Abstract
Phages are naturally occurring viruses that selectively kill bacterial species without disturbing the individual’s normal flora, averting the collateral damage of antimicrobial usage. The safety and the effectiveness of phages have been mainly confirmed in the food industry as well as in animal models. In this study, we report on the successful isolation of phages specific to Vancomycin-resistant Enterococci, including Enterococcus faecium (VREfm) and Enterococcus faecalis from sewage samples, and demonstrate their efficacy and safety for VREfm infection in the greater wax moth Galleria mellonella model. No virulence-associated genes, antibiotic resistance genes or integrases were detected in the phages’ genomes, rendering them safe to be used in an in vivo model. Phages may be considered as potential agents for therapy for bacterial infections secondary to multidrug-resistant organisms such as VREfm.
1. Introduction
By some estimates, infections from multidrug-resistant organisms (MDROs) will cause 10 million deaths per year by 2050 worldwide—more deaths than cancer [1]. Despite the efficacy of antibiotics to prevent or treat bacterial infections, their long-term use is associated with many sequelae, including the development of MDROs and the disruption of the microbiota, the gut microbiota, in particular, which may lead to the translocation of bacteria, including MDROs, into the bloodstream with a subsequent increase in complications and mortality [2]. Vancomycin-resistant Enterococcus faecium (VREfm) is a major and prevalent MDRO with considerable clinical infection control and public health implications [3,4]. The ability of VREfm clones to transfer genes encoding resistance to drugs such as vancomycin and daptomycin to other bacteria [5], their adaptation to harsh environments, and their long-term survival on high-touch surface areas contribute to an increased risk of VREfm colonization and transmission to other patients through the environment, the hands of healthcare workers, and/or equipment [6,7,8,9]. Particularly, VREfm colonization in the gastrointestinal tract can also lead to VREfm infection in high-risk individuals, such as cancer patients [2,4,10]. VREfm dominance in the gut can occur when the gut microbiome is disturbed and does not recover completely. It is hypothesized that the loss of some bacterial populations after antibiotic therapy and the disruption of the normal flora enables the expansion of VREfm and its dominance of the gut microbiome [11], leading to the loss of heterogeneity of the gut microbiome and an increased rate of morbidity and mortality in these patients [11,12].
Considering the public health concerns related to MDROs in healthcare settings and the lack of accepted and effective interventions for their eradication, new strategies to prevent or treat potential MDRO infections, thus improving clinical outcomes, are needed. Bacteriophages (or phages) are naturally occurring viruses that kill selective bacterial species. They are the most abundant organisms in the biosphere with an estimated number of 1031 phage particles [13]. Phages may constitute a good or adjunct alternative to antibiotics because of their (1) specificity to prevent or control specific bacterial species without disrupting the host’s microbiome and averting other drawbacks of antimicrobial usage [13,14,15], (2) co-evolution with the bacterial host that limits resistance, (3) safety and lack of side effects for humans [16,17], and (4) cost-effectiveness in phage production for large-scale applications [18]. Several studies have been reported in the United States on the safety and efficacy of phages and no known significant adverse events have been detected in healthy immunocompetent individuals or immunocompromised patients [19,20,21,22,23]. We recently reviewed the studies involving phage therapy against MDROs in a clinical setting [22]. These studies demonstrated good phage efficacy and safety.
In this study, we isolate VREfm phages from wastewater to design effective combinations against prevalent VREfm strains isolated in the hospital environment. Furthermore, we use the Galleria mellonella or the greater wax moth larva model as an in vivo pre-screening model, preceding the mammalian model, to study the safety and efficacy of bacteriophages in eradicating the predominant strains of VREfm. This host is an alternative and innovative model to study microbial virulence, as well as to evaluate the efficacy of antimicrobial agents such as antibiotics and phage therapy [24,25].
2. Materials and Methods
2.1. Bacterial Strains and Media
A vancomycin-susceptible enterococci (VSE) was used to isolate phages from wastewater samples. Phages were then propagated on several VREfm strains isolated from the hospital environment, rectal swabs from cancer patients, and wastewater samples, along with the VSE strain and an Enterococcus faecalis strain purchased from the Félix d’Hérelle collection (www.phages.ulaval.ca, accessed on 20 July 2022). Tryptic Soy Broth (TSB, Becton Dickinson) was used for bacterial culture and phage amplification.
2.2. Phage Recovery
A wastewater sample was obtained from a local municipal facility in Houston, Texas. The sample was centrifuged at 10,000× g for 10 min and filtered using a 0.45 µm syringe filter. The phage isolation procedure is reported elsewhere [26]. Briefly, an overnight culture of 100 µL of VSE was added to 5 mL of 2 times concentrated TSB and 5 mL of the filtered sewage sample, and incubated overnight at 37 °C as a first amplification. After several amplifications, the final filtrate was checked for the presence of VREfm-specific phages by a spot test [26,27]. Several clear phage plaques were picked, purified, and characterized.
2.3. Host Range
The recovered VREfm phages were first characterized by determining their host ranges on a panel of 12 strains of enterococci isolated from different sources: stool samples from VREfm-colonized patients and VREfm-infected patients, patient room environments, sewage samples, clinical isolates of daptomycin-resistant VREfm strains, VSE, and Enterococcus faecalis strains [28].
2.4. Electron Microscopy
A 1.5 mL sample of phage lysate (titer of at least 109 PFU/mL) was centrifuged at 23,500× g for 1 h at 4 °C. The supernatant was removed, leaving approximately 100 µL in the tube. The phage pellet was washed twice with 1.4 mL of ammonium acetate (0.1 M, pH 7.5). The residual volume (100 µL) was used to prepare the observation grid as follows: A 400-mesh carbon-coated Formvar nickel grid was glow-discharged using the PELCO easiGlow (Ted Pella, Redding, CA, USA) followed by floating onto a 10 µL droplet of provided sample for 5 min. The sample grid was then floated on a 10 µL droplet of 2% aqueous phosphotungstic acid (pH 7.0) for 30 s and examined with a FEI Tecnai G2 Spirit Twin TEM (FEI Corp., Hillsboro, OR, USA). The residual liquid was removed from the grid by touching the edge with blotting paper. Digital images were acquired with a Gatan UltraScan 1000 2k × 2k camera and Digital Micrograph software (Gatan Inc., Pleasanton, CA, USA). Phages were observed at 120 kV using a Tecnai G2 Spirit TWIN transmission electron microscope (200 nm) located at the Electron Microscopy Core of the University of Florida’s Interdisciplinary Center for Biotechnology Research [29,30].
2.5. Phage DNA Preparation and Sequencing
DNA extraction was performed on the selected phage lysates using the Phage DNA isolation kit (NORGEN Biotek) according to the manufacturer’s instructions with DNase/Rnase treatment [31]. Genome sequencing was performed using an Illumina MiSeq with 250 bp paired-end reads. The extracted DNA was further cleaned up using a QiaQuick PCR purification kit as per the manufacturer’s instructions (Qiagen). The library was prepared using a Nextera Flex kit followed by sequencing on a 250 PE run on the Illumina MiSeq [32].
2.6. Bioinformatic Analysis
Raw reads were processed using FaQCs [33] and assembled using the Geneious assembler [34,35]. For each phage, a single contig produced the final assembly. Gene calling and annotation were performed using the RASTtk [34] pipeline through PATRIC’s Genome Annotation service [35] and tRNA predictions were completed using ARAGORN [36]. Coding sequences (CDS) and reads were searched for virulence and antibiotic resistance genes by using BLAST [37] to compare assembled genomes against the Virulence Factor Database (VFDB) [38], the PATRIC Virulence Factor Database [39], the Antibiotic Resistance Gene Database (ARDB) [40], and the Comprehensive Antibiotic Resistance Database (CARD) [41]. ShortBRED [42] was used for targeted searches of coding sequences and reads for genes in VRDB, CARD, and the Resfam Antibiotic Resistance Gene Database through EDGE Bioinformatic [43,44]. Phage genus was predicted from the sequenced relatives identified by using BWA-Mem (version 0.7.9) [45] by aligning contigs to NCBI’s RefSeq database and by CDS homology using Phage Search Tool Enhanced Release (PHASTER) [46]. Phage lifestyles were predicted using PHACTs [47]. Integrases and attachment sites were searched for using PHASTER and by parsing annotated genomes for “integrase.” The percentage of total reads mapped to the host was determined by aligning reads to NCBI’s RefSeq database using BWA-Mem and determining the number of reads that mapped to an Enterococcus faecalis genome over the total number of reads. Alignments were produced using Mauve (version 2.4.0) [48].
2.7. In Vivo Efficacy of the Phages
Isolated and characterized phages were combined in a phage cocktail or phage mixture and tested for their efficacy and safety in an in vivo model. Galleria mellonella larvae were injected with VRE004, a VREfm strain isolated from stools of a VRE-infected HCT recipient at a concentration of 107 colony-forming units (CFU)/10 µL. Two isolated phages were combined in a cocktail and injected at a concentration of 2 × 106 plaque-forming units (PFU)/10 µL 1 h post-bacterial injection. An additional group of larvae received the same phage cocktail 1 h prior to VREfm injection as a prevention or prophylactic group. Control groups included larvae injected with bacteria alone, phages alone (to measure toxicity due to phage administration), sterile medium (to measure any lethal effects due to physical trauma from the injection), or without any manipulation [49]. Each group comprised 8 larvae and the experiments were replicated 5 times. The insect’s health state and mortality were observed and scored after 24 h and 48 h of incubation at 37 °C using a published health index scoring system [50]. Using STATA version 12, we conducted two mixed-effect logistic regression models, with conditions predicting the number of surviving larvae by 48 h of follow-up and controlling for replicates. For the first model, we compared a saline solution (control) to phages alone. For the second model, we compared VREfm alone to VREfm with phages. Finally, we conducted a one-way analysis of variance to compare VREfm alone to the presence of the phage cocktail and to the control group with respect to VREfm abundance. For each relevant statistically significant finding, we reported the p-value with 95% confidence intervals.
3. Results
3.1. Characterization of the Isolated Phages
Two enterococci phages (MDA1 and MDA2) targeting several VREfm and VSE strains were successfully isolated. Both phages belonged to the Caudovirales order. Phage MDA1 belongs to the Podoviridae family, whereas phage MDA2 belongs to the Myoviridae family. Podoviridae and Myoviridae can be mainly distinguished by the length of their tails; Podoviridae are characterized by their short tail and Myoviridae have a long contractile tail [29]. Phage MDA1 has an icosahedral head of 40.4 ± 1.4 nm in diameter and a non-contractile tail with a length of 18.6 ± 1.6 nm. Phage MDA2 has an icosahedral capsid of 100.8 ± 7.2 nm in diameter, and a contractile tail of 179 ± 25 nm in length and 17.2 ± 5.1 nm in width (Figure 1). Both phages were lytic and were able to eradicate at least 6 out of the 12 tested VREfm strains. The Podoviridae phages had a wider host range, infecting a total of 11/12 bacterial strains compared to 6/12 bacterial strains with the Myoviridae phages. The phage cocktail was able to eradicate all the 12 tested bacterial strains, including the daptomycin-resistant VREfm strains (Table 1).
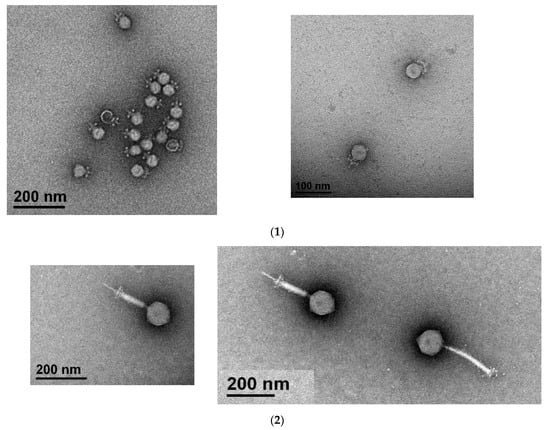
Figure 1.
Visualization of phages MDA1 (1) and MDA2 (2) under the electron microscope. Scale bars indicate 100 nm and 200 nm.

Table 1.
Activity of phages against VREfm isolated from different sources. Boxes with (+) to show the ability of the phage to lyse the VREfm strain tested, (-) indicates that the phage had no effect on the tested bacteria.
The genomes of MDA1 and MDA2 were 18,058 bp and 140,226 bp long, respectively. MDA1’s linear genome had a GC content of 33% (Figure 2). No tRNA was found in the genome. When compared to other published enterococcal genomes, phage MDA1’s genome had 82% homology with Enterococcus phage vB_Efae230P-4 (Figure S1) [51]. Phage MDA2 had a circular genome with a GC content of 35.8%, which was similar to that of other enterococcal phages and of its enterococcal hosts (38%) [51,52,53]. Six tRNAs were found in its genome. tRNAs are usually present in the phage genome as a result of variance in codon usage between their genome and their host’s genome, enabling phages to bypass codons overused in the phage genes and compensate for the extra bias [54,55]. The linearized genome of phage MDA2 is represented in Figure 3. When compared to other published enterococcal genomes, the genome of MDA2 was 70%, similar to that of phage phiEF24C [56]. Both Podoviridae vB_Efae230P-4 and Myoviridae phiEF24C were isolated from the environment in Japan and targeted vancomycin-resistant E. faecalis [51,56] (Figure S2). Of interest, no virulence-associated genes, antibiotic resistance genes or integrase genes were detected in the phages’ genomes (Tables S1 and S2).
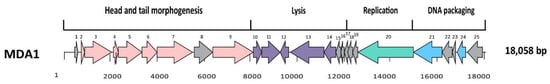
Figure 2.
Genome structure of MDA1 phage. Each arrow represents an ORF. Colors represent different genomic regions including the DNA packaging (blue), the head and tail morphogenesis genes (pink), host lysis genes (purple), and the replication–transcription region (green). Grey arrows represent genes coding for hypothetical proteins.
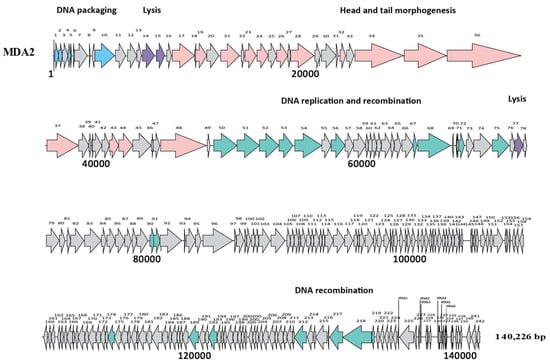
Figure 3.
Genome structure of MDA2 phage. Each arrow represents an ORF. Colors represent different genomic regions including the DNA packaging (blue), the head and tail morphogenesis genes (pink), host lysis genes (purple), and the replication–transcription region (green). Grey arrows represent genes coding for hypothetical proteins. tRNAs are represented by vertical lines.
3.2. Safety and Efficacy of the Phage Cocktail
The phage cocktail (MDA1 and MDA2) was administered to VRE-injected larvae and survival was monitored for 48 h. At 48 h, larvae injected with saline only (control) were just as likely to be alive as larvae injected with the phage cocktail only (82.5% vs. 85% alive; OR = 1.2, SE = 0.8, p = 0.75) demonstrating the safety of the phages. Additionally, by 48 h of follow-up, larvae injected with VREfm (including daptomycin-resistant VREfm) and phages were 3.7 (treatment group) and 6.5 times (prophylactic group) more likely to survive than larvae injected with VREfm only (55% vs. 25% alive; OR = 3.7, SE = 1.8, p = 0.07 and 67.5% vs. 25% alive; OR = 6.5, SE = 3.3, p < 0.001, respectively), demonstrating the efficacy of phages against VREfm in a larva model. Then, 16S analysis was performed on all the larvae to qualitatively identify VREfm abundance in each group. Larvae from the same group were pooled and homogenized with sterile media and sent for DNA extraction and 16S sequencing and analysis. The larvae groups that received sterile media or phages alone did not have any VREfm and were comparable to the control (without any manipulation). VREfm abundance was significantly higher in the group receiving VREfm alone compared to the control (p < 0.001), the group receiving VREfm and then phages (p < 0.001), or the group receiving phages and then VREfm (p < 0.001). As expected, there were no significant differences in VREfm abundance between the control, the group receiving VREfm and then phages, or the group receiving phages and then VREfm.
3.3. Data Availability
The genome sequences of MDA1 and MDA2 have been deposited in GenBank under accession numbers MW623430 and MW633168, respectively.
4. Discussion
Vancomycin-resistant enterococci have long been widespread in the community as well as in hospital settings [6,57,58,59,60]. New interventions against these pathogens are needed for prevention and treatment of resistant enterococcal infections. Phages constitute a safe and effective strategy against pathogenic bacteria. In this study, we were able to isolate two different phages from sewage samples. The genomic characterization of these phages indicated the absence of virulence, antibiotic resistance, and integrase genes, making them safe to be used in the larvae model and all in vivo models. The phages were able to eradicate multiple VREfm, including daptomycin-resistant VREfm strains isolated from different sources such as the environment, hospital rooms, and patients. Additionally, we showed the efficacy of a phage cocktail against this bacterium in vitro and in an in vivo larva model. Indeed, the VREfm-specific phage cocktail composed of the two distinct phages was shown to be effective in a larvae model, reducing VREfm abundance and increasing larval survival over a 48 h phage treatment compared to the group that did not receive phages but only VREfm. Our results highlight the feasibility and the potential success of these phages in eradicating VREfm.
Phage therapy is gaining more attention among the scientific and medical communities in Western countries for many reasons, including the slow process for new antibiotic development, and the increasing incidence of MDROs worldwide. Phages may present an approach that can potentially aid in the fight against MDRO colonization and infections. Naturally occurring phages with good safety profiles have been used in Western countries to treat infections caused by MDROs [20,22,23]. Nevertheless, there are a lack of in vivo VREfm-specific phage studies performed on VREfm-colonized and VREfm-infected patients, including daptomycin-resistant VREfm.
The efficacy and safety of antimicrobial agents such as phages are assessed in an animal model before their potential application in humans. In vivo experiments are crucial to identify safety issues and the potential loss of activity due to host factors. However, experiments using murine models is time-consuming, expensive, and could be ethically objectionable. Galleria mellonella larvae or wax moth is an alternative and innovative model to study microbial virulence, as well as to evaluate the efficacy of antimicrobial agents such as antibiotics and phages [25,61,62]. It can serve as a pre-screening in vivo experiment preceding a mammalian model. This model is inexpensive, simple, and does not require ethical approval; moreover, Galleria mellonella survives at 37 °C, and has a significantly similar innate immune system to vertebrates [25,61,62,63]. The Galleria mellonella model has been adopted to evaluate phage therapy against multiple bacterial pathogens, demonstrating the efficacy of phages in vivo in increasing bacterial clearance and survival rates [24,64,65,66,67]. To our knowledge, no study has investigated this in vivo model to evaluate the efficacy of bacteriophages against VREfm.
Few phages with activity against VREfm have been isolated [68,69,70,71,72]. To our knowledge, two studies tested VREfm phages in an in vivo model. Briefly, a single intraperitoneal injection of 3 × 108 plaque-forming units of the VREfm ENB6, isolated from raw sewage at a local municipal sewage treatment plant, administered 45 min after bacterial challenge with a clinical VREfm isolate, was enough to rescue VREfm bacteremic mice. In the absence of that phage, VREfm bacteremia was fatal within 48 h [68]. A recent report showed the successful treatment of a VREfm abdominal infection in a pediatric liver transplant patient that was failing many courses of antibiotics and surgical management. An individualized two-phage cocktail was administered intravenously over 2 h twice daily at a concentration of 8 × 107 PFU/mL over a ten-day course of 2 mL/kg bodyweight followed by another ten days of 2 mL/kg bodyweight at a concentration of 5 × 108 PFU/mL. To reduce the theoretical risk of an allergic reaction against the phage preparation, an H1-antagonist was administered before every phage application. A reduction in c-reactive protein starting the day after the first dose and constant clinical improvement were observed. There were no observed adverse events attributable to phage administration and VREfm was not detected in the routine screening from rectal swabs [72].
5. Conclusions
Phages may constitute an alternative strategy to manage resistant bacteria such as VREfm in an innovative, safe, and natural way. Our results highlight the feasibility and the potential success of these phages in inhibiting VREfm in in vitro and in vivo models. Future studies in animal models as well as clinical trials to test the ability of these phages to safely and effectively eradicate VREfm-colonized or infected animal and human hosts, such as immunocompromised cancer patients, in particular, are long overdue. In addition, the impact of phage therapy on the gut microbiota balance and the regulation of the immune system in these hosts should also be determined in future trials.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/pharmaceutics14081591/s1, Supplementary Table S1: Gene characteristics of MDA1 genome; Supplementary Table S2: Gene characteristics of MDA2 genome; Figure S1: Genome comparison between MDA1 and Enterococcus phage vB_Efae230P-4. Figure S2: Genome comparison between MDA2 and Enterococcus phage phiEF24C.
Author Contributions
Conceptualization, L.E.H. and R.F.C.; data curation, L.E.H. and J.R.C.; formal analysis, L.E.H.; funding acquisition, L.E.H. and M.S.; methodology, L.E.H., G.A., J.F.M., A.L.T., C.P.C., M.D., S.T.Y., C.P.H. and M.S.; project administration, L.E.H., C.P.H. and A.M.; resources, A.L.T., M.S. and A.M.; software, J.R.C. and A.M.; supervision, M.S. and A.M.; writing—original draft, L.E.H.; writing—review and editing, R.F.C. All authors have read and agreed to the published version of the manuscript.
Funding
Xenex Disinfection Services provided funding for laboratory analysis. L.E.H.’s research was funded by the American Cancer Society Postdoctoral award 131044-PF-18-191-01-LIB. The work was also supported by the NIH/NCI under award number P30CA016672. Electron microscopy was performed by the CCSG-funded High-Resolution Electron Microscopy Facility, NCI # CA0166772. The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Acknowledgments
We thank the TAILOR center at the Baylor College of Medicine for bacteriophage genome sequencing and analyses. We also thank Kenneth Dunner Jr. and Nicole Machi for their assistance with the electron microcopy.
Conflicts of Interest
R.F.C. has received research grants, stock options, from and acts as a consultant to Xenex Disinfection Services. M.S. is employed by Xenex Disinfection Services. All other authors report no conflict of interest.
References
- de Kraker, M.E.; Stewardson, A.J.; Harbarth, S. Will 10 Million People Die a Year due to Antimicrobial Resistance by 2050? PLoS Med. 2016, 13, e1002184. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ford, C.D.; Lopansri, B.K.; Haydoura, S.; Snow, G.; Dascomb, K.K.; Asch, J.; Bo Petersen, F.; Burke, J.P. Frequency, risk factors, and outcomes of vancomycin-resistant Enterococcus colonization and infection in patients with newly diagnosed acute leukemia: Different patterns in patients with acute myelogenous and acute lymphoblastic leukemia. Infect. Control Hosp. Epidemiol. 2015, 36, 47–53. [Google Scholar] [CrossRef] [PubMed]
- Scheich, S.; Lindner, S.; Koenig, R.; Reinheimer, C.; Wichelhaus, T.A.; Hogardt, M.; Besier, S.; Kempf, V.A.J.; Kessel, J.; Martin, H.; et al. Clinical impact of colonization with multidrug-resistant organisms on outcome after allogeneic stem cell transplantation in patients with acute myeloid leukemia. Cancer 2018, 124, 286–296. [Google Scholar] [CrossRef] [PubMed]
- Shono, Y.; Docampo, M.D.; Peled, J.U.; Perobelli, S.M.; Velardi, E.; Tsai, J.J.; Slingerland, A.E.; Smith, O.M.; Young, L.F.; Gupta, J.; et al. Increased GVHD-related mortality with broad-spectrum antibiotic use after allogeneic hematopoietic stem cell transplantation in human patients and mice. Sci. Transl. Med. 2016, 8, 339ra71. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dubin, K.; Pamer, E.G. Enterococci and Their Interactions with the Intestinal Microbiome. Microbiol. Spectr. 2014, 5, 5–6. [Google Scholar]
- Arias, C.A.; Murray, B.E. The rise of the Enterococcus: Beyond vancomycin resistance. Nat. Rev. Microbiol. 2012, 10, 266–278. [Google Scholar] [CrossRef] [Green Version]
- Chemaly, R.F.; Simmons, S.; Dale, C., Jr.; Ghantoji, S.S.; Rodriguez, M.; Gubb, J.; Stachowiak, J.; Stibich, M. The role of the healthcare environment in the spread of multidrug-resistant organisms: Update on current best practices for containment. Ther. Adv. Infect. Dis. 2014, 2, 79–90. [Google Scholar] [CrossRef]
- Montealegre, M.C.; Singh, K.V.; Murray, B.E. Gastrointestinal Tract Colonization Dynamics by Different Enterococcus faecium Clades. J. Infect. Dis 2016, 213, 1914–1922. [Google Scholar] [CrossRef] [Green Version]
- Zirakzadeh, A.; Patel, R. Vancomycin-resistant enterococci: Colonization, infection, detection, and treatment. Mayo Clin. Proc. 2006, 81, 529–536. [Google Scholar] [CrossRef] [Green Version]
- Bilinski, J.; Robak, K.; Peric, Z.; Marchel, H.; Karakulska-Prystupiuk, E.; Halaburda, K.; Rusicka, P.; Swoboda-Kopec, E.; Wroblewska, M.; Wiktor-Jedrzejczak, W.; et al. Impact of Gut Colonization by Antibiotic-Resistant Bacteria on the Outcomes of Allogeneic Hematopoietic Stem Cell Transplantation: A Retrospective, Single-Center Study. Biol. Blood Marrow Transplant 2016, 22, 1087–1093. [Google Scholar] [CrossRef] [Green Version]
- Ubeda, C.; Taur, Y.; Jenq, R.R.; Equinda, M.J.; Son, T.; Samstein, M.; Viale, A.; Socci, N.D.; van den Brink, M.R.; Kamboj, M.; et al. Vancomycin-resistant Enterococcus domination of intestinal microbiota is enabled by antibiotic treatment in mice and precedes bloodstream invasion in humans. J. Clin. Investig. 2010, 120, 4332–4341. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Xu, S.; Ren, Z.; Jiang, J.; Zheng, S. Gut microbiota and allogeneic transplantation. J. Transl. Med. 2015, 13, 275. [Google Scholar] [CrossRef] [PubMed]
- Clokie, M.R.; Millard, A.D.; Letarov, A.V.; Heaphy, S. Phages in nature. Bacteriophage 2011, 1, 31–45. [Google Scholar] [CrossRef] [Green Version]
- Salmond, G.P.; Fineran, P.C. A century of the phage: Past, present and future. Nat. Rev. Microbiol. 2015, 13, 777–786. [Google Scholar] [CrossRef] [PubMed]
- Scarpellini, E.; Ianiro, G.; Attili, F.; Bassanelli, C.; De Santis, A.; Gasbarrini, A. The human gut microbiota and virome: Potential therapeutic implications. Dig. Liver Dis. 2015, 47, 1007–1012. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Borysowski, J.; Gorski, A. Is phage therapy acceptable in the immunocompromised host? Int. J. Infect. Dis. 2008, 12, 466–471. [Google Scholar] [CrossRef] [Green Version]
- Bruttin, A.; Brussow, H. Human volunteers receiving Escherichia coli phage T4 orally: A safety test of phage therapy. Antimicrob. Agents Chemother. 2005, 49, 2874–2878. [Google Scholar] [CrossRef] [Green Version]
- Zimecki, M.; Artym, J.; Kocieba, M.; Weber-Dabrowska, B.; Borysowski, J.; Gorski, A. Prophylactic effect of bacteriophages on mice subjected to chemotherapy-induced immunosuppression and bone marrow transplant upon infection with Staphylococcus aureus. Med. Microbiol. Immunol. 2010, 199, 71–79. [Google Scholar] [CrossRef]
- Aslam, S.; Yung, G.; Dan, J.; Reed, S.; LeFebvre, M.; Logan, C.; Taplitz, R.; Law, N.; Golts, E.; Afshar, K.; et al. (373)—Bacteriophage Treatment in a Lung Transplant Recipient. J. Heart Lung Transpl. 2018, 37, S155–S156. [Google Scholar] [CrossRef]
- Chan, B.K.; Turner, P.E.; Kim, S.; Mojibian, H.R.; Elefteriades, J.A.; Narayan, D. Phage treatment of an aortic graft infected with Pseudomonas aeruginosa. Evol. Med. Public Health 2018, 2018, 60–66. [Google Scholar] [CrossRef] [Green Version]
- Dedrick, R.M.; Guerrero-Bustamante, C.A.; Garlena, R.A.; Russell, D.A.; Ford, K.; Harris, K.; Gilmour, K.C.; Soothill, J.; Jacobs-Sera, D.; Schooley, R.T.; et al. Engineered bacteriophages for treatment of a patient with a disseminated drug-resistant Mycobacterium abscessus. Nat. Med. 2019, 25, 730–733. [Google Scholar] [CrossRef] [PubMed]
- El Haddad, L.; Harb, C.P.; Gebara, M.A.; Stibich, M.A.; Chemaly, R.F. A Systematic and Critical Review of Bacteriophage Therapy against Multi-Drug Resistant ESKAPE Organisms in Humans. Clin. Infect. Dis. 2018, 69, 167–178. [Google Scholar] [CrossRef] [PubMed]
- Schooley, R.T.; Biswas, B.; Gill, J.J.; Hernandez-Morales, A.; Lancaster, J.; Lessor, L.; Barr, J.J.; Reed, S.L.; Rohwer, F.; Benler, S.; et al. Development and Use of Personalized Bacteriophage-Based Therapeutic Cocktails to Treat a Patient with a Disseminated Resistant Acinetobacter baumannii Infection. Antimicrob. Agents Chemother. 2017, 61, e00954-17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nale, J.Y.; Chutia, M.; Carr, P.; Hickenbotham, P.T.; Clokie, M.R. 'Get in Early'; Biofilm and Wax Moth (Galleria mellonella) Models Reveal New Insights into the Therapeutic Potential of Clostridium difficile Bacteriophages. Front. Microbiol. 2016, 7, 1383. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tsai, C.J.; Loh, J.M.; Proft, T. Galleria mellonella infection models for the study of bacterial diseases and for antimicrobial drug testing. Virulence 2016, 7, 214–229. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- El Haddad, L.; Moineau, S. Characterization of a Novel Panton-Valentine leukocidin (PVL)-encoding staphylococcal phage and its naturally PVL-lacking variant. Appl. Environ. Microbiol. 2013, 79, 2828–2832. [Google Scholar] [CrossRef] [Green Version]
- El Haddad, L.; Ben Abdallah, N.; Plante, P.-L.; Dumaresq, J.; Katsarava, R.; Labrie, S.; Corbeil, J.; St-Gelais, D.; Moineau, S. Improving the Safety of Staphylococcus aureus Polyvalent Phages by Their Production on a Staphylococcus xylosus Strain. PLoS ONE 2014, 9, e102600. [Google Scholar] [CrossRef] [Green Version]
- Kaur, S.; Harjai, K.; Chhibber, S. Methicillin-resistant Staphylococcus aureus phage plaque size enhancement using sublethal concentrations of antibiotics. Appl. Environ. Microbiol. 2012, 78, 8227–8233. [Google Scholar] [CrossRef] [Green Version]
- Ackermann, H.W. 5500 Phages examined in the electron microscope. Arch. Virol. 2007, 152, 227–243. [Google Scholar] [CrossRef] [PubMed]
- Clokie, M.; Kropinski, A. Bacteriophages: Methods and Protocols, Volume 1: Isolation, Characterization, and Interactions; Humana Press: Totowa, NJ, USA, 2009. [Google Scholar]
- Kim, M.S.; Myung, H. Complete Genome of Staphylococcus aureus Phage SA11. J. Virol. 2012, 86, 10232. [Google Scholar] [CrossRef] [Green Version]
- Russell, D.A. Sequencing, Assembling, and Finishing Complete Bacteriophage Genomes. In Bacteriophages: Methods and Protocols; Clokie, M.R.J., Kropinski, A.M., Lavigne, R., Eds.; Springer: New York, NY, USA, 2018; Volume 3, pp. 109–125. [Google Scholar]
- Lo, C.C.; Chain, P.S. Rapid evaluation and quality control of next generation sequencing data with FaQCs. BMC Bioinform. 2014, 15, 366. [Google Scholar] [CrossRef] [Green Version]
- Brettin, T.; Davis, J.J.; Disz, T.; Edwards, R.A.; Gerdes, S.; Olsen, G.J.; Olson, R.; Overbeek, R.; Parrello, B.; Pusch, G.D.; et al. RASTtk: A modular and extensible implementation of the RAST algorithm for building custom annotation pipelines and annotating batches of genomes. Sci. Rep. 2015, 5, 8365. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wattam, A.R.; Davis, J.J.; Assaf, R.; Boisvert, S.; Brettin, T.; Bun, C.; Conrad, N.; Dietrich, E.M.; Disz, T.; Gabbard, J.L.; et al. Improvements to PATRIC, the all-bacterial Bioinformatics Database and Analysis Resource Center. Nucleic Acids Res. 2017, 45, D535–D542. [Google Scholar] [CrossRef]
- Laslett, D.; Canback, B. ARAGORN, a program to detect tRNA genes and tmRNA genes in nucleotide sequences. Nucleic Acids Res. 2004, 32, 11–16. [Google Scholar] [CrossRef] [PubMed]
- Johnson, M.; Zaretskaya, I.; Raytselis, Y.; Merezhuk, Y.; McGinnis, S.; Madden, T.L. NCBI BLAST: A better web interface. Nucleic Acids Res. 2008, 36, W5–W9. [Google Scholar] [CrossRef]
- Chen, L.; Zheng, D.; Liu, B.; Yang, J.; Jin, Q. VFDB 2016: Hierarchical and refined dataset for big data analysis--10 years on. Nucleic Acids Res. 2016, 44, D694–D697. [Google Scholar] [CrossRef] [PubMed]
- Mao, C.; Abraham, D.; Wattam, A.R.; Wilson, M.J.; Shukla, M.; Yoo, H.S.; Sobral, B.W. Curation, integration and visualization of bacterial virulence factors in PATRIC. Bioinformatics 2015, 31, 252–258. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, B.; Pop, M. ARDB--Antibiotic Resistance Genes Database. Nucleic Acids Res. 2009, 37, D443–D447. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McArthur, A.G.; Waglechner, N.; Nizam, F.; Yan, A.; Azad, M.A.; Baylay, A.J.; Bhullar, K.; Canova, M.J.; De Pascale, G.; Ejim, L.; et al. The comprehensive antibiotic resistance database. Antimicrob. Agents Chemother. 2013, 57, 3348–3357. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kaminski, J.; Gibson, M.K.; Franzosa, E.A.; Segata, N.; Dantas, G.; Huttenhower, C. High-Specificity Targeted Functional Profiling in Microbial Communities with ShortBRED. PLoS Comput. Biol. 2015, 11, e1004557. [Google Scholar] [CrossRef] [Green Version]
- Gibson, M.K.; Forsberg, K.J.; Dantas, G. Improved annotation of antibiotic resistance determinants reveals microbial resistomes cluster by ecology. ISME J 2015, 9, 207–216. [Google Scholar] [CrossRef] [PubMed]
- Li, P.-E.; Lo, C.-C.; Anderson, J.J.; Davenport, K.W.; Bishop-Lilly, K.A.; Xu, Y.; Ahmed, S.; Feng, S.; Mokashi, V.P.; Chain, P.S.G. Enabling the democratization of the genomics revolution with a fully integrated web-based bioinformatics platform. Nucleic Acids Res. 2016, 45, 67–80. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, H. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv 2013, arXiv:1303.3997. [Google Scholar]
- Arndt, D.; Grant, J.R.; Marcu, A.; Sajed, T.; Pon, A.; Liang, Y.; Wishart, D.S. PHASTER: A better, faster version of the PHAST phage search tool. Nucleic Acids Res. 2016, 44, W16–W21. [Google Scholar] [CrossRef] [Green Version]
- McNair, K.; Bailey, B.A.; Edwards, R.A. PHACTS, a computational approach to classifying the lifestyle of phages. Bioinformatics 2012, 28, 614–618. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Darling, A.C.; Mau, B.; Blattner, F.R.; Perna, N.T. Mauve: Multiple alignment of conserved genomic sequence with rearrangements. Genome Res. 2004, 14, 1394–1403. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alghoribi, M.F.; Gibreel, T.M.; Dodgson, A.R.; Beatson, S.A.; Upton, M. Galleria mellonella infection model demonstrates high lethality of ST69 and ST127 uropathogenic E. coli. PLoS ONE 2014, 9, e101547. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Loh, J.M.; Adenwalla, N.; Wiles, S.; Proft, T. Galleria mellonella larvae as an infection model for group A streptococcus. Virulence 2013, 4, 419–428. [Google Scholar] [CrossRef] [Green Version]
- Lossouarn, J.; Briet, A.; Moncaut, E.; Furlan, S.; Bouteau, A.; Son, O.; Leroy, M.; DuBow, M.S.; Lecointe, F.; Serror, P.; et al. Enterococcus faecalis Countermeasures Defeat a Virulent Picovirinae Bacteriophage. Viruses 2019, 11, 48. [Google Scholar] [CrossRef] [Green Version]
- Nasr Azadani, D.; Zhang, D.; Hatherill, J.R.; Silva, D.; Turner, J.W. Isolation, characterization, and comparative genomic analysis of a phage infecting high-level aminoglycoside-resistant (HLAR) Enterococcus faecalis. PeerJ 2020, 8, e9171. [Google Scholar] [CrossRef] [PubMed]
- Lam, M.M.C.; Seemann, T.; Bulach, D.M.; Gladman, S.L.; Chen, H.; Haring, V.; Moore, R.J.; Ballard, S.; Grayson, M.L.; Johnson, P.D.R.; et al. Comparative analysis of the first complete Enterococcus faecium genome. J. Bacteriol. 2012, 194, 2334–2341. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Khalifa, L.; Gelman, D.; Shlezinger, M.; Dessal, A.L.; Coppenhagen-Glazer, S.; Beyth, N.; Hazan, R. Defeating Antibiotic- and Phage-Resistant Enterococcus faecalis Using a Phage Cocktail in Vitro and in a Clot Model. Front. Microbiol. 2018, 9, 326. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Prabhakaran, R.; Chithambaram, S.; Xia, X. Aeromonas phages encode tRNAs for their overused codons. Int. J. Comput. Biol. Drug Des. 2014, 7, 168–182. [Google Scholar] [CrossRef] [PubMed]
- Uchiyama, J.; Rashel, M.; Maeda, Y.; Takemura, I.; Sugihara, S.; Akechi, K.; Muraoka, A.; Wakiguchi, H.; Matsuzaki, S. Isolation and characterization of a novel Enterococcus faecalis bacteriophage phiEF24C as a therapeutic candidate. FEMS Microbiol. Lett. 2008, 278, 200–206. [Google Scholar] [CrossRef] [Green Version]
- El Haddad, L.; Hanson, B.M.; Arias, C.A.; Ghantoji, S.S.; Harb, C.P.; Stibich, M.; Chemaly, R.F. Emergence and Transmission of Daptomycin and Vancomycin-Resistant Enterococci Between Patients and Hospital Rooms. Clin. Infect. Dis. 2021, 73, 2306–2313. [Google Scholar] [CrossRef]
- Sundermann, A.J.; Babiker, A.; Marsh, J.W.; Shutt, K.A.; Mustapha, M.M.; Pasculle, A.W.; Ezeonwuka, C.; Saul, M.I.; Pacey, M.P.; Van Tyne, D.; et al. Outbreak of Vancomycin-resistant Enterococcus faecium in Interventional Radiology: Detection Through Whole-genome Sequencing-based Surveillance. Clin. Infect. Dis. Off. Publ. Infect. Dis. Soc. Am. 2020, 70, 2336–2343. [Google Scholar] [CrossRef]
- Sassi, M.; Guérin, F.; Zouari, A.; Beyrouthy, R.; Auzou, M.; Fines-Guyon, M.; Potrel, S.; Dejoies, L.; Collet, A.; Boukthir, S.; et al. Emergence of optrA-mediated linezolid resistance in enterococci from France, 2006–2016. J. Antimicrob. Chemother. 2019, 74, 1469–1472. [Google Scholar] [CrossRef] [PubMed]
- Benamu, E.; Deresinski, S. Vancomycin-resistant enterococcus infection in the hematopoietic stem cell transplant recipient: An overview of epidemiology, management, and prevention. F1000resarch 2018, 7, 3. [Google Scholar] [CrossRef] [Green Version]
- Beeton, M.L.; Alves, D.R.; Enright, M.C.; Jenkins, A.T. Assessing phage therapy against Pseudomonas aeruginosa using a Galleria mellonella infection model. Int. J. Antimicrob. Agents 2015, 46, 196–200. [Google Scholar] [CrossRef] [Green Version]
- Desbois, A.P.; Coote, P.J. Utility of Greater Wax Moth Larva (Galleria mellonella) for Evaluating the Toxicity and Efficacy of New Antimicrobial Agents. Adv. Appl. Microbiol. 2012, 78, 25–53. [Google Scholar]
- Champion, O.L.; Wagley, S.; Titball, R.W. Galleria mellonella as a model host for microbiological and toxin research. Virulence 2016, 7, 840–845. [Google Scholar] [CrossRef] [Green Version]
- Abbasifar, R.; Kropinski, A.M.; Sabour, P.M.; Chambers, J.R.; MacKinnon, J.; Malig, T.; Griffiths, M.W. Efficiency of bacteriophage therapy against Cronobacter sakazakii in Galleria mellonella (greater wax moth) larvae. Arch. Virol. 2014, 159, 2253–2261. [Google Scholar] [CrossRef] [PubMed]
- Jeon, J.; Park, J.-H.; Yong, D. Efficacy of bacteriophage treatment against carbapenem-resistant Acinetobacter baumannii in Galleria mellonella larvae and a mouse model of acute pneumonia. BMC Microbiol. 2019, 19, 70. [Google Scholar] [CrossRef] [PubMed]
- Jeon, J.; Yong, D. Two Novel Bacteriophages Improve Survival in Galleria mellonella Infection and Mouse Acute Pneumonia Models Infected with Extensively Drug-Resistant Pseudomonas aeruginosa. Appl. Environ. Microbiol. 2019, 85, e02900–e02918. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Manohar, P.; Nachimuthu, R.; Lopes, B.S. The therapeutic potential of bacteriophages targeting gram-negative bacteria using Galleria mellonella infection model. BMC Microbiol. 2018, 18, 97. [Google Scholar] [CrossRef] [Green Version]
- Biswas, B.; Adhya, S.; Washart, P.; Paul, B.; Trostel, A.N.; Powell, B.; Carlton, R.; Merril, C.R. Bacteriophage therapy rescues mice bacteremic from a clinical isolate of vancomycin-resistant Enterococcus faecium. Infect. Immun. 2002, 70, 204–210. [Google Scholar] [CrossRef] [Green Version]
- Gong, P.; Cheng, M.; Li, X.; Jiang, H.; Yu, C.; Kahaer, N.; Li, J.; Zhang, L.; Xia, F.; Hu, L.; et al. Characterization of Enterococcus faecium bacteriophage IME-EFm5 and its endolysin LysEFm5. Virology 2016, 492, 11–20. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Wang, W.; Lv, Y.; Zheng, W.; Mi, Z.; Pei, G.; An, X.; Xu, X.; Han, C.; Liu, J.; et al. Characterization and complete genome sequence analysis of novel bacteriophage IME-EFm1 infecting Enterococcus faecium. J. Gen. Virol. 2014, 95, 2565–2575. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xing, S.; Zhang, X.; Sun, Q.; Wang, J.; Mi, Z.; Pei, G.; Huang, Y.; An, X.; Fu, K.; Zhou, L.; et al. Complete genome sequence of a novel, virulent Ahjdlikevirus bacteriophage that infects Enterococcus faecium. Arch. Virol. 2017, 162, 3843–3847. [Google Scholar] [CrossRef]
- Paul, K.; Merabishvili, M.; Hazan, R.; Christner, M.; Herden, U.; Gelman, D.; Khalifa, L.; Yerushalmy, O.; Coppenhagen-Glazer, S.; Harbauer, T.; et al. Bacteriophage Rescue Therapy of a Vancomycin-Resistant Enterococcus faecium Infection in a One-Year-Old Child following a Third Liver Transplantation. Viruses 2021, 13, 1785. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).