]
- Nizet, V. Antimicrobial Peptide Resistance Mechanisms of Human Bacterial Pathogens. Curr. Issues Mol. Biol. 2006, 8, 11–26. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schauber, J.; Gallo, R.L. Antimicrobial Peptides and the Skin Immune Defense System. J. Allergy Clin. Immunol. 2008, 122, 261–266. [Google Scholar] [CrossRef] [Green Version]
- Mitta, G.; Vandenbulcke, F.; Roch, P. Original Involvement of Antimicrobial Peptides in Mussel Innate Immunity. FEBS Lett. 2000, 486, 185–190. [Google Scholar] [CrossRef] [Green Version]
- García, M.G.; Rodríguez, A.; Alba, A.; Vázquez, A.A.; Vicente, F.E.M.; Pérez-Erviti, J.; Spellerberg, B.; Stenger, S.; Grieshober, M.; Conzelmann, C.; et al. New Antibacterial Peptides from the Freshwater Mollusk Pomacea poeyana (Pilsbry, 1927). Biomolecules 2020, 10, 1473. [Google Scholar] [CrossRef]
- Kubiczek, D.; Raber, H.; Gonzalez-García, M.; Morales-Vicente, F.; Staendker, L.; Otero-Gonzalez, A.J.; Rosenau, F. Derivates of the Antifungal Peptide CM-P5 Inhibit Development of Candida auris Biofilms in Vitro. Antibiotics 2020, 9, 363. [Google Scholar] [CrossRef]
- López-Abarrategui, C.; McBeth, C.; Mandal, S.M.; Sun, Z.J.; Heffron, G.; Alba-Menéndez, A.; Migliolo, L.; Reyes-Acosta, O.; García-Villarino, M.; Nolasco, D.O.; et al. Cm-P5: An Antifungal Hydrophilic Peptide Derived from the Coastal Mollusk Cenchritis muricatus (Gastropoda: Littorinidae). FASEB J. 2015, 29, 3315–3325. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vicente, F.E.M.; González-Garcia, M.; Diaz Pico, E.; Moreno-Castillo, E.; Garay, H.E.; Rosi, P.E.; Jimenez, A.M.; Campos-Delgado, J.A.; Rivera, D.G.; Chinea, G.; et al. Design of a Helical-Stabilized, Cyclic, and Nontoxic Analogue of the Peptide Cm-P5 with Improved Antifungal Activity. ACS Omega 2019, 4, 19081–19095. [Google Scholar] [CrossRef] [Green Version]
- Amann, V.; Kissmann, A.K.; Krämer, M.; Krebs, I.; Perez-Erviti, J.A.; Otero-Gonzalez, A.J.; Morales-Vicente, F.; Rodríguez, A.; Ständker, L.; Weil, T.; et al. Increased Activities against Biofilms of the Pathogenic Yeast Candida albicans of Optimized Pom-1 Derivatives. Pharmaceutics 2022, 14, 318. [Google Scholar] [CrossRef] [PubMed]
- Gasteiger, E.; Hoogland, C.; Gattiker, A.; Duvaud, S.; Wilkins, M.R.; Appel, R.D.; Bairoch, A. Protein Identification and Analysis Tools on the ExPASy Server. In The Proteomics Protocols Handbook; Humana Press: Totowa, NJ, USA, 2005; pp. 571–607. [Google Scholar]
- M27-A3; Reference Method for Broth Dilution Antifungal Susceptibily Testing of Yeasts. Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2008; Volume 28.
- O’Toole, G.A. Microtiter Dish Biofilm Formation Assay. J. Vis. Exp. 2011, 47, e2437. [Google Scholar] [CrossRef] [PubMed]
- Lieber, M.; Todaro, G.; Smith, B.; Szakal, A.; Nelson-Rees, W. A Continuous Tumor-Cell Line from a Human Lung Carcinoma with Properties of Type II Alveolar Epithelial Cells. Int. J. Cancer 1976, 17, 62–70. [Google Scholar] [CrossRef] [PubMed]
- Okita, K.; Matsumura, Y.; Sato, Y.; Okada, A.; Morizane, A.; Okamoto, S.; Hong, H.; Nakagawa, M.; Tanabe, K.; Tezuka, K.; et al. A More Efficient Method to Generate Integration-Free Human IPS Cells. Nat. Methods 2011, 8, 409–412. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- O’Toole, G.A.; Kolter, R. Initiation of biofilm formation in Pseudomonas fluorescens WCS365 proceeds via multiple, convergent signalling pathways: A genetic analysis. Mol. Microbiol. 1998, 28, 449–461. [Google Scholar] [CrossRef]
- Harrington, R.; Kindermann, S.L.; Hou, Q.; Taylor, R.J.; Azie, N.; Horn, D.L. Candidemia and Invasive Candidiasis among Hospitalized Neonates and Pediatric Patients. Curr. Med. Res. Opin. 2017, 33, 1803–1812. [Google Scholar] [CrossRef]
- Pfaller, M.A.; Diekema, D.J. Epidemiology of Invasive Candidiasis: A Persistent Public Health Problem. Clin. Microbiol. Rev. 2007, 20, 133–163. [Google Scholar] [CrossRef] [Green Version]
- Wall, G.; Montelongo-Jauregui, D.; Vidal Bonifacio, B.; Lopez-Ribot, J.L.; Uppuluri, P. Candida albicans Biofilm Growth and Dispersal: Contributions to Pathogenesis. Curr. Opin. Microbiol. 2019, 52, 1–6. [Google Scholar] [CrossRef]
- Crump, J.A.; Collignon, P.J. Intravascular Catheter-Associated Infections. Eur. J. Clin. Microbiol. Infect. Dis. 2000, 19, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.; Mishra, B.; Epand, R.F.; Epand, R.M. High-Quality 3D Structures Shine Light on Antibacterial, Anti-Biofilm and Antiviral Activities of Human Cathelicidin LL-37 and Its Fragments. Biochim. Biophys. Acta BBA Biomembr. 2014, 1838, 2160–2172. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ridyard, K.E.; Overhage, J. The Potential of Human Peptide LL-37 as an Antimicrobial and Anti-Biofilm Agent. Antibiotics 2021, 10, 650. [Google Scholar] [CrossRef] [PubMed]
- Bin Hafeez, A.; Jiang, X.; Bergen, P.J.; Zhu, Y. Antimicrobial Peptides: An Update on Classifications and Databases. Int. J. Mol. Sci. 2021, 22, 11691. [Google Scholar] [CrossRef]
- Münch, J.; Ständker, L.; Adermann, K.; Schulz, A.; Schindler, M.; Chinnadurai, R.; Pöhlmann, S.; Chaipan, C.; Biet, T.; Peters, T.; et al. Discovery and Optimization of a Natural HIV-1 Entry Inhibitor Targeting the Gp41 Fusion Peptide. Cell 2007, 129, 263–275. [Google Scholar] [CrossRef] [Green Version]
- Di Mambro, T.; Vanzolini, T.; Bruscolini, P.; Perez-Gaviro, S.; Marra, E.; Roscilli, G.; Bianchi, M.; Fraternale, A.; Schiavano, G.F.; Canonico, B.; et al. A New Humanized Antibody Is Effective against Pathogenic Fungi In Vitro. Sci. Rep. 2021, 11, 19500. [Google Scholar] [CrossRef]
- Matthews, R.C.; Rigg, G.; Hodgetts, S.; Carter, T.; Chapman, C.; Gregory, C.; Illidge, C.; Burnie, J. Preclinical Assessment of the Efficacy of Mycograb, a Human Recombinant Antibody against Fungal HSP90. Antimicrob. Agents Chemother. 2003, 47, 2208–2216. [Google Scholar] [CrossRef] [Green Version]
- Bugli, F.; Cacaci, M.; Martini, C.; Torelli, R.; Posteraro, B.; Sanguinetti, M.; Paroni Sterbini, F. Human Monoclonal Antibody-Based Therapy in the Treatment of Invasive Candidiasis. Clin. Dev. Immunol. 2013, 2013, 403121. [Google Scholar] [CrossRef] [Green Version]
- Olsen, J.S.; Brown, C.; Capule, C.C.; Rubinshtein, M.; Doran, T.M.; Srivastava, R.K.; Feng, C.; Nilsson, B.L.; Yang, J.; Dewhurst, S. Amyloid-Binding Small Molecules Efficiently Block SEVI (Semen-Derived Enhancer of Virus Infection)- and Semen-Mediated Enhancement of HIV-1 Infection. J. Biol. Chem. 2010, 285, 35488–35496. [Google Scholar] [CrossRef] [Green Version]
- Roan, N.R.; Müller, J.A.; Liu, H.; Chu, S.; Arnold, F.; Stürzel, C.M.; Walther, P.; Dong, M.; Witkowska, H.E.; Kirchhoff, F.; et al. Peptides Released by Physiological Cleavage of Semen Coagulum Proteins Form Amyloids That Enhance HIV Infection. Cell Host Microbe 2011, 10, 541–550. [Google Scholar] [CrossRef] [Green Version]
- Dueholm, M.S.; Petersen, S.V.; Sønderkaer, M.; Larsen, P.; Christiansen, G.; Hein, K.L.; Enghild, J.J.; Nielsen, J.L.; Nielsen, K.L.; Nielsen, P.H.; et al. Functional Amyloid in Pseudomonas. Mol. Microbiol. 2010, 77, 1009–1020. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Romero, D.; Aguilar, C.; Losick, R.; Kolter, R. Amyloid Fibers Provide Structural Integrity to Bacillus Subtilis Biofilms. Proc. Natl. Acad. Sci. USA 2010, 107, 2230–2234. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Prigent-Combaret, C.; Prensier, G.; le Thi, T.T.; Vidal, O.; Lejeune, P.; Dorel, C. Developmental Pathway for Biofilm Formation in Curli-Producing Escherichia coli Strains: Role of Flagella, Curli and Colanic Acid. Environ. Microbiol. 2000, 2, 450–464. [Google Scholar] [CrossRef]
- Otoo, H.N.; Lee, K.G.; Qiu, W.; Lipke, P.N. Candida albicans Als Adhesins Have Conserved Amyloid-Forming Sequences. Eukaryot. Cell 2008, 7, 776–782. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yan, H.; Hancock, R.E.W. Synergistic Interactions between Mammalian Antimicrobial Defense Peptides. Antimicrob. Agents Chemother. 2001, 45, 1558–1560. [Google Scholar] [CrossRef] [Green Version]
- Marquette, A.; Bechinger, B. Biophysical Investigations Elucidating the Mechanisms of Action of Antimicrobial Peptides and Their Synergism. Biomolecules 2018, 8, 18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gordya, N.; Yakovlev, A.; Kruglikova, A.; Tulin, D.; Potolitsina, E.; Suborova, T.; Bordo, D.; Rosano, C.; Chernysh, S. Natural Antimicrobial Peptide Complexes in the Fighting of Antibiotic Resistant Biofilms: Calliphora vicina Medicinal Maggots. PLoS ONE 2017, 12, e0173559. [Google Scholar] [CrossRef] [Green Version]
- Berditsch, M.; Jäger, T.; Strempel, N.; Schwartz, T.; Overhage, J.; Ulrich, A.S. Synergistic Effect of Membrane-Active Peptides Polymyxin B and Gramicidin S on Multidrug-Resistant Strains and Biofilms of Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 2015, 59, 5288–5296. [Google Scholar] [CrossRef] [Green Version]
- Girmenia, C.; Venditti, M.; Martino, P. Fluconazole in Combination with Flucytosine in the Treatment of Fluconazole-Resistant Candida Infections. Diagn. Microbiol. Infect. Dis. 2003, 46, 227–231. [Google Scholar] [CrossRef]
- Zhang, Q.; Liu, F.; Zeng, M.; Mao, Y.; Song, Z. Drug Repurposing Strategies in the Development of Potential Antifungal Agents. Appl. Microbiol. Biotechnol. 2021, 105, 5259–5279. [Google Scholar] [CrossRef]




Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Häring, M.; Amann, V.; Kissmann, A.-K.; Herberger, T.; Synatschke, C.; Kirsch-Pietz, N.; Perez-Erviti, J.A.; Otero-Gonzalez, A.J.; Morales-Vicente, F.; Andersson, J.; et al. Combination of Six Individual Derivatives of the Pom-1 Antibiofilm Peptide Doubles Their Efficacy against Invasive and Multi-Resistant Clinical Isolates of the Pathogenic Yeast Candida albicans. Pharmaceutics 2022, 14, 1332. https://doi.org/10.3390/pharmaceutics14071332
Häring M, Amann V, Kissmann A-K, Herberger T, Synatschke C, Kirsch-Pietz N, Perez-Erviti JA, Otero-Gonzalez AJ, Morales-Vicente F, Andersson J, et al. Combination of Six Individual Derivatives of the Pom-1 Antibiofilm Peptide Doubles Their Efficacy against Invasive and Multi-Resistant Clinical Isolates of the Pathogenic Yeast Candida albicans. Pharmaceutics. 2022; 14(7):1332. https://doi.org/10.3390/pharmaceutics14071332
Chicago/Turabian StyleHäring, Michelle, Valerie Amann, Ann-Kathrin Kissmann, Tilmann Herberger, Christopher Synatschke, Nicole Kirsch-Pietz, Julio A. Perez-Erviti, Anselmo J. Otero-Gonzalez, Fidel Morales-Vicente, Jakob Andersson, and et al. 2022. "Combination of Six Individual Derivatives of the Pom-1 Antibiofilm Peptide Doubles Their Efficacy against Invasive and Multi-Resistant Clinical Isolates of the Pathogenic Yeast Candida albicans" Pharmaceutics 14, no. 7: 1332. https://doi.org/10.3390/pharmaceutics14071332
APA StyleHäring, M., Amann, V., Kissmann, A.-K., Herberger, T., Synatschke, C., Kirsch-Pietz, N., Perez-Erviti, J. A., Otero-Gonzalez, A. J., Morales-Vicente, F., Andersson, J., Weil, T., Stenger, S., Rodríguez, A., Ständker, L., & Rosenau, F. (2022). Combination of Six Individual Derivatives of the Pom-1 Antibiofilm Peptide Doubles Their Efficacy against Invasive and Multi-Resistant Clinical Isolates of the Pathogenic Yeast Candida albicans. Pharmaceutics, 14(7), 1332. https://doi.org/10.3390/pharmaceutics14071332






