Mg2+ Ions Regulating 3WJ-PRNA to Construct Controllable RNA Nanoparticle Drug Delivery Platforms
Abstract
:1. Introduction
2. Materials and Methods
3. Results and Discussion
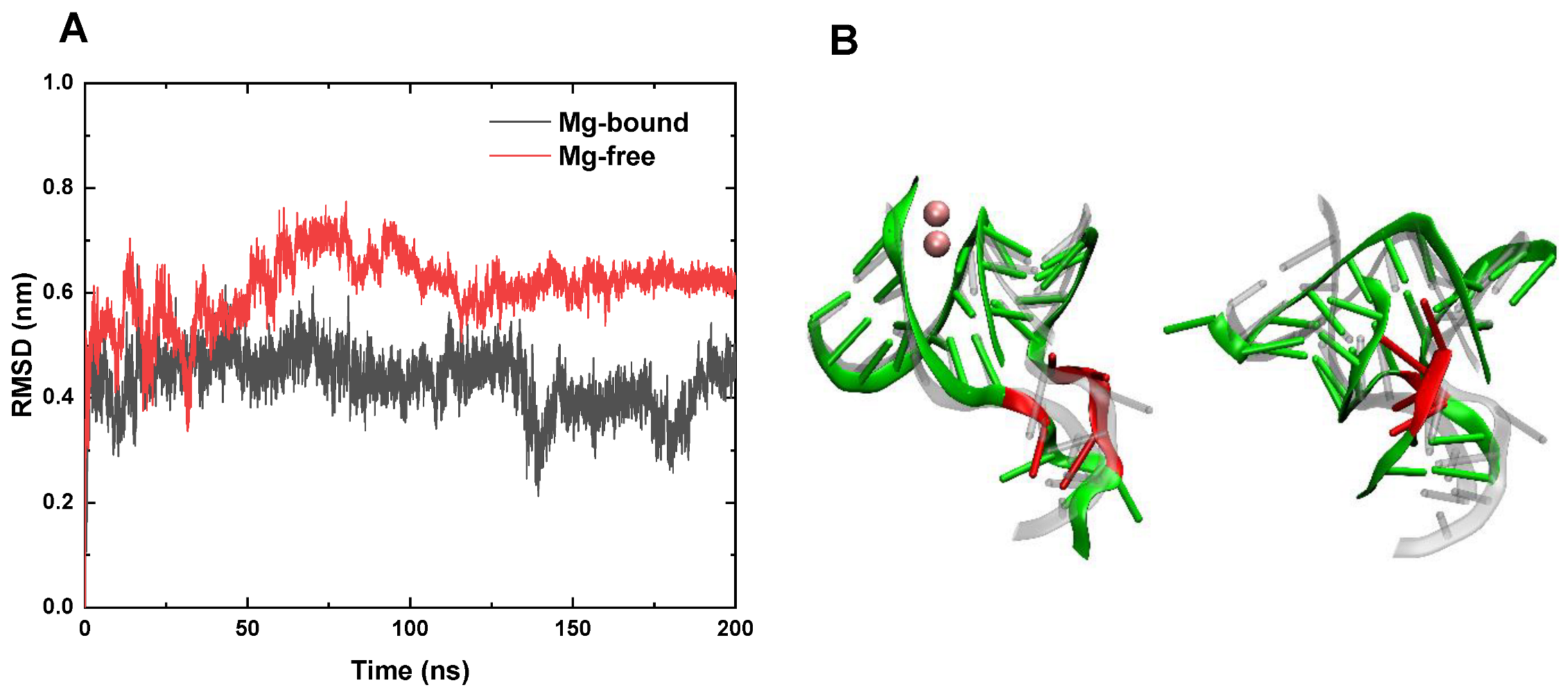
3.1. RMSD and Conformation Behaviors at Room Temperature
3.2. RMSD and Conformation Behaviors at High Temperature
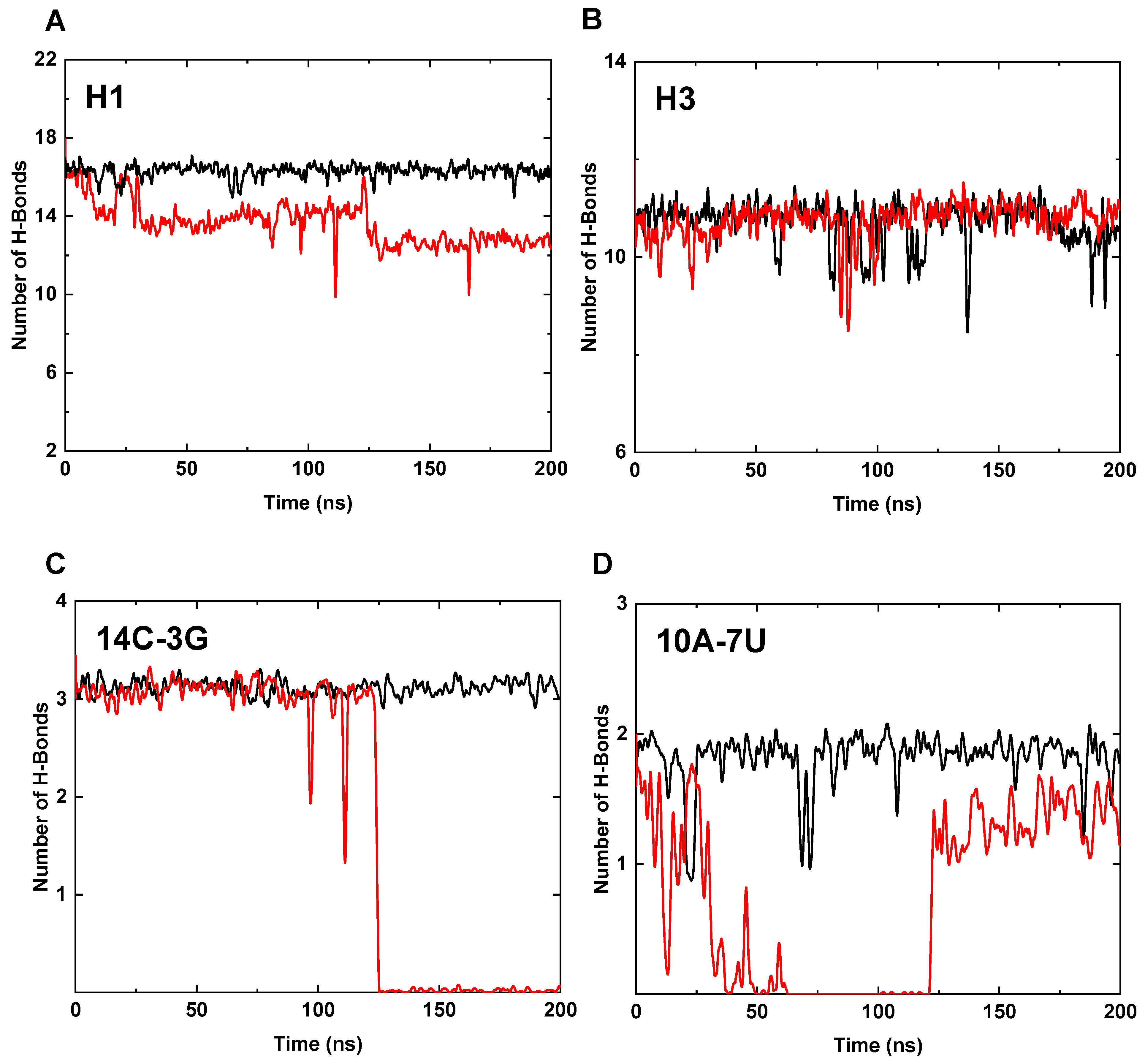
3.3. Interaction Analysis of the Core Region
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Jasinski, D.; Haque, F.; Binzel, D.W.; Guo, P. Advancement of the Emerging Field of RNA Nanotechnology. ACS Nano 2017, 11, 1142–1164. [Google Scholar] [CrossRef]
- Binzel, D.W.; Li, X.; Burns, N.; Khan, E.; Lee, W.J.; Chen, L.C.; Ellipilli, S.; Miles, W.; Ho, Y.S.; Guo, P. Thermostability, Tunability, and Tenacity of RNA as Rubbery Anionic Polymeric Materials in Nanotechnology and Nanomedicine-Specific Cancer Targeting with Undetectable Toxicity. Chem. Rev. 2021, 121, 7398–7467. [Google Scholar] [CrossRef]
- Lee, T.J.; Haque, F.; Shu, D.; Yoo, J.Y.; Li, H.; Yokel, R.A.; Horbinski, C.; Kim, T.H.; Kim, S.H.; Kwon, C.H.; et al. RNA nanoparticle as a vector for targeted siRNA delivery into glioblastoma mouse model. Oncotarget 2015, 6, 14766–14776. [Google Scholar] [CrossRef] [Green Version]
- Haque, F.; Xu, C.; Jasinski, D.L.; Li, H.; Guo, P. Using Planar Phi29 pRNA Three-Way Junction to Control Size and Shape of RNA Nanoparticles for Biodistribution Profiling in Mice. Methods Mol. Biol. 2017, 1632, 359–380. [Google Scholar]
- Guo, S.; Xu, C.; Yin, H.; Hill, J.; Pi, F.; Guo, P. Tuning the size, shape and structure of RNA nanoparticles for favorable cancer targeting and immunostimulation. Wiley Interdiscip. Rev. Nanomed. Nanobiotechnol. 2020, 12, e1582. [Google Scholar] [CrossRef]
- Ghimire, C.; Wang, H.; Li, H.; Vieweger, M.; Xu, C.; Guo, P. RNA Nanoparticles as Rubber for Compelling Vessel Extravasation to Enhance Tumor Targeting and for Fast Renal Excretion to Reduce Toxicity. ACS Nano 2020, 14, 13180–13191. [Google Scholar] [CrossRef]
- Xu, C.; Haque, F.; Jasinski, D.L.; Binzel, D.W.; Shu, D.; Guo, P. Favorable biodistribution, specific targeting and conditional endosomal escape of RNA nanoparticles in cancer therapy. Cancer Lett. 2018, 414, 57–70. [Google Scholar] [CrossRef]
- Hosseinkhani, H.; Domb, A.J. Biodegradable polymers in gene-silencing technology. Polym. Adv. Technol. 2019, 30, 2647–2655. [Google Scholar] [CrossRef]
- Yabu, H.; Higuchi, T.; Ijiro, K.; Shimomura, M. Spontaneous formation of polymer nanoparticles by good-solvent evaporation as a nonequilibrium process. Chaos 2005, 15, 047505. [Google Scholar] [CrossRef]
- Guo, S.; Li, H.; Ma, M.; Fu, J.; Dong, Y.; Guo, P. Size, Shape, and Sequence-Dependent Immunogenicity of RNA Nanoparticles. Mol. Ther. Nucleic Acids 2017, 9, 399–408. [Google Scholar] [CrossRef] [Green Version]
- Satalkar, P.; Elger, B.S.; Hunziker, P.; Shaw, D. Challenges of clinical translation in nanomedicine: A qualitative study. Nanomedicine 2016, 12, 893–900. [Google Scholar] [CrossRef] [Green Version]
- Shi, J.; Kantoff, P.W.; Wooster, R.; Farokhzad, O.C. Cancer nanomedicine: Progress, challenges and opportunities. Nat. Rev. Cancer 2017, 17, 20–37. [Google Scholar] [CrossRef]
- Patra, J.K.; Das, G.; Fraceto, L.F.; Campos, E.V.R.; Rodriguez-Torres, M.D.P.; Acosta-Torres, L.S.; Diaz-Torres, L.A.; Grillo, R.; Swamy, M.K.; Sharma, S.; et al. Nano based drug delivery systems: Recent developments and future prospects. J. Nanobiotechnol. 2018, 16, 71. [Google Scholar] [CrossRef] [Green Version]
- Shin, M.D.; Shukla, S.; Chung, Y.H.; Beiss, V.; Chan, S.K.; Ortega-Rivera, O.A.; Wirth, D.M.; Chen, A.; Sack, M.; Pokorski, J.K.; et al. COVID-19 vaccine development and a potential nanomaterial path forward. Nat. Nanotechnol. 2020, 15, 646–655. [Google Scholar] [CrossRef]
- Kowalski, P.S.; Rudra, A.; Miao, L.; Anderson, D.G. Delivering the Messenger: Advances in Technologies for Therapeutic mRNA Delivery. Mol. Ther. 2019, 27, 710–728. [Google Scholar] [CrossRef] [Green Version]
- Piao, X.; Wang, H.; Binzel, D.W.; Guo, P. Assessment and comparison of thermal stability of phosphorothioate-DNA, DNA, RNA, 2′-F RNA, and LNA in the context of Phi29 pRNA 3WJ. RNA 2018, 24, 67–76. [Google Scholar] [CrossRef] [Green Version]
- Li, H.; Zhang, K.; Binzel, D.W.; Shlyakhtenko, L.S.; Lyubchenko, Y.L.; Chiu, W.; Guo, P. RNA nanotechnology to build a dodecahedral genome of single-stranded RNA virus. RNA Biol. 2021, 18, 2390–2400. [Google Scholar] [CrossRef]
- Hill, A.C.; Schroeder, S.J. Thermodynamic stabilities of three-way junction nanomotifs in prohead RNA. RNA 2017, 23, 521–529. [Google Scholar] [CrossRef] [Green Version]
- Shu, D.; Shu, Y.; Haque, F.; Abdelmawla, S.; Guo, P. Thermodynamically stable RNA three-way junction for constructing multifunctional nanoparticles for delivery of therapeutics. Nat. Nanotechnol. 2011, 6, 658–667. [Google Scholar] [CrossRef] [Green Version]
- Zhuo, B.; Ou, X.; Li, J. Structure and Mechanical Stabilities of the Three-Way Junction Motifs in Prohead RNA. J. Phys. Chem. B 2021, 125, 12125–12134. [Google Scholar] [CrossRef]
- Cai, R.; Price, I.R.; Ding, F.; Wu, F.; Chen, T.; Zhang, Y.; Liu, G.; Jardine, P.J.; Lu, C.; Ke, A. ATP/ADP modulates gp16-pRNA conformational change in the Phi29 DNA packaging motor. Nucleic Acids Res. 2019, 47, 9818–9828. [Google Scholar] [CrossRef]
- Guo, S.; Piao, X.; Li, H.; Guo, P. Methods for construction and characterization of simple or special multifunctional RNA nanoparticles based on the 3WJ of phi29 DNA packaging motor. Methods 2018, 143, 121–133. [Google Scholar] [CrossRef]
- Erill, I.; Caruso, S.M.; Hunters, U.P. Complete Genome Sequences of Three phi29-like Bacillus cereus Group Podoviridae. Genome Announc 2017, 5, e00701-17. [Google Scholar] [CrossRef] [Green Version]
- Liu, X.; Duan, D.; Wang, Y.; Liu, J.; Duan, D. Advancements in 3WJ-based RNA nanotechnology and its application for cancer diagnosis and therapy. Front. Biosci. (Landmark Ed.) 2022, 27, 61. [Google Scholar] [CrossRef]
- Haque, F.; Pi, F.; Zhao, Z.; Gu, S.; Hu, H.; Yu, H.; Guo, P. RNA versatility, flexibility, and thermostability for practice in RNA nanotechnology and biomedical applications. Wiley Interdiscip Rev. RNA 2018, 9, e1452. [Google Scholar] [CrossRef]
- Xu, Z.; Sun, Y.; Weber, J.K.; Cao, Y.; Wang, W.; Jasinski, D.; Guo, P.; Zhou, R.; Li, J. Directional mechanical stability of Bacteriophage phi29 motor’s 3WJ-pRNA: Extraordinary robustness along portal axis. Sci. Adv. 2017, 3, e1601684. [Google Scholar] [CrossRef] [Green Version]
- Fischer, N.M.; Poleto, M.D.; Steuer, J.; van der Spoel, D. Influence of Na+ and Mg2+ ions on RNA structures studied with molecular dynamics simulations. Nucleic Acids Res. 2018, 46, 4872–4882. [Google Scholar] [CrossRef] [Green Version]
- Barros, S.A.; Chenoweth, D.M. Recognition of nucleic acid junctions using triptycene-based molecules. Angew. Chem. Int. Ed. Engl. 2014, 53, 13746–13750. [Google Scholar] [CrossRef] [Green Version]
- Binzel, D.W.; Khisamutdinov, E.F.; Guo, P. Entropy-driven one-step formation of Phi29 pRNA 3WJ from three RNA fragments. Biochemistry 2014, 53, 2221–2231. [Google Scholar] [CrossRef]
- Tan, Z.J.; Chen, S.J. RNA helix stability in mixed Na+/Mg2+ solution. Biophys. J. 2007, 92, 3615–3632. [Google Scholar] [CrossRef] [Green Version]
- Zhang, H.; Endrizzi, J.A.; Shu, Y.; Haque, F.; Sauter, C.; Shlyakhtenko, L.S.; Lyubchenko, Y.; Guo, P.; Chi, Y.I. Crystal structure of 3WJ core revealing divalent ion-promoted thermostability and assembly of the Phi29 hexameric motor pRNA. RNA 2013, 19, 1226–1237. [Google Scholar] [CrossRef] [Green Version]
- Man, V.H.; Wu, X.; He, X.; Xie, X.Q.; Brooks, B.R.; Wang, J. Determination of van der Waals Parameters Using a Double Exponential Potential for Nonbonded Divalent Metal Cations in TIP3P Solvent. J. Chem. Theory Comput. 2021, 17, 1086–1097. [Google Scholar] [CrossRef]
- Allner, O.; Nilsson, L.; Villa, A. Magnesium Ion-Water Coordination and Exchange in Biomolecular Simulations. J. Chem. Theory Comput. 2012, 8, 1493–1502. [Google Scholar] [CrossRef] [Green Version]
- Joung, I.S.; Cheatham, T.E., 3rd. Determination of alkali and halide monovalent ion parameters for use in explicitly solvated biomolecular simulations. J. Phys. Chem. B 2008, 112, 9020–9041. [Google Scholar] [CrossRef] [Green Version]
- Zgarbova, M.; Otyepka, M.; Sponer, J.; Mladek, A.; Banas, P.; Cheatham, T.E., 3rd; Jurecka, P. Refinement of the Cornell et al. Nucleic Acids Force Field Based on Reference Quantum Chemical Calculations of Glycosidic Torsion Profiles. J. Chem. Theory Comput. 2011, 7, 2886–2902. [Google Scholar] [CrossRef]
- Hess, B.; Kutzner, C.; van der Spoel, D.; Lindahl, E. GROMACS 4: Algorithms for Highly Efficient, Load-Balanced, and Scalable Molecular Simulation. J. Chem. Theory Comput. 2008, 4, 435–447. [Google Scholar] [CrossRef] [Green Version]
- Perez, A.; Marchan, I.; Svozil, D.; Sponer, J.; Cheatham, T.E., 3rd; Laughton, C.A.; Orozco, M. Refinement of the AMBER force field for nucleic acids: Improving the description of alpha/gamma conformers. Biophys. J. 2007, 92, 3817–3829. [Google Scholar] [CrossRef] [Green Version]
- Bhadra, P.; Siu, S.W.I. Refined Empirical Force Field to Model Protein-Self-Assembled Monolayer Interactions Based on AMBER14 and GAFF. Langmuir 2019, 35, 9622–9633. [Google Scholar] [CrossRef]
- Hess, B.; Bekker, H.; Berendsen, H.J.C.; Fraaije, J.G.E.M. LINCS: A linear constraint solver for molecular simulations. J. Comput. Chem. 1997, 18, 1463–1472. [Google Scholar] [CrossRef]
- Darden, T.; York, D.; Pedersen, L. Particle mesh Ewald: AnN⋅log(N) method for Ewald sums in large systems. J. Chem. Phys. 1993, 98, 10089–10092. [Google Scholar] [CrossRef] [Green Version]
- Essmann, U.; Perera, L.; Berkowitz, M.L.; Darden, T.; Lee, H.; Pedersen, L.G. A smooth particle mesh Ewald method. J. Chem. Phys. 1995, 103, 8577–8593. [Google Scholar] [CrossRef] [Green Version]
- Venable, R.M.; Kramer, A.; Pastor, R.W. Molecular Dynamics Simulations of Membrane Permeability. Chem. Rev. 2019, 119, 5954–5997. [Google Scholar] [CrossRef] [PubMed]
- Abraham, M.J.; van der Spoel, D.; Lindahl, E.; Hess, B. GROMACS User Manual Version 5.1.2; Uppsala University: Uppsala, Sweden, 2016. [Google Scholar]
- Goddard, T.D.; Huang, C.C.; Meng, E.C.; Pettersen, E.F.; Couch, G.S.; Morris, J.H.; Ferrin, T.E. UCSF ChimeraX: Meeting modern challenges in visualization and analysis. Protein Sci. 2018, 27, 14–25. [Google Scholar] [CrossRef]
- Laureanti, J.; Brandi, J.; Offor, E.; Engel, D.; Rallo, R.; Ginovska, B.; Martinez, X.; Baaden, M.; Baker, N.A. Visualizing biomolecular electrostatics in virtual reality with UnityMol-APBS. Protein Sci. 2020, 29, 237–246. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Simpson, A.A.; Tao, Y.; Leiman, P.G.; Badasso, M.O.; He, Y.; Jardine, P.J.; Olson, N.H.; Morais, M.C.; Grimes, S.; Anderson, D.L.; et al. Structure of the bacteriophage phi29 DNA packaging motor. Nature 2000, 408, 745–750. [Google Scholar] [CrossRef]
- Varani, G.; McClain, W.H. The G x U wobble base pair. A fundamental building block of RNA structure crucial to RNA function in diverse biological systems. EMBO Rep. 2000, 1, 18–23. [Google Scholar] [CrossRef]
- Spencer, M. The stereochemistry of deoxyribonucleic acid. II. Hydrogen-bonded pairs of bases. Acta Crystallogr. 1959, 12, 66–71. [Google Scholar] [CrossRef]

| Area | Type and Label of Nucleobases | Hydrogen Bonds |
|---|---|---|
| Junction | Chain A: 9U Chain C: 10U 11U 12U | 0 |
| Helix 1 | Chain A: 13G 14C 15C 16A 17U 8G Chain C: 14C 13G 12C 11U 10A 9C | 16 |
| Helix 2 | Chain A: 10G 11U Chain B: 19C 18A | 5 |
| Helix 3 | Chain B: 13G 14U 15U 16G 17A Chain C: 18U 17A 16A 15C 14U | 11 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, L.; Li, J. Mg2+ Ions Regulating 3WJ-PRNA to Construct Controllable RNA Nanoparticle Drug Delivery Platforms. Pharmaceutics 2022, 14, 1413. https://doi.org/10.3390/pharmaceutics14071413
Chen L, Li J. Mg2+ Ions Regulating 3WJ-PRNA to Construct Controllable RNA Nanoparticle Drug Delivery Platforms. Pharmaceutics. 2022; 14(7):1413. https://doi.org/10.3390/pharmaceutics14071413
Chicago/Turabian StyleChen, Le, and Jingyuan Li. 2022. "Mg2+ Ions Regulating 3WJ-PRNA to Construct Controllable RNA Nanoparticle Drug Delivery Platforms" Pharmaceutics 14, no. 7: 1413. https://doi.org/10.3390/pharmaceutics14071413
APA StyleChen, L., & Li, J. (2022). Mg2+ Ions Regulating 3WJ-PRNA to Construct Controllable RNA Nanoparticle Drug Delivery Platforms. Pharmaceutics, 14(7), 1413. https://doi.org/10.3390/pharmaceutics14071413





