Acetic Acid Mediated for One-Pot Synthesis of Novel Pyrazolyl s-Triazine Derivatives for the Targeted Therapy of Triple-Negative Breast Tumor Cells (MDA-MB-231) via EGFR/PI3K/AKT/mTOR Signaling Cascades
Abstract
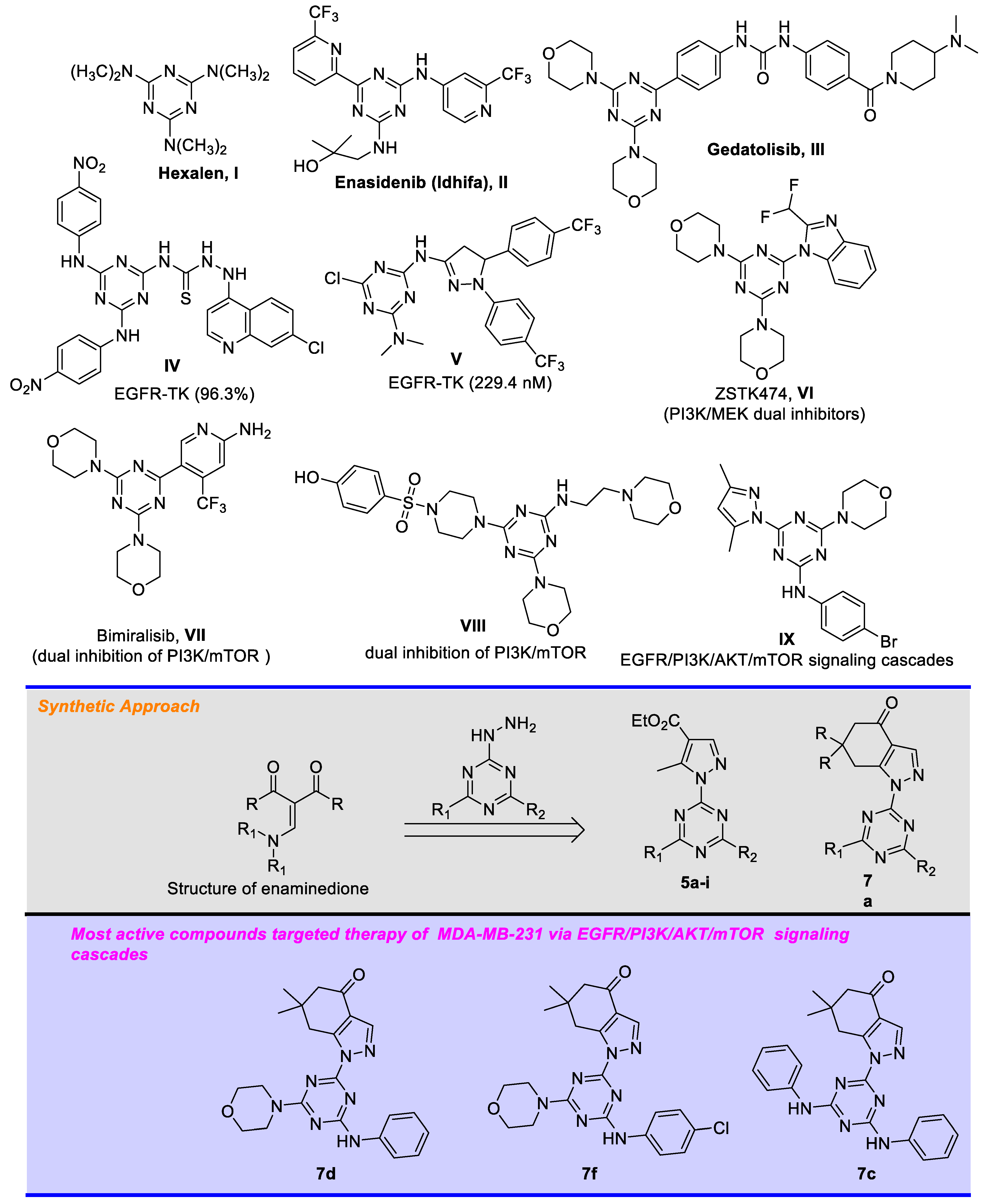
:1. Introduction
2. Materials and Methods
2.1. Chemistry
2.1.1. Materials and Methods
General Procedure for the Synthesis of 2-Hydrazino-6-Substituted s-Triazine Derivatives, 3a–l
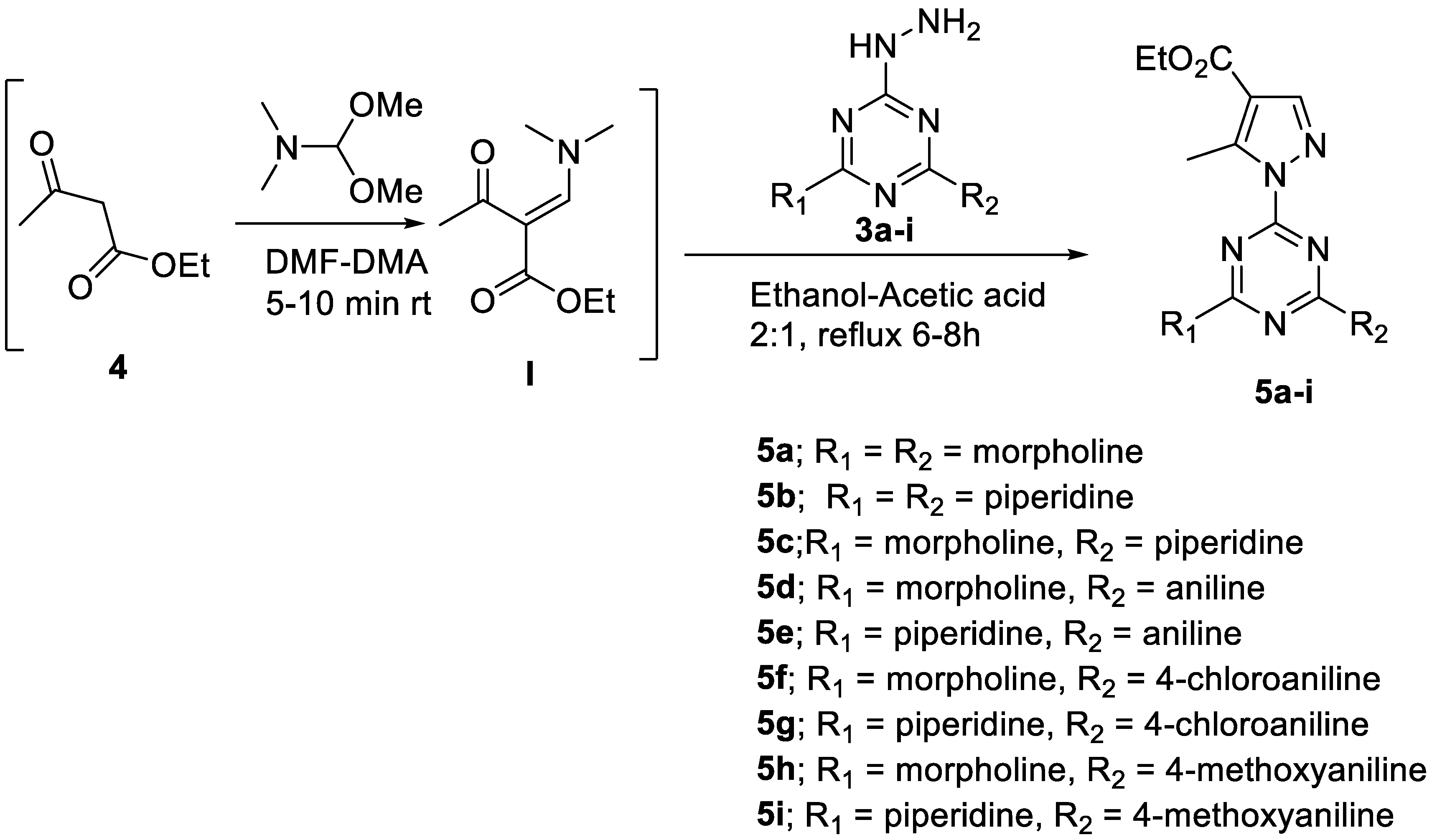
General Procedure for the Synthesis of 5a–i
- Ethyl 1-(4,6-dimorpholino-1,3,5-triazin-2-yl)-5-methyl-1H-pyrazole-4-carboxylate, 5a
- 2.
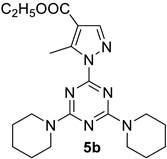
- Ethyl 1-(4,6-di(piperidin-1-yl)-1,3,5-triazin-2-yl)-5-methyl-1H-pyrazole-4-carboxylate, 5b
- 3.
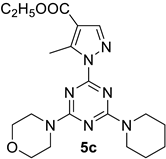
- Ethyl 5-methyl-1-(4-morpholino-6-(piperidin-1-yl)-1,3,5-triazin-2-yl)-1H-pyrazole-4-carboxylate, 5c
- 4.
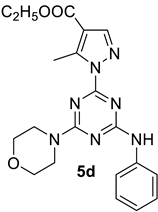
- Ethyl 5-methyl-1-(4-morpholino-6-(phenylamino)-1,3,5-triazin-2-yl)-1H-pyrazole-4-carboxylate, 5d
- 5.
- Ethyl 5-methyl-1-(4-(phenylamino)-6-(piperidin-1-yl)-1,3,5-triazin-2-yl)-1H-pyrazole-4-carboxylate, 5e
- 6.
- Ethyl 1-(4-((4-chlorophenyl)amino)-6-morpholino-1,3,5-triazin-2-yl)-5-methyl-1H-pyrazole-4-carboxylate, 5f
- 7.
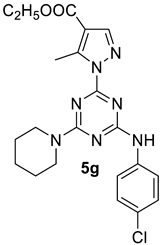
- Ethyl 1-(4-((4-chlorophenyl)amino)-6-(piperidin-1-yl)-1,3,5-triazin-2-yl)-5-methyl-1H-pyrazole-4-carboxylate, 5g
- 8.
- Ethyl 1-(4-((4-methoxyphenyl)amino)-6-morpholino-1,3,5-triazin-2-yl)-5-methyl-1H-pyrazole-4-carboxylate, 5h
- 9.
- Ethyl 1-(4-((4-methoxyphenyl)amino)-6-(piperidin-1-yl)-1,3,5-triazin-2-yl)-5-methyl-1H-pyrazole-4-carboxylate, 5i
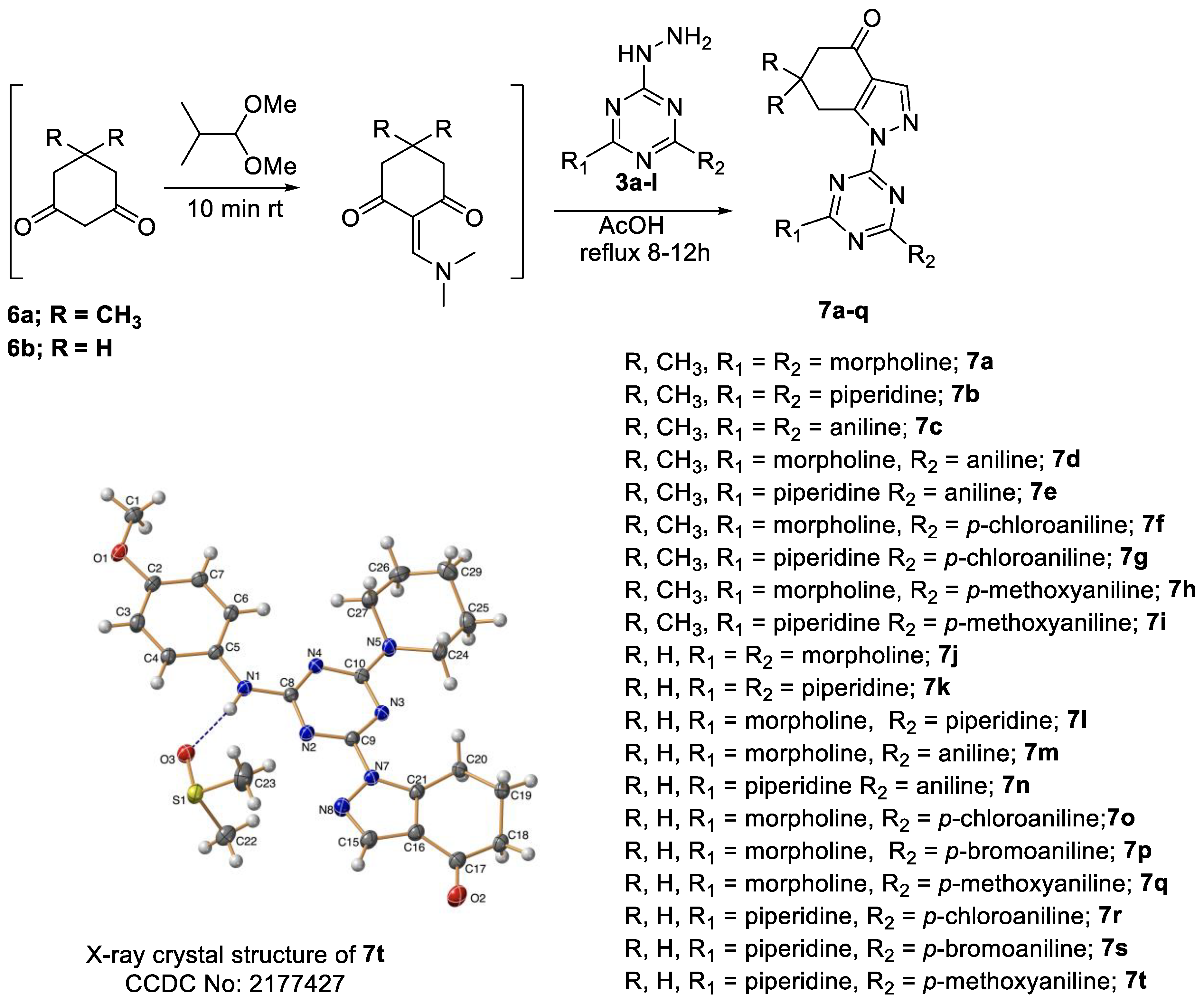
General Procedure for the Synthesis of s-Triazine Derivatives 7a–t
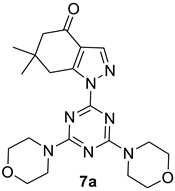
- 1-(4,6-Dimorpholino-1,3,5-triazin-2-yl)-6,6-dimethyl-1,5,6,7-tetrahydro-4H-indazol-4-one, 7a
- 2.
- 1-(4,6-Di(piperidin-1-yl)-1,3,5-triazin-2-yl)-6,6-dimethyl-1,5,6,7-tetrahydro-4H-indazol-4-one, 7b
- 3.
- 1-(4,6-Bis(phenylamino)-1,3,5-triazin-2-yl)-6,6-dimethyl-1,5,6,7-tetrahydro-4H-indazol-4-one, 7c
- 4.
- 6,6-Dimethyl-1-(4-morpholino-6-(phenylamino)-1,3,5-triazin-2-yl)-1,5,6,7-tetrahydro-4H-indazol-4-one, 7d
- 5.
- 6,6-Dimethyl-1-(4-(phenylamino)-6-(piperidin-1-yl)-1,3,5-triazin-2-yl)-1,5,6,7-tetrahydro-4H-indazol-4-one, 7e
- 6.
- 1-(4-((4-Chlorophenyl)amino)-6-morpholino-1,3,5-triazin-2-yl)-6,6-dimethyl-1,5,6,7-tetrahydro-4H-indazol-4-one, 7f
- 7.
- 1-(4-((4-Chlorophenyl)amino)-6-(piperidin-1-yl)-1,3,5-triazin-2-yl)-6,6-dimethyl-1,5,6,7-tetrahydro-4H-indazol-4-one, 7g
- 8.
- 1-(4-((4-Methoxyphenyl)amino)-6-morpholino-1,3,5-triazin-2-yl)-6,6-dimethyl-1,5,6,7-tetrahydro-4H-indazol-4-one, 7h
- 9.
- 1-(4-((4-Methoxyphenyl)amino)-6-(piperidin-1-yl)-1,3,5-triazin-2-yl)-6,6-dimethyl-1,5,6,7-tetrahydro-4H-indazol-4-one, 7i
- 10.
- 1-(4,6-Dimorpholino-1,3,5-triazin-2-yl)-1,5,6,7-tetrahydro-4H-indazol-4-one, 7j
- 11.
- 1-(4,6-Di(piperidin-1-yl)-1,3,5-triazin-2-yl)-1,5,6,7-tetrahydro-4H-indazol-4-one, 7k
- 12.
- 1-(4-Morpholino-6-(piperidin-1-yl)-1,3,5-triazin-2-yl)-1,5,6,7-tetrahydro-4H-indazol-4-one, 7l
- 13.
- 1-(4-Morpholino-6-(phenylamino)-1,3,5-triazin-2-yl)-1,5,6,7-tetrahydro-4H-indazol-4-one, 7m
- 14.
- 1-(4-(Phenylamino)-6-(piperidin-1-yl)-1,3,5-triazin-2-yl)-1,5,6,7-tetrahydro-4H-indazol-4-one, 7n
- 15.
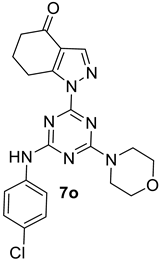
- 1-(4-((4-Chlorophenyl)amino)-6-morpholino-1,3,5-triazin-2-yl)-1,5,6,7-tetrahydro-4H-indazol-4-one, 7o
- 16.
- 1-(4-((4-Bromophenyl)amino)-6-morpholino-1,3,5-triazin-2-yl)-1,5,6,7-tetrahydro-4H-indazol-4-one, 7p
- 17.
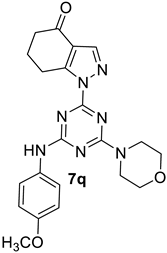
- 1-(4-((4-Methoxyphenyl)amino)-6-morpholino-1,3,5-triazin-2-yl)-1,5,6,7-tetrahydro-4H-indazol-4-one, 7q
- 18.
- 1-(4-((4-Chlorophenyl)amino)-6-(piperidin-1-yl)-1,3,5-triazin-2-yl)-1,5,6,7-tetrahydro-4H-indazol-4-one, 7r
- 19.
- 1-(4-((4-Bromophenyl)amino)-6-(piperidin-1-yl)-1,3,5-triazin-2-yl)-1,5,6,7-tetrahydro-4H-indazol-4-one, 7s
- 20.
- 1-(4-((4-Methoxyphenyl)amino)-6-(piperidin-1-yl)-1,3,5-triazin-2-yl)-1,5,6,7-tetrahydro-4H-indazol-4-one, 7t
2.2. Biology
2.2.1. Cell Culture
2.2.2. Cell Viability Assay (MTT)
2.2.3. EGFR Protein Kinase (PK) Inhibition
2.2.4. PI3K/AKT/mTOR Downstream Signaling Pathway
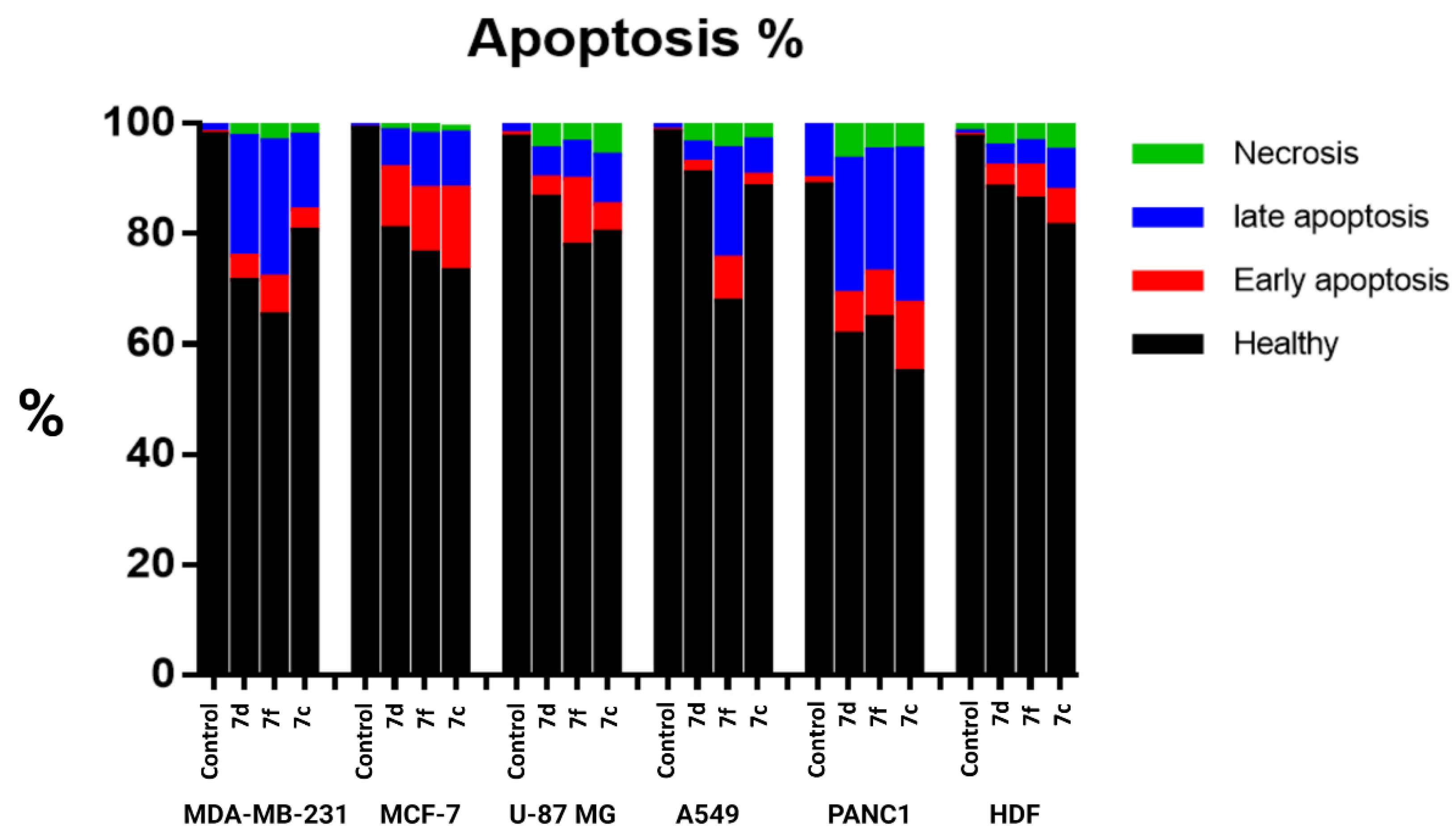
2.2.5. Apoptosis by Flow Cytometry
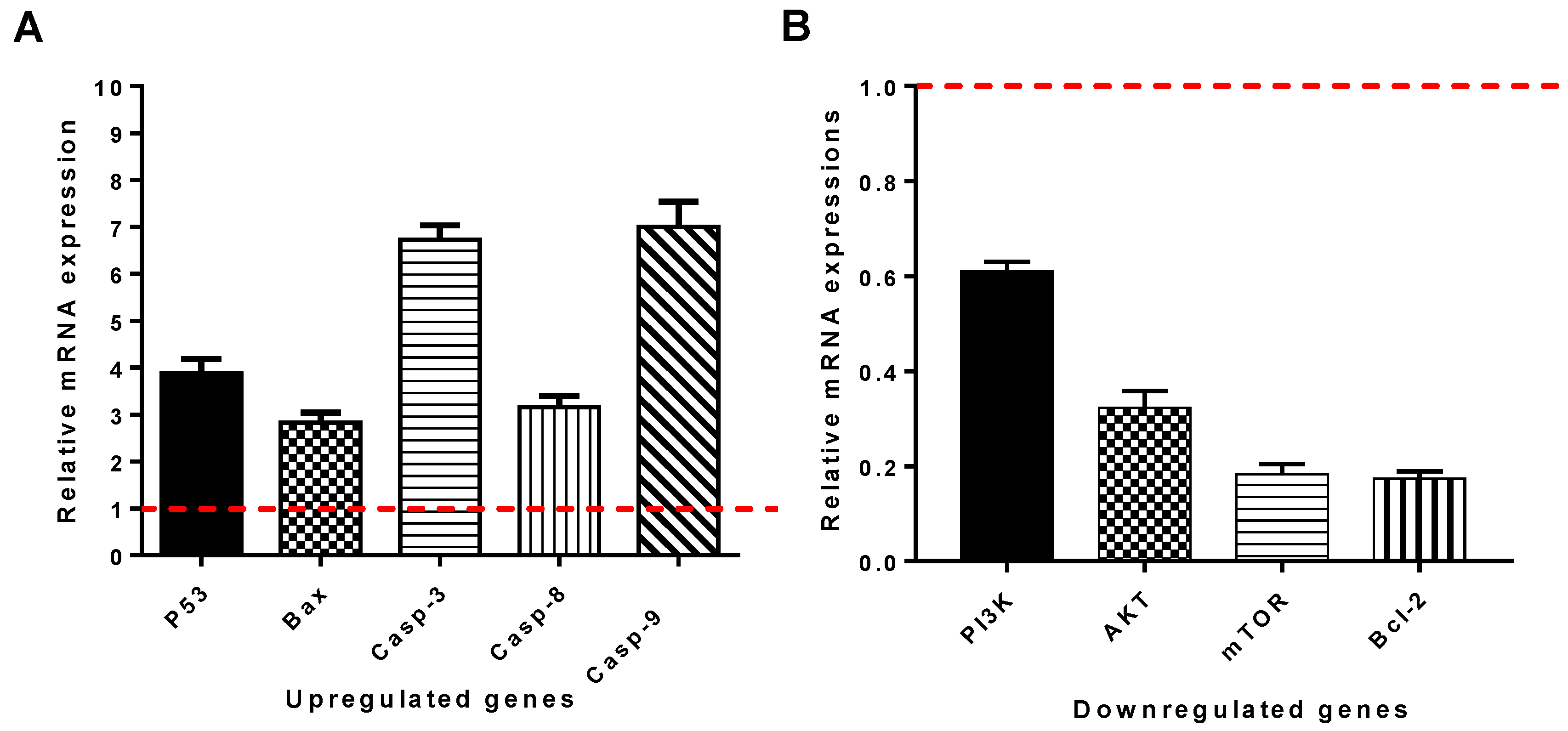
2.2.6. Gene Expression Analysis Using RT-qPCR
2.2.7. CDOCKER Docking
2.2.8. Scoring of Docked Poses
3. Results and Discussion
3.1. Chemistry

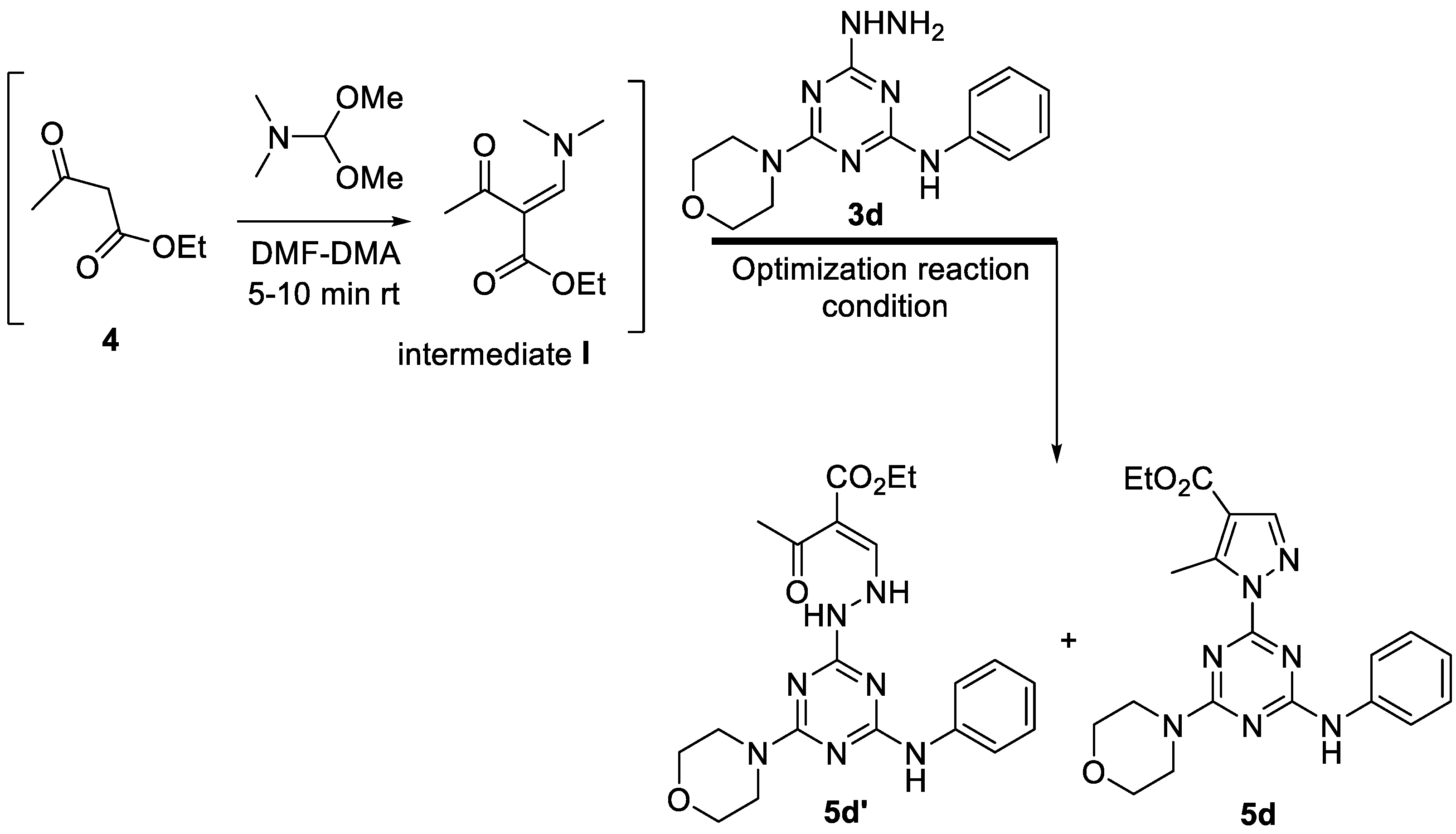
| Entry | Solvent | Time (h) | 5d’ % | 5d% |
|---|---|---|---|---|
| 1 | 5% AcOH-EtOH | 4 | 50 | 50 |
| 2 | 5% AcOH-EtOH | 8 | 50 | 50 |
| 3 | 10% AcOH-EtOH | 8 | 40 | 60 |
| 4 | 20% AcOH-EtOH | 6 | ~20 | ~80 |
| 5 | AcOH-EtOH (1:2) | 6 | traces | ~90 |
| 6 | AcOH-EtOH (1:2) | 8 | traces | ~95 |
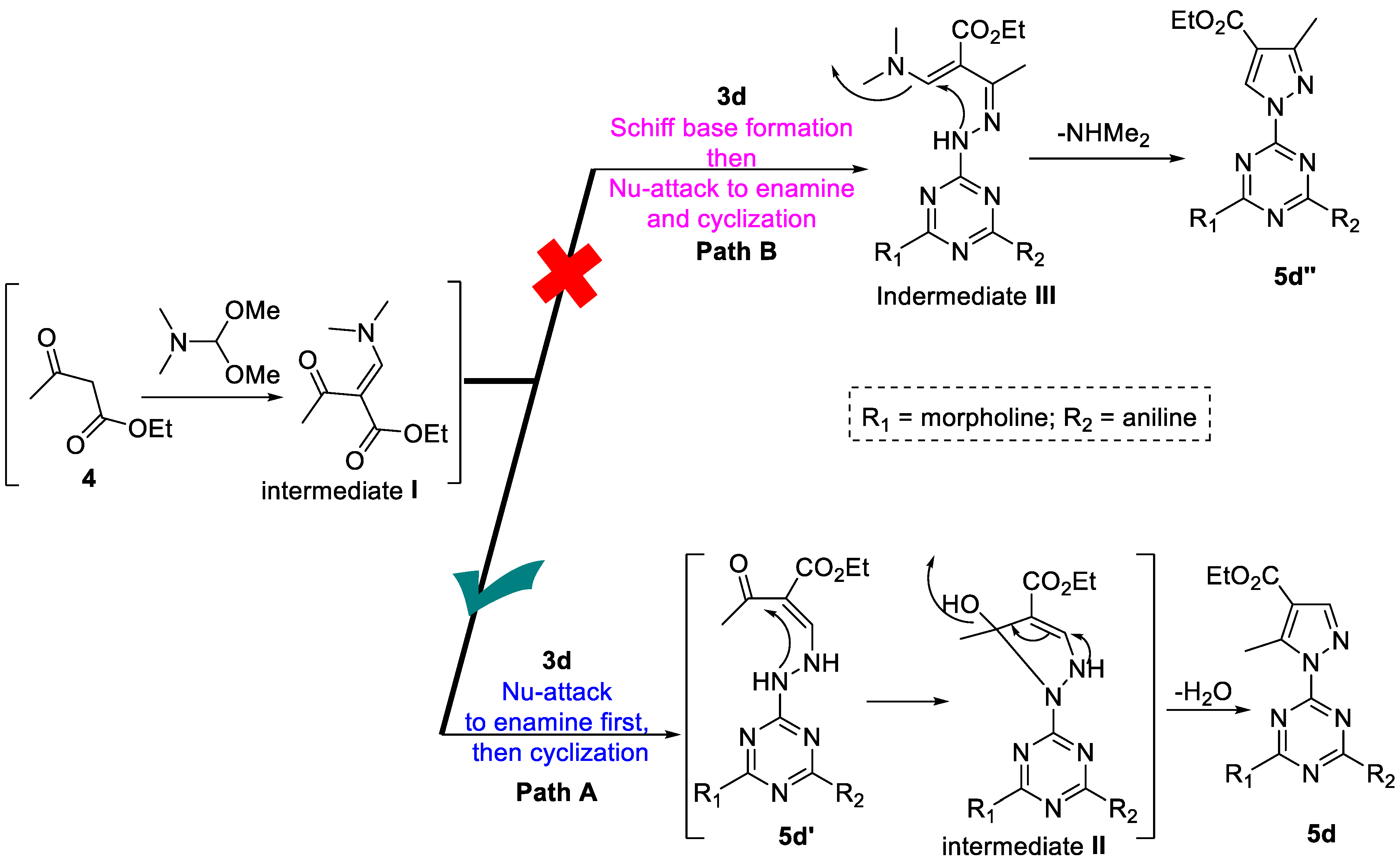
3.2. Biology
3.2.1. Cytotoxicity
3.2.2. EGFR Enzymatic Assay
3.2.3. PI3K/AKT/mTOR Downstream Signaling Pathway
3.2.4. Apoptosis by Flow Cytometry
3.2.5. Compound 7f Upregulated Pro-Apoptotic Genes and Downregulated Anti-Apoptotic Ones
3.2.6. Molecular Docking Study
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Szabó, G.; Fischer, J.; Kis-Varga, Á.; Gyires, K. New celecoxib derivatives as anti-inflammatory agents. J. Med. Chem. 2008, 51, 142–147. [Google Scholar] [CrossRef] [PubMed]
- Popova, G.; Ladds, M.; Johansson, L.; Aljona, S.; Johanna, L.; Lars, S.; Sara, H.S.; Weixing, Q.; Hjalmar, G.; Neeraj, G.; et al. Optimization of tetrahydroindazoles as inhibitors of human dihydroorotate dehydrogenase and evaluation of their activity and in vitro metabolic stability. J. Med. Chem. 2020, 63, 3915–3934. [Google Scholar] [CrossRef] [PubMed]
- Hammad, A.; Abutaleb, N.S.; Elsebaei, M.M.; Allison, B.N.; Mohamed, A.; Alsagher, A.; Jelan, A.A.; Abdelaziz, A.; Sammar, A.; Humaira, J.; et al. From phenylthiazoles to phenylpyrazoles: Broadening the antibacterial spectrum toward Carbapenem-resistant bacteria. J. Med. Chem. 2019, 62, 7998–8010. [Google Scholar] [CrossRef]
- Kunitomo, J.; Yoshikawa, M.; Fushimi, M.; Kawada, A.; Quinn, J.F.; Oki, H.; Kokubo, H.; Kondo, M.; Nakashima, K.; Kamiguchi, N.; et al. Discovery of 1-[2-Fluoro-4-(1 H -pyrazol-1-yl)phenyl]-5-methoxy-3-(1-phenyl-1 H -pyrazol-5-yl)pyridazin-4(1 H)-one (TAK-063), a highly potent, selective, and orally active phosphodiesterase 10A (PDE10A) inhibitor. J. Med. Chem. 2014, 57, 9627–9643. [Google Scholar] [CrossRef] [PubMed]
- Salerno, S.; Barresi, E.; Amendola, G.; Berrino, E.; Milite, C.; Marini, A.M.; Da Settimo, F.; Novellino, E.; Supuran, C.T.; Cosconati, S.; et al. 4-Substituted benzenesulfonamides incorporating bi/tricyclic moieties act as potent and isoform-selective carbonic anhydrase II/IX inhibitors. J. Med. Chem. 2018, 61, 5765–5770. [Google Scholar] [CrossRef] [PubMed]
- Prakash, O.; Hussain, K.; Kumar, R.; Wadhwa, D.; Sharma, C.; Aneja, K.R. Synthesis and antimicrobial evaluation of new 1,4-dihydro-4-pyrazolylpyridines and 4-pyrazolylpyridines. Org. Med. Chem. Lett. 2011, 1, 5. [Google Scholar] [CrossRef] [Green Version]
- Li, M.; Zhao, B.X. Progress of the synthesis of condensed pyrazole derivatives (from 2010 to mid-2013). Eur. J. Med. Chem. 2014, 85, 311–340. [Google Scholar] [CrossRef] [PubMed]
- Ningaiah, S.; Bhadraiah, U.K.; Doddaramappa, S.D.; Keshavamurthy, S.; Javarasetty, C. Novel pyrazole integrated 1,3,4-oxadiazoles: Synthesis, characterization and antimicrobial evaluation. Bioorganic Med. Chem. Lett. 2014, 24, 245–248. [Google Scholar] [CrossRef] [PubMed]
- El-Sayed, M.A.A.; Abdel-Aziz, N.I.; Abdel-Aziz, A.A.M.; El-Azab, A.S.; Eltahir, K.E.H. Synthesis, biological evaluation and molecular modeling study of pyrazole and pyrazoline derivatives as selective COX-2 inhibitors and anti-inflammatory agents. Part 2. Bioorganic Med. Chem. 2012, 20, 3306–3316. [Google Scholar] [CrossRef]
- Rathelot, P.; Azas, N.; El-Kashef, H.; Delmas, F.; Di Giorgio, C.; Timon-David, P.; Maldonado, J.; Vanelle, P. 1,3-Diphenylpyrazoles: Synthesis and antiparasitic activities of azomethine derivatives. Eur. J. Med. Chem. 2002, 37, 671–679. [Google Scholar] [CrossRef]
- Abdel-Aziz, M.; Abuo-Rahma, G.; Hassan, A.A. Synthesis of novel pyrazole derivatives and evaluation of their antidepressant and anticonvulsant activities. Eur. J. Med. Chem. 2009, 44, 3480–3487. [Google Scholar] [CrossRef]
- Hashem, A.I.; Youssef, A.S.A.; Kandeel, K.A.; Abou-Elmagd, W.S.I. Conversion of some 2(3H)-furanones bearing a pyrazolyl group into other heterocyclic systems with a study of their antiviral activity. Eur. J. Med. Chem. 2007, 42, 934–939. [Google Scholar] [CrossRef]
- Mert, S.; Kasimoǧullari, R.; Iça, T.; Çolak, F.; Altun, A.; Ok, S. Synthesis, structure-activity relationships, and in vitro antibacterial and antifungal activity evaluations of novel pyrazole carboxylic and dicarboxylic acid derivatives. Eur. J. Med. Chem. 2014, 78, 86–96. [Google Scholar] [CrossRef] [PubMed]
- Lv, P.C.; Li, H.Q.; Sun, J.; Zhou, Y.; Zhu, H.L. Synthesis and biological evaluation of pyrazole derivatives containing thiourea skeleton as anticancer agents. Bioorganic Med. Chem. 2010, 18, 4606–4614. [Google Scholar] [CrossRef] [PubMed]
- Khloya, P.; Celik, G.; Ram, S.; Vullo, D.; Supuran, C.T.; Sharma, P.K. 4-Functionalized 1,3-diarylpyrazoles bearing benzenesulfonamide moiety as selective potent inhibitors of the tumor associated carbonic anhydrase isoforms IX and XII. Eur. J. Med. Chem. 2014, 76, 284–290. [Google Scholar] [CrossRef] [PubMed]
- Penning, T.D.; Talley, J.J.; Bertenshaw, S.R.; Carter, J.S.; Collins, P.W.; Docter, S.; Graneto, M.J.; Lee, L.F.; Malecha, J.W.; Miyashiro, J.M.; et al. Synthesis and biological evaluation of the 1,5-diarylpyrazole class of cyclooxygenase-2 inhibitors: Identification of 4-[5-(4-methylphenyl)- 3(trifluoromethyl)-1h-pyrazol-1-yl]benzenesulfonamide (sc-58635, celecoxib). J. Med. Chem. 1997, 40, 1347–1365. [Google Scholar] [CrossRef] [PubMed]
- Terrett, N.K.; Bell, A.S.; Brown, D.; Ellis, P. Sildenafil (Viagra(TM)), a potent and selective inhibitor of type 5 CGMP phosphodiesterase with utility for the treatment of male erectile dysfunction. Bioorg. Med. Chem. Lett. 1996, 6, 1819–1824. [Google Scholar] [CrossRef]
- Seltzman, H.H.; Carroll, F.I.; Burgess, J.P.; Wyrick, C.D.; Burch, D.F. Synthesis, spectral studies and tritiation of the cannabinoid antagonist SR141716A. J. Chem. Soc. Chem. Commun. 1995, 15, 1549–1550. [Google Scholar] [CrossRef]
- Salem, M.E.; Darweesh, A.F.; Mekky, A.E.M.; Farag, A.M.; Elwahy, A.H.M. 2-Bromo-1-(1H-pyrazol-4-yl)ethanone: Versatile precursor for novel mono- and bis[pyrazolylthiazoles]. J. Heterocycl. Chem. 2017, 54, 226–234. [Google Scholar] [CrossRef]
- Rafique, R.; Khan, K.M.; Arshia; Chigurupati, S.; Wadood, A.; Rehman, A.U.; Salar, U.; Venugopal, V.; Shamim, S.; Taha, M.; et al. Synthesis, in vitro α-amylase inhibitory, and radicals (DPPH & ABTS) scavenging potentials of new N-sulfonohydrazide substituted indazoles. Bioorg. Chem. 2020, 94, 103410. [Google Scholar] [CrossRef] [PubMed]
- Fustero, S.; Sánchez-Roselló, M.; Barrio, P.; Simón-Fuentes, A. From 2000 to mid-2010: A fruitful decade for the synthesis of pyrazoles. Chem. Rev. 2011, 111, 6984–7034. [Google Scholar] [CrossRef]
- Dadiboyena, S.; Nefzi, A. Synthesis of functionalized tetrasubstituted pyrazolyl heterocycles—A review. Eur. J. Med. Chem. 2011, 46, 5258–5275. [Google Scholar] [CrossRef] [PubMed]
- Janin, Y.L. Preparation and chemistry of 3/5-halogenopyrazoles. Chem. Rev. 2012, 112, 3924–3958. [Google Scholar] [CrossRef] [PubMed]
- Alinezhad, H.; Tajbakhsh, M.; Zare, M. Catalyst-free one-pot synthesis of 1,4,5-trisubstituted pyrazoles in 2,2,2-trifluoroethanol. J. Fluor. Chem. 2011, 132, 995–1000. [Google Scholar] [CrossRef]
- Kennedy, L.J. An expedient synthesis of regioisomeric pyrazole-fused cycloalkanones. Synlett 2008, 4, 600–604. [Google Scholar] [CrossRef]
- Schenone, P.; Mosti, L.; Menozzi, G. Reaction of 2-dimethylaminomethylene-1,3-diones with dinucleophiles. I. Synthesis of 1,5-disubstituted 4-acylpyrazoles. J. Heterocycl. Chem. 1982, 19, 1355–1361. [Google Scholar] [CrossRef]
- Menear, K.A.; Gomez, S.; Malagu, K.; Bailey, C.; Blackburn, K.; Cockcroft, X.-L.; Ewen, S.; Fundo, A.; Le Gall, A.; Hermann, G.; et al. Identification and optimisation of novel and selective small molecular weight kinase inhibitors of mTOR. Bioorg. Med. Chem. Lett. 2009, 19, 5898–5901. [Google Scholar] [CrossRef]
- Shah, D.R.; Modh, R.P.; Chikhalia, K.H. Privileged s-triazines: Structure and pharmacological applications. Future Med. Chem. 2014, 6, 463–477. [Google Scholar] [CrossRef] [PubMed]
- Liu, B. A Systematic review on antitumor agents with 1, 3, 5-triazines. Med. Chem. 2015, 5, 131–148. [Google Scholar] [CrossRef] [Green Version]
- Perspicace, E.; Jouan-Hureaux, V.; Ragno, R.; Ballante, F.; Sartini, S.; La Motta, C.; Da Settimo, F.; Chen, B.; Kirsch, G.; Schneider, S.; et al. Design, synthesis and biological evaluation of new classes of thieno[3,2-d]pyrimidinone and thieno[1,2,3]triazine as inhibitor of vascular endothelial growth factor receptor-2 (VEGFR-2). Eur. J. Med. Chem. 2013, 63, 765–781. [Google Scholar] [CrossRef]
- Alhameed, R.A.; Almarhoon, Z.; Sholkamy, E.N.; Khan, S.A.; Albericio, F.; El-faham, A. Novel 4,6-Disubstituted s-Triazin-2-yl Amino Acid Derivatives as Promising Antifungal Agents. J. Fungi 2020, 6, 237–251. [Google Scholar] [CrossRef] [PubMed]
- Al Rasheed, H.; Malebari, A.M.; Dahlous, K.; Fayne, D.; El-Faham, A. Synthesis, anti-proliferative activity, and molecular docking study of new series of 1,3-5-triazine Schiff base derivatives. Molecules 2020, 25, 4065. [Google Scholar] [CrossRef] [PubMed]
- Al Rasheed, H.; Dahlous, K.; Sharma, A.; Sholkamy, E.; El-Faham, A.; de la Torre, B.G.; Albericio, F. Barbiturate-and thiobarbituarte-based s-triazine hydrazone derivatives with promising antiproliferative activities. ACS Omega 2020, 5, 15805–15811. [Google Scholar] [CrossRef]
- De la Hoz, A.; Diaz-Ortiz, A.; Elguero, J.; Martinez, L.; Moreno, A.; Sanchez-Migallon, A. Solvent-free preparation of tris-pyrazolyl-1,3,5-triazines. Tetrahedron 2001, 57, 4403. [Google Scholar] [CrossRef]
- de la Hoz, A.; Sánchez-Migallón, A. Green synthesis of 1,3,5-triazines with applications in supramolecular and materials chemistry. Targets Heterocycl. Syst. 2016, 20, 139–173. [Google Scholar] [CrossRef]
- Ayyangar, N.R.; Lahoti, R.J.; Lugade, A.G.; Otiv, S.R. Synthesis of monoazo disperse dyes from 5-amino-3-methyl-1-(3′,5′-disubstituted) s-triazinylpyrazoles and a study of their visible absorption and dyeing properties. J. Soc. Dye. Colour. 1986, 102, 176–181. [Google Scholar] [CrossRef]
- Mikhaylichenko, S.N.; Patel, S.M.; Dalili, S.; Chesnyuk, A.A.; Zaplishny, V.N. Synthesis and structure of new 1,3,5-triazine-pyrazole derivatives. Tetrahedron Lett. 2009, 50, 2505–2508. [Google Scholar] [CrossRef]
- Sharma, A.; Ghabbour, H.; Khan, S.T.; de la Torre, B.G.; Albericio, F.; El-Faham, A. Novel pyrazolyl-s-triazine derivatives, molecular structure and antimicrobial activity. J. Mol. Struct. 2017, 1145, 244–253. [Google Scholar] [CrossRef]
- Farooq, M.; Sharma, A.; Almarhoon, Z.; Al-Dhfyan, A.; El-Faham, A.; Taha, N.A.; Wadaan, M.A.M.; Torre, B.; Albericio, F. Design and synthesis of mono-and di-pyrazolyl-s-triazine derivatives, their anticancer profile in human cancer cell lines, and in vivo toxicity in zebrafish embryos. Bioorg. Chem. 2019, 87, 457–464. [Google Scholar] [CrossRef]
- Keldsen, N.; Havsteen, H.; Vergote, I.; Bertelsen, K.; Jakobsen, A. Altretamine (hexamethylmelamine) in the treatment of platinum-resistant ovarian cancer: A phase II study. Gynecol. Oncol. 2003, 88, 118–122. [Google Scholar] [CrossRef]
- Kim, E.S. Enasidenib: First Global Approval. Drugs 2017, 77, 1705–1711. [Google Scholar] [CrossRef] [PubMed]
- Shor, R.E.; Dai, J.; Lee, S.; Pisarsky, L.; Matei, I.; Lucotti, S.; Lyden, D.; Bissell, M.J.; Ghajar, C.M. The PI3K/mTOR inhibitor Gedatolisib eliminates dormant breast cancer cells in organotypic culture, but fails to prevent metastasis in preclinical settings. Mol. Oncol. 2022, 16, 130–147. [Google Scholar] [CrossRef] [PubMed]
- Bhat, H.R.; Masih, A.; Shakya, A.; Ghosh, S.K.; Singh, U.P. Design, synthesis, anticancer, antibacterial, and antifungal evaluation of 4-aminoquinoline-1,3,5-triazine derivatives. J. Heterocycl. Chem. 2019, 57, 390–399. [Google Scholar] [CrossRef]
- Raghu, M.S.; Kumar, C.; Prashanth, M.K.; Kumar, K.Y. Novel 1,3,5-triazine-based pyrazole derivatives as potential antitumor agents and EFGR kinase inhibitors: Synthesis, cytotoxicity, DNA binding, molecular docking and DFT studies. New J. Chem. 2021, 45, 13909–13924. [Google Scholar] [CrossRef]
- Van-Dort, M.E.; Galbán, S.; Wang, H.; Sebolt-Leopold, J.; Whitehead, C.; Hong, H.; Rehemtulla, A.; Ross, B.D. Dual inhibition of allosteric mitogen-activated protein kinase (MEK) and phosphatidylinositol 3-kinase (PI3K) oncogenic targets with a bifunctional inhibitor. Bioorganic Med. Chem. 2015, 23, 1386–1394. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Y.; Tortorella, M. Molecular design of dual inhibitors of PI3K and potential molecular target of cancer for its treatment: A review. Eur. J. Med. Chem. 2022, 228, 114039. [Google Scholar] [CrossRef]
- Zoncu, R.; Efeyan, A.; Sabatini, D.M. MTOR: From growth signal integration to cancer, diabetes and ageing. Nat. Rev. Mol. Cell Biol. 2011, 12, 21–35. [Google Scholar] [CrossRef] [Green Version]
- Beaufils, F.; Cmiljanovic, N.; Cmiljanovic, V.; Bohnacker, T.; Melone, A.; Marone, R.; Jackson, E.; Zhang, X.; Sele, A.; Borsari, C.; et al. 5-(4,6-Dimorpholino-1,3,5-triazin-2-yl)-4-(trifluoromethyl)pyridin-2-amine (PQR309), a Potent, Brain-Penetrant, Orally Bioavailable, Pan-Class i PI3K/mTOR Inhibitor as Clinical Candidate in Oncology. J. Med. Chem. 2017, 60, 7524–7538. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hu, J.; Zhang, Y.; Tan, N.; Lu, Y.; Guo, P.; Huang, Z. Discovery of novel 1,3,5-triazine derivatives as potent inhibitor of cervical cancer via dual inhibition of PI3K/mTOR. Bioorganic Med. Chem. 2021, 32, 115997. [Google Scholar] [CrossRef]
- Shawish, I.; Barakat, A.; Aldalbahi, A.; Malebari, A.; Nafie, M.S.; Bekhit, A.A.; Albohy, A.; Khan, A.; Ul-Haq, Z.; Haukka, M.; et al. Synthesis and antiproliferative activity of a new series of mono- and bis(dimethylpyrazolyl)-s-triazine derivatives targeting EGFR/PI3K/AKT/mTOR signaling cascades. ACS Omega 2022, 7, 24858–24870. [Google Scholar] [CrossRef]
- Mosmann, T. Rapid colorimetric assay for cellular growth and survival: Application to proliferation and cytotoxicity assays. J. Immunol. Methods 1983, 65, 55–63. [Google Scholar] [CrossRef]
- Hisham, M.; Youssif, B.; Osman, E.; Hayallah, A.M.; Abdel-Aziz, M. Synthesis and biological evaluation of novel xanthine derivatives as potential apoptotic antitumor agents. Eur. J. Med. Chem. 2019, 176, 117–128. [Google Scholar] [CrossRef] [PubMed]
- Nafie, M.S.; Arafa, K.; Sedky, N.K.; Alakhdar, A.A.; Arafa, R.K. Triaryl dicationic DNA minor-groove binders with antioxidant activity display cytotoxicity and induce apoptosis in breast cancer: Dicationic compounds as anticancer and apoptotic inducing agents. Chem.-Biol. Interact. 2020, 324, 109087. [Google Scholar] [CrossRef] [PubMed]
- Sarhan, A.; Boraei, A.; Barakat, A.; Nafie, M.S. Discovery of hydrazide-based pyridazino[4,5-b]indole scaffold as a new phosphoinositide 3-kinase (PI3K) inhibitor for breast cancer therapy. RSC Adv. 2020, 10, 19534–19541. [Google Scholar] [CrossRef] [PubMed]
- Nafie, M.S.; Mahgoub, S.; Amer, A.M. Antimicrobial and antiproliferative activities of novel synthesized 6-(quinolin-2-ylthio) pyridine derivatives with molecular docking study as multi-targeted JAK2/STAT3 inhibitors. Chem. Biol. Drug Des. 2021, 97, 553–564. [Google Scholar] [CrossRef] [PubMed]
- Wu, G.; Robertson, D.H.; Iii, C.; Vieth, M. Detailed analysis of grid-based molecular docking: A case study of CDOCKER-A CHARMm-based MD docking algorithm. J. Comput. Chem. 2003, 24, 1549–1562. [Google Scholar] [CrossRef]
- Toviwek, B.; Gleeson, D.; Gleeson, M. QM/MM and molecular dynamics investigation of the mechanism of covalent inhibition of TAK1 kinase. Org. Biomol. Chem. 2021, 19, 1412–1425. [Google Scholar] [CrossRef]
- Jain, A.N. Scoring noncovalent protein-ligand interactions: A continuous differentiable function tuned to compute binding affinities. J. Comput.-Aided Mol. Des. 1996, 10, 427–440. [Google Scholar] [CrossRef]
- Venkatachalam, C.M.; Jiang, X.; Oldfield, T.; Waldman, M. LigandFit: A novel method for the shape-directed rapid docking of ligands to protein active sites. J. Mol. Graph. Model. 2003, 21, 289–307. [Google Scholar] [CrossRef]
- Muegge, I.; Martin, Y.C. A general and fast scoring function for protein-ligand interactions: A simplified potential approach. J. Med. Chem. 1999, 42, 791–804. [Google Scholar] [CrossRef]
- Muegge, I. A knowledge-based scoring function for protein-ligand interactions: Probing the reference state. Perspect. Drug Discov. Des. 2002, 20, 99–114. [Google Scholar] [CrossRef]
- Clark, R.D.; Strizhev, A.; Leonard, J.M.; Blake, J.F.; Matthew, J.B. Consensus scoring for ligand/protein interactions. J. Mol. Graph. Model. 2002, 20, 281–295. [Google Scholar] [CrossRef]
- Feher, M. Consensus scoring for protein-ligand interactions. Drug Discov. Today. 2006, 11, 421–428. [Google Scholar] [CrossRef] [PubMed]
- Ullrich, T.; Dersch, C.M.; Rothman, R.B.; Jacobson, A.E.; Rice, K.C. Derivatives of 17-(2-methylallyl)-substituted noroxymorphone: Variation of the delta address and its effects on affinity and selectivity for the delta opioid receptor. Bioorg. Med. Chem. Lett. 2001, 11, 2883–2885. [Google Scholar] [CrossRef]
- Olivera, R.; SanMartin, R.; Dominguez, E. Dibenzoxepino [4, 5-d] pyrazoles: A facile approach via the Ullmann-ether reaction. Tetrahedron Lett. 2000, 41, 4353–4356. [Google Scholar] [CrossRef]
- Rikagu Oxford Diffraction. CrysAlisPro; Agilent Technologies Inc.: Oxfordshire, UK, 2018. [Google Scholar]
- Sheldrick, G.M. SHELXT-Integrated Space-Group and Crystal-Structure Determination. Acta Crystallogr. Sect. A Found. Adv. 2015, 71, 3–8. [Google Scholar] [CrossRef] [Green Version]
- Sheldrick, G.M. Crystal Structure Refinement with SHELXL. Acta Crystallogr. Sect. C Struct. Chem. 2015, 71, 3–8. [Google Scholar] [CrossRef]
- Hübschle, C.B.; Sheldrick, G.M.; Dittrich, B. ShelXle: A Qt graphical user interface for SHELXL. J. Appl. Crystallogr. 2011, 44, 1281–1284. [Google Scholar] [CrossRef] [Green Version]
- Hassan, B.; Akcakanat, A.; Holder, A.M.; Meric-Bernstam, F. Targeting the PI3-Kinase/Akt/mTOR Signaling Pathway. Surg. Oncol. Clin. N. Am. 2013, 22, 641–664. [Google Scholar] [CrossRef] [Green Version]
- Lee, J.X.; Loh, K.; Yap, Y.S. PI3K/Akt/mTOR inhibitors in breast cancer. Cancer Biol. Med. 2015, 12, 342–354. [Google Scholar] [CrossRef]
- ElZahabi, H.; Nafie, M.S.; Osman, D.; Elghazawy, N.H.; Soliman, D.H.; EL-Helby, A.; Arafa, R.K. Design, synthesis and evaluation of new quinazolin-4-one derivatives as apoptotic enhancers and autophagy inhibitors with potent antitumor activity. Eur. J. Med. Chem. 2021, 222, 113609. [Google Scholar] [CrossRef] [PubMed]
- Nafie, M.S.; Khodair, A.; Hassan, H.; El-Fadeal, N.; Bogari, H.; Elhady, S.S.; Ahmed, S.A. Evaluation of 2-thioxoimadazolidin-4-one derivatives as potent anti-cancer agents through apoptosis induction and antioxidant activation: In vitro and in vivo Approaches. Molecules 2021, 27, 83. [Google Scholar] [CrossRef]



| Compound | Anticancer Activity in Human Cancer Cells Lines IC50 ± STDEV (µM/mL) | |||||
|---|---|---|---|---|---|---|
| MDA-MB-231 | U-87 MG | PANC-1 | A549 | MCF-7 | HDFs | |
 | 69.4 ± 2.5 | 65.4 ± 4.2 | 78.3 ± 7.7 | 74.6 ± 8.7 | 57.3 ± 6.2 | 74.6 ± 9.4 |
 | 307.1 ± 64.8 | 355.2 ± 8.5 | 311.6 ± 30.4 | 503.4 ± 27.5 | 306.1 ± 47.6 | 351.2 ± 52.8 |
 | 313.8 ± 13.9 | 840.4 ± 333.8 | 403.0 ± 47.8 | 404.0 ± 49.6 | 249.6 ± 26.7 | 356.2 ± 14.7 |
 | 39.4 ± 1.9 | 42.2 ± 3.4 | 73.8 ± 21.0 | 26.4 ± 2.7 | 10.5 ± 2.4 | 33.4 ± 3.7 |
 | 74.7 ± 10.3 | 193.8 ± 55.5 | 436.3 ± 59.9 | 608.4 ± 143.1 | 242.9 ± 87.9 | 140.6 ± 5.9 |
 | Not converged | 10.4 ± 6.1 | 28.7 ± 7.7 | Not converged | 207.4 ± 9.5 | 11.5 ± 2.7 |
 | 93.1 ± 7.7 | 206.2 ± 19.7 | 169.0 ± 10.9 | 202.5 ± 9.3 | 186.9 ± 34.8 | 155.5 ± 19.5 |
 | 29.7 ± 1.8 | 76.7 ± 26.4 | 57.6 ± 9.8 | 592.1 ± 31.9 | 46.6 ± 11.1 | 126.1 ± 29.6 |
 | 110.7 ± 20.8 | 330.5 ± 84.6 | 183.1 ± 30.2 | 249.4 ± 87.5 | 328.5 ± 92.6 | 170.3 ± 30.6 |
 | Not converged | Not converged | Not converged | Not converged | 431.9 ± 58.5 | Not converged |
 | 338.4 ± 114.5 | 292.8 ± 26.1 | 472.9 ± 73.3 | 228.5 ± 24.7 | 199.5 ± 13.2 | 3779.9 ± 627.3 |
 | 28.2 ± 4.4 | 17.1 ± 4.2 | 16.4 ± 11.8 | 49.8 ± 4.5 | 15.7 ± 3.1 | 245.6 ± 22.8 |
 | 97.3 ± 9.3 | 10.9 ± 4.1 | 26.7 ± 8.3 | 12.4 ± 6.4 | 8.3 ± 2.1 | 38.9 ± 8.3 |
 | 47.4 ± 13.2 | 73.1 ± 32.6 | Not converged | 89.8 ± 22.8 | 132.2 ± 28.0 | 47.4 ± 8.9 |
 | 21.1 ± 2.6 | 41.6 ± 14.9 | 32.8 ± 12.3 | 52.0 ± 10.8 | 27.0 ± 11.0 | 65.0 ± 15.6 |
 | Not converged | Not converged | Not converged | Not converged | 175.0 ± 93.6 | 4.2 ± 2.0 |
 | 402.2 ± 11.6 | Not converged | 37.7 ± 7.1 | Not converged | 206.2 ± 15.8 | 316.8 ± 23.4 |
 | 567.9 ± 67.5 | 348.4 ± 126.6 | 939.3 ± 204.1 | 579.6 ± 285.1 | 25.8 ± 89.4 | 2922.4 ± 251.1 |
 | 256.6 ± 51.1 | Not converged | 339.6 ± 67.2 | 507.7 ± 57.3 | 529.8 ± 50.3 | 1326.6 ± 128.2 |
 | 437.5 ± 105.1 | Not converged | 515.9 ± 79.2 | 379.8 ± 90.2 | 290.2 ± 37.0 | 1337.7 ± 144.4 |
 | Not converged | Not converged | Not converged | 1744.1 ± 126.7 | 654.0 ± 144.2 | 638.7 ± 78.0 |
 | 90.6 ± 13.0 | 74.7 ± 21.4 | 61.3 ± 16.9 | 43.4 ± 12.5 | 34.2 ± 4.1 | 117.0 ± 28.4 |
 | 126.3 ± 41.9 | 185.4 ± 52.9 | Not converged | 92.9 ± 12.1 | 368.7 ± 42.1 | 189.5 ± 50.3 |
 | 464.2 ± 105.7 | 51.4 ± 26.3 | Not converged | 85.7 ± 47.9 | 228.2 ± 100.0 | 54.5 ± 26.3 |
 | 175.4 ± 64.0 | 345.1 ± 58.0 | 4.46 ± 2.3 | Not converged | 17.0 ± 56.3 | 93.8 ± 40.2 |
 | 845.2 ± 205.0 | Not converged | Not converged | 179.4 ± 78.8 | 3430.9 ± 481.7 | Not converged |
 | 16.1 ± 2.5 | 44.8 ± 3.5 | 60.0 ± 8.4 | 83.9 ± 12.1 | 2182.0 ± 221.7 | 20.7 ± 6.3 |
 | 40.3 ± 9.3 | 71.5 ± 15.7 | 47.5 ± 2.6 | 18.6 ± 2.9 | 27.0 ± 4.7 | 74.7 ± 9.9 |
 | 60.2 ± 12.7 | 269.0 ± 33.8 | 807.6 ± 165.3 | 12.4 ± 1.1 | Not converged | 122.9 ± 16.7 |
| Doxorubicin | 0.287 ± 0.035 | 0.057 ± 0.033 | 2.99 ± 1.56 | 0.278 ± 0.107 | 0.055 ± 0.0183 | 0.423 ± 0.270 |
| Compound | EGFR-PK Inhibition, IC50 [nM] *,# |
|---|---|
| 7d | 70.3 ± 1.34 |
| 7f | 59.24 ± 1.16 |
| 7c | 81.6 ± 1.67 |
| Tamoxifen | 69.1 ± 1.39 |
| Compound | PI3K (ng/mL) | AKT (ng/mL) | mToR (ng/mL) |
|---|---|---|---|
| Control | 6.64 ± 0.15 | 45.39 ± 0.68 | 86.39 ± 2.1 |
| 7d | 4.39 ± 0.16 | 37.3 ± 0.69 | 69.3 ± 1.98 |
| 7f | 2.39 ± 0.12 | 25.34 ± 0.39 | 57.6 ± 1.23 |
| 7c | 6.1 ± 0.36 | 42.6.3 ± 0.49 | 86.3 ± 1.67 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shawish, I.; Barakat, A.; Aldalbahi, A.; Alshaer, W.; Daoud, F.; Alqudah, D.A.; Al Zoubi, M.; Hatmal, M.M.; Nafie, M.S.; Haukka, M.; et al. Acetic Acid Mediated for One-Pot Synthesis of Novel Pyrazolyl s-Triazine Derivatives for the Targeted Therapy of Triple-Negative Breast Tumor Cells (MDA-MB-231) via EGFR/PI3K/AKT/mTOR Signaling Cascades. Pharmaceutics 2022, 14, 1558. https://doi.org/10.3390/pharmaceutics14081558
Shawish I, Barakat A, Aldalbahi A, Alshaer W, Daoud F, Alqudah DA, Al Zoubi M, Hatmal MM, Nafie MS, Haukka M, et al. Acetic Acid Mediated for One-Pot Synthesis of Novel Pyrazolyl s-Triazine Derivatives for the Targeted Therapy of Triple-Negative Breast Tumor Cells (MDA-MB-231) via EGFR/PI3K/AKT/mTOR Signaling Cascades. Pharmaceutics. 2022; 14(8):1558. https://doi.org/10.3390/pharmaceutics14081558
Chicago/Turabian StyleShawish, Ihab, Assem Barakat, Ali Aldalbahi, Walhan Alshaer, Fadwa Daoud, Dana A. Alqudah, Mazhar Al Zoubi, Ma’mon M. Hatmal, Mohamed S. Nafie, Matti Haukka, and et al. 2022. "Acetic Acid Mediated for One-Pot Synthesis of Novel Pyrazolyl s-Triazine Derivatives for the Targeted Therapy of Triple-Negative Breast Tumor Cells (MDA-MB-231) via EGFR/PI3K/AKT/mTOR Signaling Cascades" Pharmaceutics 14, no. 8: 1558. https://doi.org/10.3390/pharmaceutics14081558












