A Quality by Design Approach in Pharmaceutical Development of Non-Viral Vectors with a Focus on miRNA
Abstract
:1. Introduction
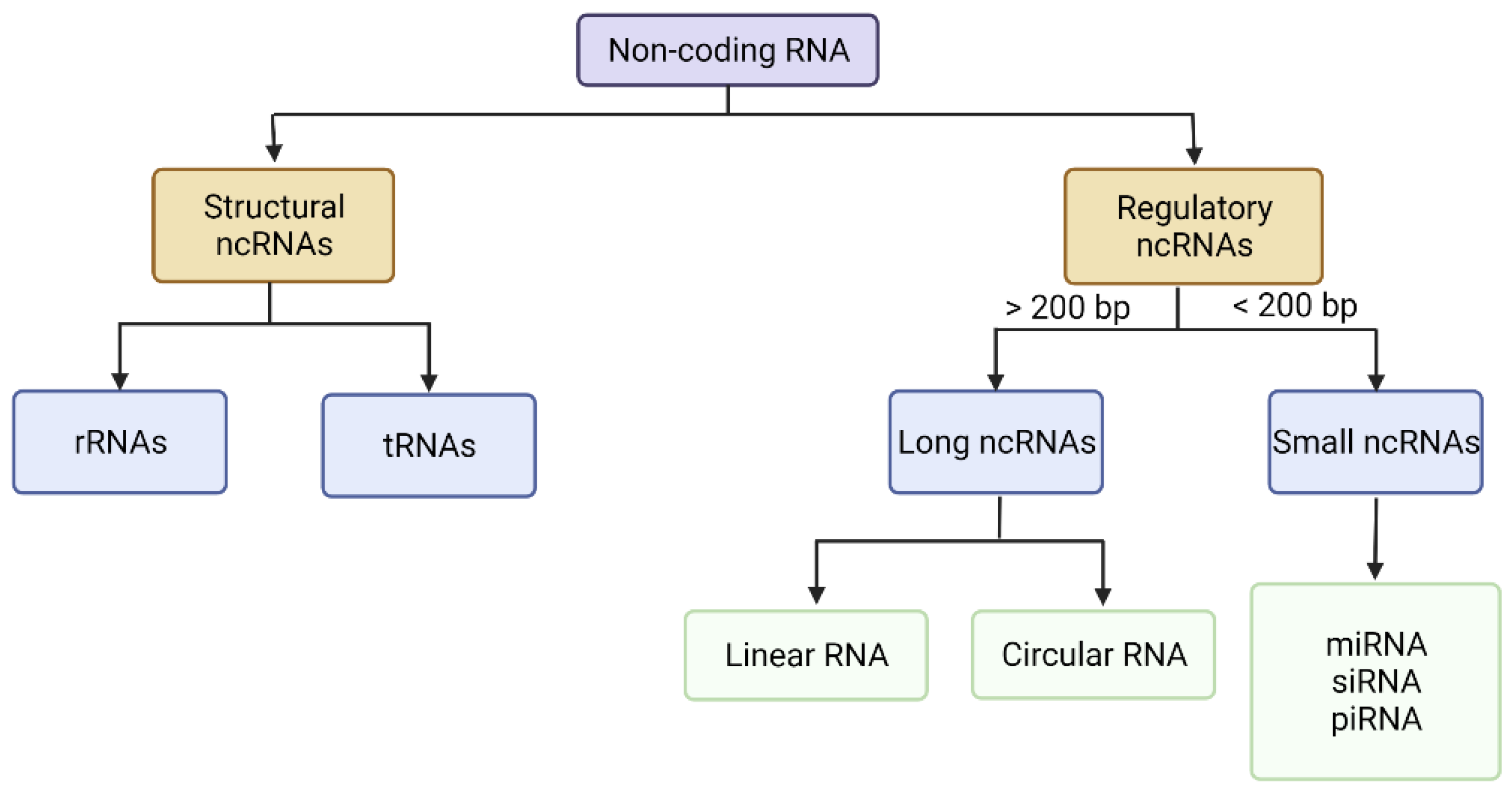
2. The Genetic Material Used in Cancer Gene Therapy
2.1. Long Non-Coding RNAs (lncRNAs)
2.2. Small Non-Coding RNAs
2.2.1. miRNA
2.2.2. siRNA
2.2.3. piRNA
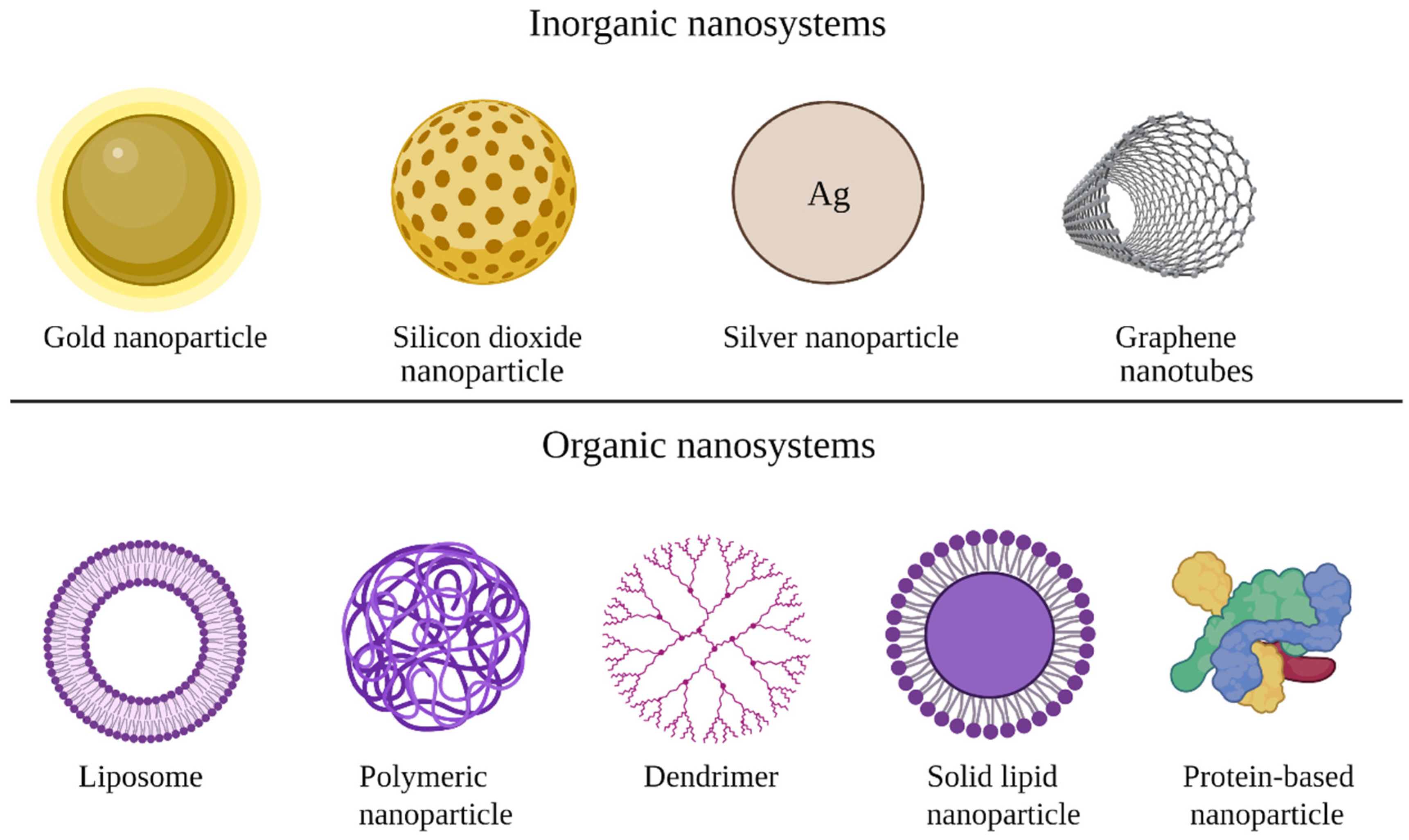
3. Types of Non-Viral Vectors
| Nanosystem | Advantages | Limitations | Ref. |
|---|---|---|---|
| Silica nanoparticles | Biocompatibility Biodegradability High surface area Versatility Surface charge control Free dispersion throughout the body | Low transfection efficiency | [17,53] |
| Gold nanoparticles | Uniformity in size, shape, and biodistribution Tuned pharmacokinetics Increased surface area Biocompatibility Easy surface modification Controlled drug release Stability Strong gene carrying ability | Toxicity issues | [54,55,56] |
| Dendrimers | Presence of surface functional groups make them suitable for modification Good long-term stability | Expensive option | [9] |
| Polymeric nanoparticles | Biocompatible Biodegradable Non-immunogenic and non-toxic Easily fabricated in large quantities Low-cost Long-term stability | Prone to degradation Potential antigenicity Low transfection efficiency The need for surface modification | [9,57] |
| Liposomes | Biocompatibility Longer circulation time Amphiphilic | Expensive option Stability issues | [9,54] |
| Solid lipid nanoparticles | Biocompatibility Low toxicity Feasible to scale up Easy to sterilize | Low incorporation rates resulting from the crystalline structure of the solid lipid Lipid particle growth | [58,59] |
3.1. Organic Nanoparticles
3.2. Inorganic Nanoparticles
4. Quality by Design (QbD) Approach in Non-Viral Vector Development
4.1. Quality Target Product Profile (QTPP) of Non-Viral Vectors
4.2. Critical Quality Attributes (CQAs) and Their Evaluation
4.2.1. Critical Quality Attributes (CQAs)
4.2.2. Nanoparticle Physical Characterization Methods and Their Transfection Efficiency Determination
| Characteristics of Nanoparticles | Method | Principle | Ref. |
|---|---|---|---|
| Particle size | DLS | Measures particle size using Brownian motion | [79,86,87] |
| PDI | |||
| Surface charge | Laser Doppler electrophoresis | Measures the particles’ frequency, obtaining electrophoretic mobility of the charged particles | [88] |
| Particle shape, morphology | Electron microscopy | Detection of reflected electrons, or transmission of electrons that pass through the sample | [82] |
| Cellular uptake | Flow cytometry | It uses fluorescence emission, which occurs as light from a laser beam strikes the moving particles. Based on the median fluorescence intensity, the area under the curve is calculated | [81,87,89,90] |
| Cell lines fluorescence microscopy | After a predefined treatment, the cells are observed by fluorescence microscopy | ||
| Intracellular localization | Confocal microscopy | The illumination and detection optics are focused on the same diffraction-limited spot in the sample, which is the only spot imaged by the detector during a confocal scan | [79,91] |
| Transfection efficiency | Cell lines RT-PCR | Nanoparticles are incubated with the cells and the level of nucleic acid is measured by RT-PCR | [79,81] |
| Flow cytometry | The mean fluorescence intensity values correspond to the approximate number of fluorescent molecules associated with a cell | [83] | |
| Cytotoxicity | Colony formation assay | Cells are treated after a predefined protocol, colored and the number of colonies is counted via an optical microscope | [81,92] |
| Cell viability | Cells are treated after a predefined protocol with the MTT solution. The cell viability is expressed as the percentage of the absorbance of the sample to that of the untreated cells | ||
| EE | Measurement followed by calculation | Determination of the percentage of genetic material encapsulated into non-viral vectors to the initial amount of genetic material included in the formulation | [38,79] |
4.2.3. Quantitative and Qualitative Evaluation of Encapsulated Genetic Material
| Objective | Method | Ref. |
|---|---|---|
| RNA or DNA quantification | UV-Vis spectrophotometry | [93,96,97] |
| Fluorescence spectrophotometry | [79] | |
| Target-specific fluorescence detection | [93,96] | |
| Capillary electrophoresis separation of fluorescently labelled nucleic acids | [93,96] | |
| qRT-PCR | [97,98,99] | |
| Gel electrophoresis on 1% agarose gel | [87] | |
| miRNA expression profiles | Provides 100% coverage of the miRNAs in the miRBase database | [96] |
4.3. Risk Assesment
4.4. Fabrication Process Understanding and Critical Process Parameters (CPPs)
4.5. Excipients Used and Critical Material Attributes (CMAs)
4.6. DoE and Linking CQAs to CMAs and CPPs
5. Conclusions and Future Perspectives
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Coelho, J.F.; Ferreira, P.C.; Alves, P.; Cordeiro, R.; Fonseca, A.C.; Góis, J.R.; Gil, M.H. Drug Delivery Systems: Advanced Technologies Potentially Applicable in Personalized Treatments. EPMA J. 2010, 1, 164–209. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- International Conference of Harmonisation. ICH Guideline S12 on Nonclinical Biodistribution Considerations for Gene Therapy Products. 2021. Available online: https://www.ema.europa.eu/en/documents/regulatory-procedural-guideline/ich-guideline-s12-nonclinical-biodistribution-considerations-gene-therapy-products-step-2b_en.pdf (accessed on 19 May 2022).
- FDA. Long Term Follow-Up after Administration of Human Gene Therapy Products—Guidance for Industry; FDA: Silver Spring, MD, USA, 2020; pp. 1–37.
- Ramamoorth, M.; Narvekar, A. Non Viral Vectors in Gene Therapy—An Overview. J. Clin. Diagn. Res. 2015, 9, GE01–GE06. [Google Scholar] [CrossRef] [PubMed]
- de Matos, M.B.C.; Miranda, B.S.; Rizky Nuari, Y.; Storm, G.; Leneweit, G.; Schiffelers, R.M.; Kok, R.J. Liposomes with Asymmetric Bilayers Produced from Inverse Emulsions for Nucleic Acid Delivery. J. Drug Target. 2019, 27, 681–689. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chaudhary, V.; Jangra, S.; Yadav, N.R. Nanotechnology Based Approaches for Detection and Delivery of MicroRNA in Healthcare and Crop Protection. J. Nanobiotechnol. 2018, 16, 40. [Google Scholar] [CrossRef] [Green Version]
- Bleiziffer, O.; Eriksson, E.; Yao, F.; Horch, R.E.; Kneser, U. Gene Transfer Strategies in Tissue Engineering: Tissue Engineering Review Series. J. Cell. Mol. Med. 2007, 11, 206–223. [Google Scholar] [CrossRef] [Green Version]
- Guo, S.; Huang, Y.; Jiang, Q.; Sun, Y.; Deng, L.; Liang, Z.; Du, Q.; Xing, J.; Zhao, Y.; Wang, P.C.; et al. Enhanced Gene Delivery and SiRNA Silencing by Gold Nanoparticles Coated with Charge-Reversal Polyelectrolyte. ACS Nano 2010, 4, 5505–5511. [Google Scholar] [CrossRef] [Green Version]
- Eftekhari, R.B.; Maghsoudnia, N.; Samimi, S.; Zamzami, A.; Dorkoosh, F.A. Co-Delivery Nanosystems for Cancer Treatment: A Review. Pharm. Nanotechnol. 2019, 7, 90–112. [Google Scholar] [CrossRef]
- Dunbar, C.E.; High, K.A.; Joung, J.K.; Kohn, D.B.; Ozawa, K.; Sadelain, M. Gene Therapy Comes of Age. Science 2018, 359, eaan4672. [Google Scholar] [CrossRef] [Green Version]
- Ginn, S.L.; Amaya, A.K.; Alexander, I.E.; Edelstein, M.; Abedi, M.R. Gene Therapy Clinical Trials Worldwide to 2017: An Update. J. Gene Med. 2018, 20, e3015. [Google Scholar] [CrossRef]
- Berg, J.; Tymoczko, J.; Stryer, L. Biochemistry, 5th ed.; WH and Company: Bürmoos, Austria, 2002. [Google Scholar]
- Bhise, K.; Kashaw, S.K.; Sau, S.; Iyer, A.K. Nanostructured Lipid Carriers Employing Polyphenols as Promising Anticancer Agents: Quality by Design (QbD) Approach. Int. J. Pharm. 2017, 526, 506–515. [Google Scholar] [CrossRef]
- Fukuda, I.M.; Pinto, C.F.F.; Moreira, C.D.S.; Saviano, A.M.; Lourenço, F.R. Design of Experiments (DoE) Applied to Pharmaceutical and Analytical Quality by Design (QbD). Braz. J. Pharm. Sci. 2018, 54, e01006. [Google Scholar] [CrossRef]
- Spiegel, J.; Adhikari, S.; Balasubramanian, S. The Structure and Function of DNA G-Quadruplexes. Trends Chem. 2020, 2, 123–136. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brázda, V.; Fojta, M. The Rich World of P53 DNA Binding Targets: The Role of Dna Structure. Int. J. Mol. Sci. 2019, 20, 5605. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Romano, G.; Veneziano, D.; Acunzo, M.; Croce, C.M. Small Non-Coding RNA and Cancer. Carcinogenesis 2017, 38, 485–491. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jarroux, J.; Morillon, A.; Pinskaya, M. History, Discovery, and Classification of LncRNAs. In Long Non Coding RNA Biology; Springer: Singapore, 2017; Volume 1008, ISBN 9789811052033. [Google Scholar]
- Amin, N.; McGrath, A.; Chen, Y.P.P. Evaluation of Deep Learning in Non-Coding RNA Classification. Nat. Mach. Intell. 2019, 1, 246–256. [Google Scholar] [CrossRef]
- Li, J.; Liu, C. Coding or Noncoding, the Converging Concepts of RNAs. Front. Genet. 2019, 10, 496. [Google Scholar] [CrossRef]
- Dahariya, S.; Paddibhatla, I.; Kumar, S.; Raghuwanshi, S.; Pallepati, A.; Gutti, R.K. Long Non-Coding RNA: Classification, Biogenesis and Functions in Blood Cells. Mol. Immunol. 2019, 112, 82–92. [Google Scholar] [CrossRef]
- Chan, J.J.; Tay, Y. Noncoding RNA: RNA Regulatory Networks in Cancer. Int. J. Mol. Sci. 2018, 19, 1310. [Google Scholar] [CrossRef] [Green Version]
- Anastasiadou, E.; Jacob, L.S.; Slack, F.J. Non-Coding RNA Networks in Cancer. Nat. Rev. Cancer 2017, 18, 5–18. [Google Scholar] [CrossRef]
- Costa, M.C.; Leitão, A.L.; Enguita, F.J. Biogenesis and Mechanism of Action of Small Non-Coding RNAs: Insights from the Point of View of Structural Biology. Int. J. Mol. Sci. 2012, 13, 10268–10295. [Google Scholar] [CrossRef] [Green Version]
- Chao, Y.; Li, L.; Girodat, D.; Förstner, K.U.; Said, N.; Corcoran, C.; Śmiga, M.; Papenfort, K.; Reinhardt, R.; Wieden, H.J.; et al. In Vivo Cleavage Map Illuminates the Central Role of RNase E in Coding and Non-Coding RNA Pathways. Mol. Cell 2017, 65, 39–51. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Si, W.; Shen, J.; Zheng, H.; Fan, W. The Role and Mechanisms of Action of MicroRNAs in Cancer Drug Resistance. Clin. Epigenet. 2019, 11, 25. [Google Scholar] [CrossRef]
- Asghariazar, V.; Sakhinia, E.; Mansoori, B.; Mohammadi, A.; Baradaran, B. Tumor Suppressor MicroRNAs in Lung Cancer: An Insight to Signaling Pathways and Drug Resistance. J. Cell. Biochem. 2019, 120, 19274–19289. [Google Scholar] [CrossRef] [PubMed]
- Svoronos, A.A.; Engelman, D.M.; Slack, F.J. OncomiR or Tumor Suppressor? The Duplicity of MicroRNAs in Cancer. Cancer Res. 2016, 76, 3666–3670. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Malumbres, M. miRNAs and Cancer: An Epigenetics View. Mol. Asp. Med. 2013, 34, 863–874. [Google Scholar] [CrossRef] [Green Version]
- Catto, J.W.F.; Alcaraz, A.; Bjartell, A.S.; De Vere White, R.; Evans, C.P.; Fussel, S.; Hamdy, F.C.; Kallioniemi, O.; Mengual, L.; Schlomm, T.; et al. MicroRNA in Prostate, Bladder, and Kidney Cancer: A Systematic Review. Eur. Urol. 2011, 59, 671–681. [Google Scholar] [CrossRef] [PubMed]
- Xue, J.; Yang, J.; Luo, M.; Cho, W.C.; Liu, X. MicroRNA-Targeted Therapeutics for Lung Cancer Treatment. Expert Opin. Drug Discov. 2017, 12, 141–157. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Gao, D.Y.; Huang, L. In Vivo Delivery of miRNAs for Cancer Therapy: Challenges and Strategies. Adv. Drug Deliv. Rev. 2015, 81, 128–141. [Google Scholar] [CrossRef] [Green Version]
- Saito, Y.; Jones, P.A. Epigenetic Activation of Tumor Suppressor MicroRNAs in Human Cancer Cells. Cell Cycle 2006, 5, 2220–2222. [Google Scholar] [CrossRef]
- Bonneau, E.; Neveu, B.; Kostantin, E.; Tsongalis, G.J.; De Guire, V. How Close Are MiRNAs from Clinical Practice? A Perspective on the Diagnostic and Therapeutic Market. Electron. J. Int. Fed. Clin. Chem. Lab. Med. 2019, 30, 114–127. [Google Scholar]
- Ganju, A.; Khan, S.; Hafeez, B.B.; Behrman, S.W.; Yallapu, M.M.; Chauhan, S.C.; Jaggi, M. miRNA Nanotherapeutics for Cancer. Drug Discov. Today 2017, 22, 424–432. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, S.W.L.; Paoletti, C.; Campisi, M.; Osaki, T.; Adriani, G.; Kamm, R.D.; Mattu, C.; Chiono, V. MicroRNA Delivery through Nanoparticles. J. Control Release 2019, 313, 80–95. [Google Scholar] [CrossRef] [PubMed]
- Hu, B.; Zhong, L.; Weng, Y.; Peng, L.; Huang, Y.; Zhao, Y.; Liang, X.J. Therapeutic SiRNA: State of the Art. Signal Transduct. Target. Ther. 2020, 5, 101. [Google Scholar] [CrossRef] [PubMed]
- Bochicchio, S.; Dapas, B.; Russo, I.; Ciacci, C.; Piazza, O.; De Smedt, S.; Pottie, E.; Barba, A.A.; Grassi, G. In Vitro and Ex Vivo Delivery of Tailored SiRNA-Nanoliposomes for E2F1 Silencing as a Potential Therapy for Colorectal Cancer. Int. J. Pharm. 2017, 525, 377–387. [Google Scholar] [CrossRef]
- Zhang, Z.; Zhang, J.; Diao, L.; Han, L. Small Non-Coding RNAs in Human Cancer: Function, Clinical Utility, and Characterization. Oncogene 2021, 40, 1570–1577. [Google Scholar] [CrossRef]
- Shim, G.; Kim, D.; Le, Q.-V.; Park, G.T.; Kwon, T.; Oh, Y.-K. Nonviral Delivery Systems For Cancer Gene Therapy: Strategies And Challenges. Curr. Gene Ther. 2018, 18, 3–20. [Google Scholar] [CrossRef]
- Ghosh, S.; Brown, A.M.; Jenkins, C.; Campbell, K. Viral Vector Systems for Gene Therapy: A Comprehensive Literature Review of Progress and Biosafety Challenges. Appl. Biosaf. 2020, 25, 7–18. [Google Scholar] [CrossRef]
- Atkinson, H.; Chalmers, R. Delivering the Goods: Viral and Non-Viral Gene Therapy Systems and the Inherent Limits on Cargo DNA and Internal Sequences. Genetica 2010, 138, 485–498. [Google Scholar] [CrossRef]
- Ahlawat, J.; Guillama Barroso, G.; Narayan, M.; Alvarado, M.; Armendariz, I.; Bernal, J.; Carabaza, X.; Chavez, S.; Cruz, P.; Escalante, V.; et al. Nanocarriers as Potential Drug Delivery Candidates for Overcoming the Blood-Brain Barrier: Challenges and Possibilities. ACS Omega 2020, 5, 12583–12595. [Google Scholar] [CrossRef]
- Kimura, S.; Harashima, H. Current Status and Challenges Associated with CNS-Targeted Gene Delivery across the BBB. Pharmaceutics 2020, 12, 1216. [Google Scholar] [CrossRef]
- Lombardo, S.M.; Schneider, M.; Türeli, A.E.; Türeli, N.G. Key for Crossing the BBB with Nanoparticles: The Rational Design. Beilstein J. Nanotechnol. 2020, 11, 866–883. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z. Development of A Liposomal Formulation for Brain Targeting. Ph.D. Thesis, The University of Mississippi, Oxford, MS, USA, 2020. [Google Scholar]
- Zhi, D.; Zhang, S.; Cui, S.; Zhao, Y.; Wang, Y.; Zhao, D. The Headgroup Evolution of Cationic Lipids for Gene Delivery. Bioconjug. Chem. 2013, 24, 487–519. [Google Scholar] [CrossRef] [PubMed]
- Pardridge, W.M. Blood-Brain Barrier and Delivery of Protein and Gene Therapeutics to Brain. Front. Aging Neurosci. 2020, 11, 1373. [Google Scholar] [CrossRef] [PubMed]
- Roma-Rodrigues, C.; Rivas-garcía, L.; Baptista, P.V.; Fernandes, A.R. Gene Therapy in Cancer Treatment: Why Go Nano? Pharmaceutics 2020, 12, 233. [Google Scholar] [CrossRef] [Green Version]
- Awasthi, R.; Rathbone, M.J.; Hansbro, P.M.; Bebawy, M.; Dua, K. Therapeutic Prospects of MicroRNAs in Cancer Treatment through Nanotechnology. Drug Deliv. Transl. Res. 2018, 8, 97–110. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- He, Z.; Liu, K.; Byrne, H.J.; Cullen, P.J.; Tian, F.; Curtin, J.F. Combination Strategies for Targeted Delivery of Nanoparticles for Cancer Therapy. In Applications of Targeted Nano Drugs and Delivery Systems, 1st ed.; Mohapatra, S.S., Ranjan, S., Dasgupta, N., Mishra, R.K., Thomas, S., Eds.; Elsevier: Amsterdam, The Netherlands, 2019; pp. 191–219. ISBN 9780128140291. [Google Scholar]
- Zhang, M.; Liu, E.; Cui, Y.; Huang, Y. Nanotechnology-Based Combination Therapy for Overcoming Multidrug-Resistant Cancer. Cancer Biol. Med. 2017, 14, 212–227. [Google Scholar] [CrossRef] [Green Version]
- Bharti, C.; Gulati, N.; Nagaich, U.; Pal, A. Mesoporous Silica Nanoparticles in Target Drug Delivery System: A Review. Int. J. Pharm. Investig. 2015, 5, 124. [Google Scholar] [CrossRef] [Green Version]
- Lamberti, M.; Zappavigna, S.; Sannolo, N.; Porto, S.; Caraglia, M. Advantages and Risks of Nanotechnologies in Cancer Patients and Occupationally Exposed Workers. Expert Opin. Drug Deliv. 2014, 11, 1087–1101. [Google Scholar] [CrossRef]
- Ghosn, Y.; Kamareddine, M.H.; Tawk, A.; Elia, C.; El Mahmoud, A.; Terro, K.; El Harake, N.; El-Baba, B.; Makdessi, J.; Farhat, S. Inorganic Nanoparticles as Drug Delivery Systems and Their Potential Role in the Treatment of Chronic Myelogenous Leukaemia. Technol. Cancer Res. Treat. 2019, 18, 1–12. [Google Scholar] [CrossRef]
- Sun, N.-F.; Liu, Z.-A.; Huang, W.-B.; Tian, A.-L.; Hu, S.-Y. The Research of Nanoparticles as Gene Vector for Tumor Gene Therapy. Crit. Rev. Oncol. Hematol. 2014, 89, 352–357. [Google Scholar] [CrossRef]
- Bhatia, S. Natural Polymer Drug Delivery Systems: Nanoparticles, Plants, and Algae, 1st ed.; Springer: Berlin/Heidelberg, Germany, 2016. [Google Scholar]
- Dey, S.; Datta, S.; Dasgupta, S.; Mazumder, B.; Pathak, Y.V. Lipid Nanoparticles for Topical Application of Drugs for Skin Diseases; Elsevier Inc.: Amsterdam, The Netherlands, 2016; ISBN 9780323428910. [Google Scholar]
- Naseri, N.; Valizadeh, H.; Zakeri-Milani, P. Solid Lipid Nanoparticles and Nanostructured Lipid Carriers: Structure Preparation and Application. Adv. Pharm. Bull. 2015, 5, 305–313. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nagavarma, B.V.N.; Yadav, H.K.S.; Ayaz, A.; Vasudha, L.S.; Shivakumar, H.G. Different Techniques for Preparation of Polymeric Nanoparticles—A Review. Asian J. Pharm. Clin. Res. 2012, 5, 16–23. [Google Scholar]
- Mittal, P.; Saharan, A.; Verma, R.; Altalbawy, F.M.A.; Alfaidi, M.A.; Batiha, G.E.S.; Akter, W.; Gautam, R.K.; Uddin, M.S.; Rahman, M.S. Dendrimers: A New Race of Pharmaceutical Nanocarriers. Biomed. Res. Int. 2021, 2021, 8844030. [Google Scholar] [CrossRef]
- Dzmitruk, V.; Apartsin, E.; Ihnatsyeu-Kachan, A.; Abashkin, V.; Shcharbin, D.; Bryszewska, M. Dendrimers Show Promise for SiRNA and Microrna Therapeutics. Pharmaceutics 2018, 10, 126. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barba, A.A.; Bochicchio, S.; Dalmoro, A.; Lamberti, G. Lipid Delivery Systems for Nucleic-Acid-Based-Drugs: From Production to Clinical Applications. Pharmaceutics 2019, 11, 360. [Google Scholar] [CrossRef] [Green Version]
- Crespo-Barreda, A.; Encabo-Berzosa, M.M.; González-Pastor, R.; Ortíz-Teba, P.; Iglesias, M.; Serrano, J.L.; Martin-Duque, P. Viral and Nonviral Vectors for In Vivo and Ex Vivo Gene Therapies. Transl. Regen. Med. Clin. 2016, 1, 155–177. [Google Scholar] [CrossRef]
- European Medicines Agency. ICH Guideline Q8 (R2) on Pharmaceutical Development; International Conference of Harmonisation; European Medicines Agency: Amsterdam, The Netherlands, 2017.
- Singh, H.; Khurana, L.K.; Singh, R. Pharmaceutical Development; Elsevier Inc.: Amsterdam, The Netherlands, 2017; ISBN 9780128021033. [Google Scholar]
- Beg, S.; Hasmain, S. Pharmaceutical Quality by Design; Elsevier Inc.: Amsterdam, The Netherlands, 2019; ISBN 9780128157992. [Google Scholar]
- Dhoble, S.; Patravale, V. Development of Anti-Angiogenic Erlotinib Liposomal Formulation for Pulmonary Hypertension: A QbD Approach. Drug Deliv. Transl. Res. 2019, 9, 980–996. [Google Scholar] [CrossRef]
- Food and Drug Administration. Guidance for Industry, PAT-A Framework for Innovative Pharmaceutical Development, Manufacturing and Quality Assurance; Food and Drug Administration: Silver Spring, MD, USA, 2004.
- Waghule, T.; Dabholkar, N.; Gorantla, S.; Rapalli, V.K.; Saha, R.N.; Singhvi, G. Quality by Design (QbD) in the Formulation and Optimization of Liquid Crystalline Nanoparticles (LCNPs): A Risk Based Industrial Approach. Biomed. Pharmacother. 2021, 141, 111940. [Google Scholar] [CrossRef]
- Stagner, W.C.; Haware, R.V. QbD Innovation Through Advances in PAT, Data Analysis Methodologies, and Material Characterization. AAPS PharmSciTech 2019, 20, 295. [Google Scholar] [CrossRef] [Green Version]
- Besseling, R.; Damen, M.; Wijgergangs, J.; Hermes, M.; Wynia, G.; Gerich, A. New Unique PAT Method and Instrument for Real-Time Inline Size Characterization of Concentrated, Flowing Nanosuspensions. Eur. J. Pharm. Sci. 2019, 133, 205–213. [Google Scholar] [CrossRef]
- Pandey, A.P.; Karande, K.P.; Sonawane, R.O.; Deshmukh, P.K. Applying Quality by Design (QbD) Concept for Fabrication of Chitosan Coated Nanoliposomes. J. Liposome Res. 2014, 24, 37–52. [Google Scholar] [CrossRef]
- Tanaka, T.; Hanaoka, H.; Sakurai, S. Optimization of the Quality by Design Approach for Gene Therapy Products: A Case Study for Adeno-Associated Viral Vectors. Eur. J. Pharm. Biopharm. 2020, 155, 88–102. [Google Scholar] [CrossRef] [PubMed]
- Rathore, A.S. Roadmap for Implementation of Quality by Design (QbD) for Biotechnology Products. Trends Biotechnol. 2009, 27, 546–553. [Google Scholar] [CrossRef] [PubMed]
- Gan, L.; Li, Z.; Lv, Q.; Huang, W. Rabies Virus Glycoprotein (RVG29)-Linked MicroRNA-124-Loaded Polymeric Nanoparticles Inhibit Neuroinflammation in a Parkinson’s Disease Model. Int. J. Pharm. 2019, 567, 118449. [Google Scholar] [CrossRef] [PubMed]
- Mohamed, A.; Kunda, N.K.; Ross, K.; Hutcheon, G.A.; Saleem, I.Y. Polymeric Nanoparticles for the Delivery of MiRNA to Treat Chronic Obstructive Pulmonary Disease (COPD). Eur. J. Pharm. Biopharm. 2019, 136, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Hong, S.T.; Lin, H.; Wang, C.S.; Chang, C.H.; Lin, A.M.Y.; Yang, J.C.H.; Lo, Y.L. Improving the Anticancer Effect of Afatinib and MicroRNA by Using Lipid Polymeric Nanoparticles Conjugated with Dual PH-Responsive and Targeting Peptides. J. Nanobiotechnol. 2019, 17, 89. [Google Scholar] [CrossRef] [Green Version]
- Yan, Y.; Li, X.Q.; Duan, J.L.; Bao, C.J.; Cui, Y.N.; Su, Z.B.; Xu, J.R.; Luo, Q.; Chen, M.; Xie, Y.; et al. Nanosized Functional MiRNA Liposomes and Application in the Treatment of TNBC by Silencing Slug Gene. Int. J. Nanomed. 2019, 14, 3645–3667. [Google Scholar] [CrossRef] [Green Version]
- Shi, S.J.; Zhong, Z.R.; Liu, J.; Zhang, Z.R.; Sun, X.; Gong, T. Solid Lipid Nanoparticles Loaded with Anti-MicroRNA Oligonucleotides (AMOs) for Suppression of MicroRNA-21 Functions in Human Lung Cancer Cells. Pharm. Res. 2012, 29, 97–109. [Google Scholar] [CrossRef]
- Cui, X.; Song, K.; Lu, X.; Feng, W.; Di, W. Liposomal Delivery of MicroRNA-7 Targeting EGFR to Inhibit the Growth, Invasion, and Migration of Ovarian Cancer. ACS Omega 2021, 6, 11669–11678. [Google Scholar] [CrossRef]
- Aliofkhazraei, M. Handbook of Nanoparticles; Springer International Publishing: Cham, Switzerland, 2015; ISBN 9783319153384. [Google Scholar]
- Homann, S.; Hofmann, C.; Gorin, A.M.; Nguyen, H.C.X.; Huynh, D.; Hamid, P.; Maithel, N.; Yacoubian, V.; Mu, W.; Kossyvakis, A.; et al. A Novel Rapid and Reproducible Flow Cytometric Method for Optimization of Transfection Efficiency in Cells. PLoS ONE 2017, 12, e0182941. [Google Scholar] [CrossRef] [Green Version]
- Du, Z.; Munye, M.M.; Tagalakis, A.D.; Manunta, M.D.I.; Hart, S.L. The Role of the Helper Lipid on the DNA Transfection Efficiency of Lipopolyplex Formulations. Sci. Rep. 2014, 4, 4–9. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ibuki, Y.; Toyooka, T. Nanoparticle Uptake Measured by Flow Cytometry. Methods Mol. Biol. 2012, 926, 157–166. [Google Scholar] [CrossRef] [PubMed]
- Maroof, H.; Islam, F.; Dong, L.; Ajjikuttira, P.; Gopalan, V.; McMillan, N.; Lam, A. Liposomal Delivery of MiR-34b-5p Induced Cancer Cell Death in Thyroid Carcinoma. Cells 2018, 7, 265. [Google Scholar] [CrossRef] [Green Version]
- Lee, H.Y.; Mohammed, K.A.; Kaye, F.; Sharma, P.; Moudgil, B.M.; Clapp, W.L.; Nasreen, N. Targeted Delivery of Let-7a MicroRNA Encapsulated Ephrin-A1 Conjugated Liposomal Nanoparticles Inhibit Tumor Growth in Lung Cancer. Int. J. Nanomed. 2013, 8, 4481–4494. [Google Scholar] [CrossRef] [Green Version]
- Ja’afar, F.; Leow, C.H.; Garbin, V.; Sennoga, C.A.; Tang, M.X.; Seddon, J.M. Surface Charge Measurement of Sonovue, Definity and Optison: A Comparison of Laser Doppler Electrophoresis and Micro-Electrophoresis. Ultrasound Med. Biol. 2015, 41, 2990–3000. [Google Scholar] [CrossRef]
- Adan, A.; Alizada, G.; Kiraz, Y.; Baran, Y.; Nalbant, A. Flow Cytometry: Basic Principles and Applications. Crit. Rev. Biotechnol. 2017, 37, 163–176. [Google Scholar] [CrossRef]
- Blakney, A.K.; Deletic, P.; McKay, P.F.; Bouton, C.R.; Ashford, M.; Shattock, R.J.; Sabirsh, A. Effect of Complexing Lipids on Cellular Uptake and Expression of Messenger RNA in Human Skin Explants. J. Control Release 2021, 330, 1250–1261. [Google Scholar] [CrossRef]
- Elliott, A.D. Confocal Microscopy: Principles and Modern Practices. Curr. Protoc. Cytom. 2020, 92, e68. [Google Scholar] [CrossRef] [PubMed]
- Qin, L.; Cao, D.; Huang, H.; Ji, G.; Feng, M.; Chen, J.; Pan, S. Improvement of Cellular Uptake and Transfection Ability of PDNA Using α-Cyclodextrin-Polyamidoamine Conjugates as Gene Delivery System. J. Biomed. Nanotechnol. 2016, 12, 261–273. [Google Scholar] [CrossRef]
- Wright, K.; de Silva, K.; Purdie, A.C.; Plain, K.M. Comparison of Methods for MiRNA Isolation and Quantification from Ovine Plasma. Sci. Rep. 2020, 10, 825. [Google Scholar] [CrossRef]
- Liu, J.; Meng, T.; Yuan, M.; Wen, L.J.; Cheng, B.L.; Liu, N.; Huang, X.; Hong, Y.; Yuan, H.; Hu, F.Q. MicroRNA-200c Delivered by Solid Lipid Nanoparticles Enhances the Effect of Paclitaxel on Breast Cancer Stem Cell. Int. J. Nanomed. 2016, 11, 6713–6725. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shi, S.; Han, L.; Deng, L.; Zhang, Y.; Shen, H.; Gong, T.; Zhang, Z.; Sun, X. Dual Drugs (MicroRNA-34a and Paclitaxel)-Loaded Functional Solid Lipid Nanoparticles for Synergistic Cancer Cell Suppression. J. Control. Release 2014, 194, 228–237. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Elias, A.; Alloza, L.; Puigdecanet, E.; Nonell, L.; Tajes, M.; Curado, J.; Enjuanes, C.; Díaz, O.; Bruguera, J.; Martí-Almor, J.; et al. Defining Quantification Methods and Optimizing Protocols for Microarray Hybridization of Circulating MicroRNAs. Sci. Rep. 2017, 7, 7725. [Google Scholar] [CrossRef] [PubMed]
- Merhautová, J.; Vychytilová-Faltejsková, P.; Demlová, R.; Slabý, O. Systemic Administration of MiRNA Mimics by Liposomal Delivery System in Animal Model of Colorectal Carcinoma. Physiol. Res. 2016, 65, S481–S488. [Google Scholar] [CrossRef] [PubMed]
- Schuster, C.; Budczies, J.; Faber, C.; Kirchner, T.; Hlubek, F. MicroRNA Expression Profiling of Specific Cells in Complex Archival Tissue Stained by Immunohistochemistry. Lab. Investig. 2011, 91, 157–165. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Binderup, H.G.; Madsen, J.S.; Helweg Heegaard, N.H.; Houlind, K.; Andersen, R.F.; Brasen, C.L. Quantification of MicroRNA Levels in Plasma—Impact of Preanalytical and Analytical Conditions. PLoS ONE 2018, 13, e0201069. [Google Scholar] [CrossRef]
- International Conference Harmonisation. ICH Guideline Q9 (R1) on Quality Risk Management. 2021. Available online: https://www.ema.europa.eu/en/documents/scientific-guideline/draft-international-conference-harmonisation-technical-requirements-registration-pharmaceuticals_en-1.pdf (accessed on 19 May 2022).
- Tefas, L.R.; Rus, L.M.; Achim, M.; Vlase, L.; Tomuță, I. Application of the Quality by Design Concept in the Development of Quercetin-Loaded Polymeric Nanoparticles. Farmacia 2018, 66, 798–810. [Google Scholar] [CrossRef]
- Mannan, S. Lees’ Loss Prevention in the Process Industries; Butterworth-Heinemann: Oxfordb, UK, 2012; ISBN 9780123971890. [Google Scholar]
- Hakemeyer, C.; McKnight, N.; John, R.S.; Meier, S.; Trexler-Schmidt, M.; Kelley, B.; Zettl, F.; Puskeiler, R.; Kleinjans, A.; Lim, F.; et al. Process Characterization and Design Space Definition. Biologicals 2016, 44, 306–318. [Google Scholar] [CrossRef]
- Csaszar, E.; Mills, S.; Zandstra, P.W. Process Evolution in Cell and Gene Therapy from Discovery to Commercialization. Can. J. Chem. Eng. 2021, 99, 2517–2524. [Google Scholar] [CrossRef]
- Worsham, R.D.; Thomas, V.; Farid, S.S. Potential of Continuous Manufacturing for Liposomal Drug Products. Biotechnol. J. 2019, 14, e1700740. [Google Scholar] [CrossRef] [Green Version]
- Elbakry, A.; Zaky, A.; Liebl, R.; Rachel, R.; Goepferich, A.; Breunig, M. Layer-by-Layer Assembled Gold Nanoparticles for Sirna Delivery. Nano Lett. 2009, 9, 2059–2064. [Google Scholar] [CrossRef] [PubMed]
- Gamrad, L.; Rehbock, C.; Westendorf, A.M.; Buer, J.; Barcikowski, S.; Hansen, W. Efficient Nucleic Acid Delivery to Murine Regulatory T Cells by Gold Nanoparticle Conjugates. Sci. Rep. 2016, 6, 28709. [Google Scholar] [CrossRef] [PubMed]
- Sun, D.; Tan, S.; Xiong, Y.; Pu, W.; Li, J.; Wei, W.A.; Huang, C.; Wei, Y.Q.; Peng, Y. MicroRNA Biogenesis Is Enhanced by Liposome-Encapsulated PIN1 Inhibitor in Hepatocellular Carcinoma. Theranostics 2019, 9, 4704–4716. [Google Scholar] [CrossRef] [PubMed]
- Vakhshiteh, F.; Khabazian, E.; Atyabi, F.; Ostad, S.N.; Madjd, Z.; Dinarvand, R. Peptide-Conjugated Liposomes for Targeted MiR-34a Delivery to Suppress Breast Cancer and Cancer Stem-like Population. J. Drug Deliv. Sci. Technol. 2020, 57, 101687. [Google Scholar] [CrossRef]
- Şalva, E.; Turan, S.Ö.; Eren, F.; Akbuʇa, J. The Enhancement of Gene Silencing Efficiency with Chitosan-Coated Liposome Formulations of SiRNAs Targeting HIF-1α and VEGF. Int. J. Pharm. 2015, 478, 147–154. [Google Scholar] [CrossRef]
- Shen, P.; Zhao, G.; Liu, Y.; Ge, Q.; Sun, Q. Liposomal Spherical Nucleic Acid Scaffolded Site-Selective Hybridization of Nanoparticles for Visual Detection of MicroRNAs. ACS Appl. Bio Mater. 2020, 3, 1656–1665. [Google Scholar] [CrossRef]
- Janeczek, A.A.; Scarpa, E.; Horrocks, M.H.; Tare, R.S.; Rowland, C.A.; Jenner, D.; Newman, T.A.; Oreffo, R.O.C.; Lee, S.F.; Evans, N.D. PEGylated Liposomes Associate with Wnt3A Protein and Expand Putative Stem Cells in Human Bone Marrow Populations. Nanomedicine 2017, 12, 845–863. [Google Scholar] [CrossRef] [Green Version]
- Pinnapireddy, S.R.; Duse, L.; Strehlow, B.; Schäfer, J.; Bakowsky, U. Composite Liposome-PEI/Nucleic Acid Lipopolyplexes for Safe and Efficient Gene Delivery and Gene Knockdown. Colloids Surf. B Biointerfaces 2017, 158, 93–101. [Google Scholar] [CrossRef]
- Tang, W.; Cao, Z.; Han, M.; Wang, Z.; Chen, J.; Sun, W.; Wu, Y.; Xiao, W.; Liu, S.; Chen, E.; et al. Hydroxychloroquine in Patients with Mainly Mild to Moderate Coronavirus Disease 2019: Open Label, Randomised Controlled Trial. BMJ 2020, 369, m1849. [Google Scholar] [CrossRef]
- Wang, L.; Liang, T.T. CD59 Receptor Targeted Delivery of MiRNA-1284 and Cisplatin-Loaded Liposomes for Effective Therapeutic Efficacy against Cervical Cancer Cells. AMB Express 2020, 10, 54. [Google Scholar] [CrossRef] [Green Version]
- Lujan, H.; Griffin, W.C.; Taube, J.H.; Sayes, C.M. Synthesis and Characterization of Nanometer-Sized Liposomes for Encapsulation and Microrna Transfer to Breast Cancer Cells. Int. J. Nanomed. 2019, 14, 5159–5173. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Deng, X.; Cao, M.; Zhang, J.; Hu, K.; Yin, Z.; Zhou, Z.; Xiao, X.; Yang, Y.; Sheng, W.; Wu, Y.; et al. Hyaluronic Acid-Chitosan Nanoparticles for Co-Delivery of MiR-34a and Doxorubicin in Therapy against Triple Negative Breast Cancer. Biomaterials 2014, 35, 4333–4344. [Google Scholar] [CrossRef] [PubMed]
- Balazs, D.A.; Godbey, W. Liposomes for Use in Gene Delivery. J. Drug Deliv. 2011, 2011, 326497. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lv, H.; Zhang, S.; Wang, B.; Cui, S.; Yan, J. Toxicity of Cationic Lipids and Cationic Polymers in Gene Delivery. J. Control Release 2006, 114, 100–109. [Google Scholar] [CrossRef]
- Cui, L.; Kittipongpittaya, K.; McClements, D.J.; Decker, E.A. Impact of Phosphoethanolamine Reverse Micelles on Lipid Oxidation in Bulk Oils. JAOCS J. Am. Oil Chem. Soc. 2014, 91, 1931–1937. [Google Scholar] [CrossRef]
- Scollo, F.; Tempra, C.; Lolicato, F.; Sciacca, M.F.M.; Raudino, A.; Milardi, D.; La Rosa, C. Phospholipids Critical Micellar Concentrations Trigger Different Mechanisms of Intrinsically Disordered Proteins Interaction with Model Membranes. J. Phys. Chem. Lett. 2018, 9, 5125–5129. [Google Scholar] [CrossRef]
- Shin, J.H.; Shin, D.H.; Kim, J.S. Let-7 MiRNA and CDK4 SiRNA Co-Encapsulated in Herceptin-Conjugated Liposome for Breast Cancer Stem Cells. Asian J. Pharm. Sci. 2020, 15, 472–481. [Google Scholar] [CrossRef]
- Fukushige, K.; Tagami, T.; Naito, M.; Goto, E.; Hirai, S.; Hatayama, N.; Yokota, H.; Yasui, T.; Baba, Y.; Ozeki, T. Developing Spray-Freeze-Dried Particles Containing a Hyaluronic Acid-Coated Liposome–Protamine–DNA Complex for Pulmonary Inhalation. Int. J. Pharm. 2020, 583, 119338. [Google Scholar] [CrossRef]
- Shaker, S.; Gardouh, A.; Ghorab, M. Factors Affecting Liposomes Particle Size Prepared by Ethanol Injection Method. Res. Pharm. Sci. 2017, 12, 346–352. [Google Scholar] [CrossRef]
- Yang, T.; Zhao, P.; Rong, Z.; Li, B.; Xue, H.; You, J.; He, C.; Li, W.; He, X.; Lee, R.J.; et al. Anti-Tumor Efficiency of Lipid-Coated Cisplatin Nanoparticles Co-Loaded with MicroRNA-375. Theranostics 2016, 6, 142–154. [Google Scholar] [CrossRef]
- Marto, J.; Gouveia, L.; Jorge, I.M.; Duarte, A.; Gonçalves, L.M.; Silva, S.M.C.; Antunes, F.; Pais, A.A.C.C.; Oliveira, E.; Almeida, A.J.; et al. Starch-Based Pickering Emulsions for Topical Drug Delivery: A QbD Approach. Colloids Surf. B Biointerfaces 2015, 135, 183–192. [Google Scholar] [CrossRef] [PubMed]
- Jain, A.; Hurkat, P.; Jain, S.K. Development of Liposomes Using Formulation by Design: Basics to Recent Advances. Chem. Phys. Lipids 2019, 224, 104764. [Google Scholar] [CrossRef] [PubMed]
| Element | Target | Justification |
|---|---|---|
| Administration route | Intravenous | To improve the efficacy and bioavailability; direct availability in the bloodstream |
| Dosage form | Injection | Low volume production allows customisation to client/quantities |
| Delivery system element | Non-viral vector | Provides safer and more effective delivery of the genetic material |
| pH | 7.35–7.45 | To prevent or reduce vascular complications |
| Osmolarity | 290–310 mOsm/L | To ensure tolerability |
| Particle size | Below 200 nm | To ensure penetration in the cell |
| Homogeneity | Monodisperse | To ensure system’s homogeneity |
| Enhanced therapeutic activity | High transfection efficiency (over 80%) | To improve system’s effectiveness |
| Storage condition | −60 °C ± 20 °C | To guarantee the stability of the genetic material |
| Improved safety | Lack of cytotoxicity, lack of haemolytic activity | To ensure appropriate biological requirements |
| Microbiological quality | Sterile and pyrogen-free | To avoid contamination with microorganisms; to ensure patient safety |
| In vitro release | Prolonged release | To ensure release according to a predefined release pattern, or to ensure spatio-temporal release of the payload |
| CQA | Target | Is It Critical? | Justification |
|---|---|---|---|
| Particle size | 100–400 nm | Yes | Internalization in tumor cells |
| PDI | 0.1–0.5 | Yes | Narrow size distribution; homogeneity of the nanosystem in terms of size |
| ZP | 5–30 mV | Yes | Formation of electrostatic bonds between the vector and the cell environment |
| Surface modifications | Hyaluronic acid, transferrin, PEG | Yes | Decreased opsonization and phagocytosis; prolonged circulation |
| Cytotoxicity | High IC50 | Yes | To ensure nanosystem safety |
| Cellular uptake | Efficient cellular uptake | Yes | To ensure penetration in the cell |
| Transfection efficiency | Over 80% | Yes | To ensure the desired biological effect |
| Nanoparticle | Method | CPPs | Ref. |
|---|---|---|---|
| Gold nanoparticles | Layer-by-layer | Stirring speed and time | [8,106] |
| Polyelectrolyte concentration | |||
| Laser ablation in liquid | Stirring speed and time | [107] | |
| Ultracentrifugation speed and time | |||
| Liposomes | Film dispersion method | Incubation time, temperature | [79] |
| Thin film hydration method | Evaporation time, pressure, temperature | [38,108,109,110,111,112,113,114,115] | |
| Hydration time, temperature | |||
| Ethanol injection method | Injection rate | [116] | |
| Polymeric nanoparticles | o/w single emulsion method | Mixing speed, temperature | [77,78] |
| Double-emulsion method | Sonication time, amplitude | [76] | |
| Stirring time, temperature | |||
| SLN | Solvent diffusion method | Sonication time | [80,94] |
| Agitation time, temperature, speed | |||
| Film-ultrasonic method | Sonication time | [95] |
| Type of Phospholipid | Name | |
|---|---|---|
| Cationic | Monovalent | 1,2-dioleoyl-3-trimethylammonium-propane (DOTAP) |
| 1,2-di-O-octadecenyl-3-trimethylammonium propane (DOTMA) | ||
| Multivalent | Dioctadecylamidoglycylspermine (DOGS) | |
| 2,3-dioleyloxy-N-[2(sperminecarboxamido) ethyl]-N,N-dimethyl-l-propanaminium trifluoroacetate (DOSPA) | ||
| Neutral | 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine (DOPE) | |
| 1,2-dioleoyl-sn-glycero-3-phosphocholine (DOPC) | ||
| Phosphatidylcholine | ||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Toma, I.; Porfire, A.S.; Tefas, L.R.; Berindan-Neagoe, I.; Tomuță, I. A Quality by Design Approach in Pharmaceutical Development of Non-Viral Vectors with a Focus on miRNA. Pharmaceutics 2022, 14, 1482. https://doi.org/10.3390/pharmaceutics14071482
Toma I, Porfire AS, Tefas LR, Berindan-Neagoe I, Tomuță I. A Quality by Design Approach in Pharmaceutical Development of Non-Viral Vectors with a Focus on miRNA. Pharmaceutics. 2022; 14(7):1482. https://doi.org/10.3390/pharmaceutics14071482
Chicago/Turabian StyleToma, Ioana, Alina Silvia Porfire, Lucia Ruxandra Tefas, Ioana Berindan-Neagoe, and Ioan Tomuță. 2022. "A Quality by Design Approach in Pharmaceutical Development of Non-Viral Vectors with a Focus on miRNA" Pharmaceutics 14, no. 7: 1482. https://doi.org/10.3390/pharmaceutics14071482
APA StyleToma, I., Porfire, A. S., Tefas, L. R., Berindan-Neagoe, I., & Tomuță, I. (2022). A Quality by Design Approach in Pharmaceutical Development of Non-Viral Vectors with a Focus on miRNA. Pharmaceutics, 14(7), 1482. https://doi.org/10.3390/pharmaceutics14071482









