Does Secondary Plant Metabolite Ursolic Acid Exhibit Antibacterial Activity against Uropathogenic Escherichia coli Living in Single- and Multispecies Biofilms?
Abstract
:1. Introduction
2. Materials and Methods
2.1. Bacterial Strains
2.2. Cultural Media and Chemicals
2.3. Antimicrobial Agent
2.4. Preparation of Bacterial Suspension
2.5. Preparation of Biofilm Cultures
2.6. Biofilm Formation on Microtiter Plates and Count of Live Bacteria in Biofilm
2.7. Biofilm Formation Assay and Quantification
2.8. Determination of the Metabolic Activity of Bacteria in a Biofilm by Spectrophotometry
2.9. Effect of UA on Bacterial Morphology
2.10. Statistical Analysis
3. Results
3.1. Survival of E. coli in Single-, Dual-, and Triple-Species Biofilms
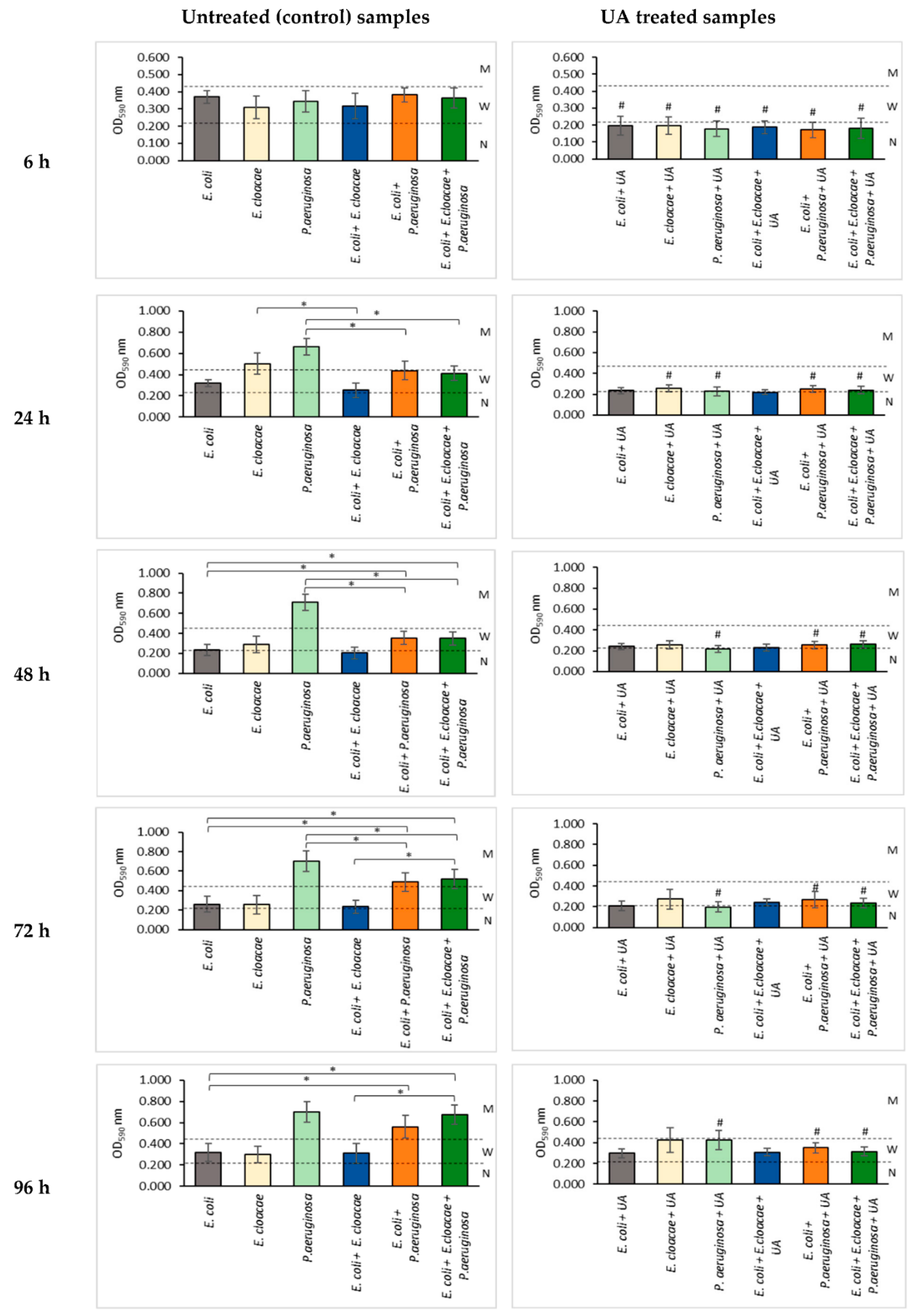
3.2. Formation of Biofilm Mass by Uropathogenic Rods in Nontreated (Control) and UA-Treated Samples
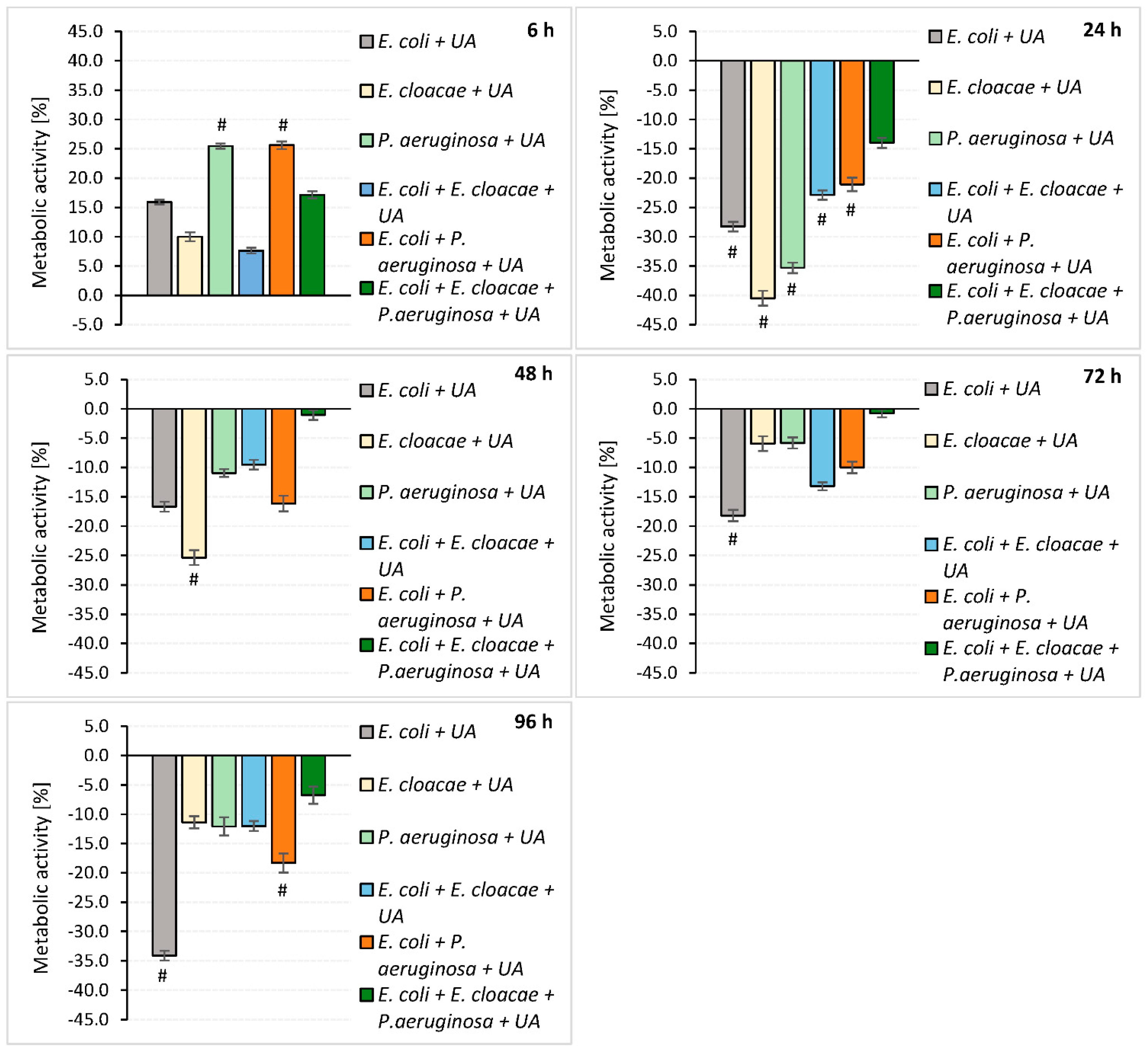
3.3. Determination of the Metabolic Activity of Bacteria Living in Mono-, Dual- and Triple-Species Biofilms
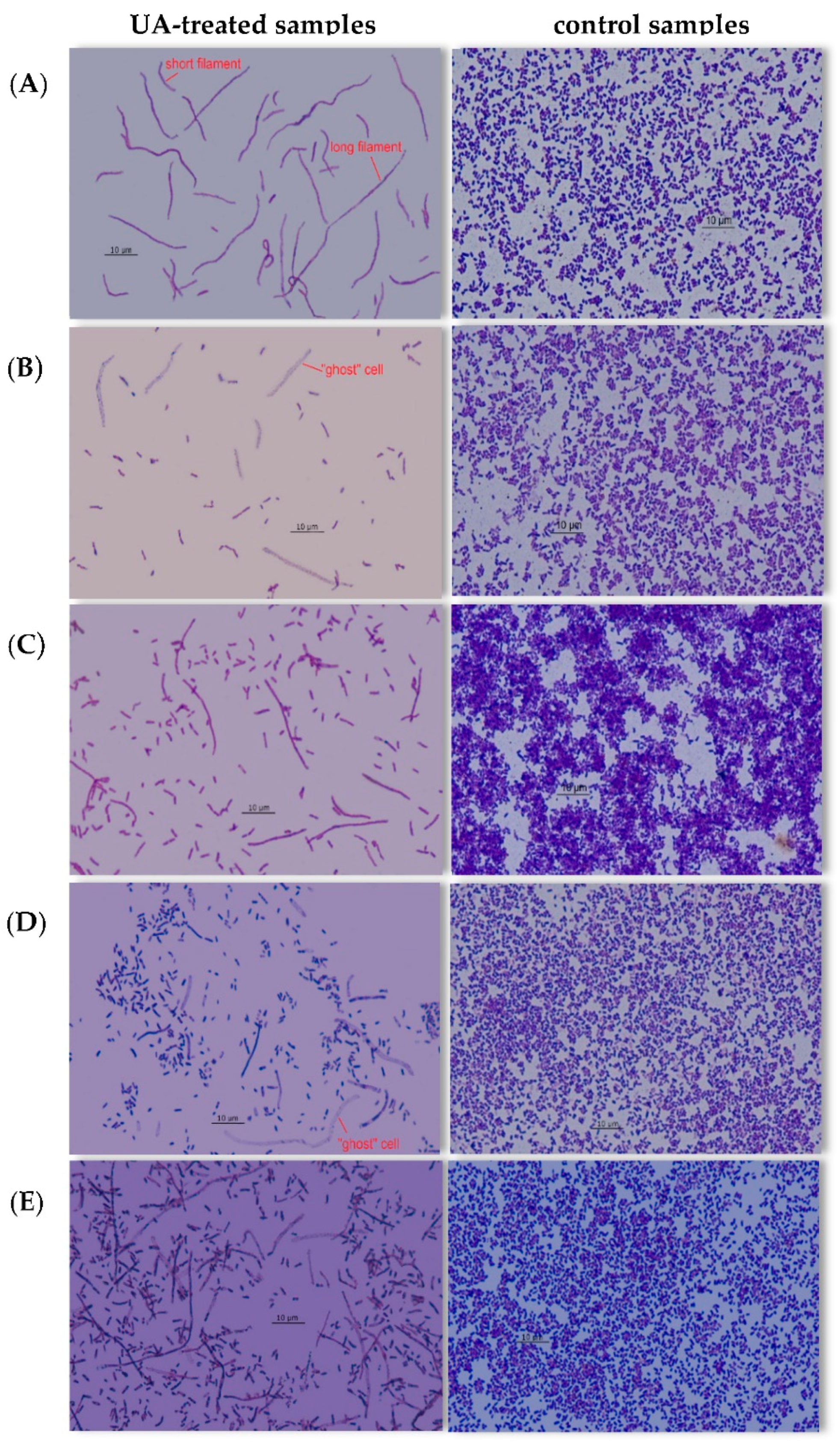
3.4. Effect of UA on Bacterial Morphology
4. Discussion
4.1. Survival of the Escherichia coli Rods in Biofilms Cultured in the Absence of UA
4.2. Biofilm Formation and Metabolic Activity of Bacteria Growing in the Absence of UA
4.3. Survival, Biomass Formation, and Metabolic Activity of Bacteria in Biofilms Treated with UA
4.4. Changes in Cell Morphology of Bacteria Treated with UA
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Jaroniewski, W. Medicinal plants of Polish forests. Bearberry Arctostaphylos uva-ursi (L.) Sprengl. Wszechświat 1986, 87, 135–136. (In Polish) [Google Scholar]
- Jäger, S.; Trojan, H.; Kopp, T.; Laszczyk, M.N.; Scheffler, A. Pentacyclic triterpene distribution in various plants—Rich sources for a new group of multi-potent plant extracts. Molecules 2009, 14, 2016–2031. [Google Scholar] [CrossRef] [PubMed]
- Mohd Azman, N.A.; Gallego, M.G.; Segovia, F.; Abdullah, S.; Shaarani, S.M.; Almajano Pablos, M.P. Study of the properties of bearberry leaf extract as a natural antioxidant in model foods. Antioxidants 2016, 5, 11. [Google Scholar] [CrossRef]
- Chaika, N.; Koshovyi, O.; Ain, R.; Kireyev, I.; Zupanets, A.; Odyntsova, V. Phytochemical profile and pharmacological activity of the dry extract from Arctostaphylos uva-ursi leaves modified with phenylalanine. ScienceRise Pharm. Sci. 2020, 6, 74–84. [Google Scholar] [CrossRef]
- Wolska, K.I.; Grudniak, A.M.; Fiecek, B.; Kraczkiewicz-Dowjat, A.; Kurek, A. Antibacterial activity of oleanolic and ursolic acids and their derivatives. Cent. Eur. J. Biol. 2010, 5, 543–553. [Google Scholar] [CrossRef]
- Jesus, J.A.; Lago, J.H.; Laurenti, M.D.; Yamamoto, E.S.; Passero, L.F. Antimicrobial activity of oleanolic and ursolic acids: An update. Evid. Based Complement. Alternat. Med. 2015, 2015, 620472. [Google Scholar] [CrossRef] [PubMed]
- Woźniak, Ł.; Skąpska, S.; Marszałek, K. Ursolic acid—A pentacyclic triterpenoid with a wide spectrum of pharmacological activities. Molecules 2015, 20, 20614. [Google Scholar] [CrossRef]
- Kashyap, D.; Tuli, H.S.; Sharma, A.K. Ursolic acid (UA): A metabolite with promising therapeutic potential. Life Sci. 2016, 146, 201–213. [Google Scholar] [CrossRef] [PubMed]
- Navina, R.; Lee, Y.G.; Kim, S.M. Molecular biological roles of ursolic acid in the treatment of human diseases. Curr. Bioact. Compd. 2017, 13, 177–185. [Google Scholar] [CrossRef]
- López-Hortas, L.; Pérez-Larrán, P.; González-Muñoz, M.J.; Falqué, E.; Domínguez, H. Recent developments on the extraction and application of ursolic acid. A review. Food Res. Int. 2018, 103, 130–149. [Google Scholar] [CrossRef] [PubMed]
- Pironi, A.M.; de Araújo, P.R.; Fernandes, M.A.; Salgado, H.R.N.; Chorilli, M. Characteristics, biological properties and analytical methods of ursolic acid: A review. Crit. Rev. Anal. Chem. 2018, 48, 86–93. [Google Scholar] [CrossRef] [PubMed]
- Mlala, S.; Oyedeji, A.O.; Gondwe, M.; Oyedeji, O.O. Ursolic acid and its derivatives as bioactive agents. Molecules 2019, 24, 2751. [Google Scholar] [CrossRef]
- Aazam, E.S.; Zaheer, Z. Growth of Ag-nanoparticles in an aqueous solution and their antimicrobial activities against Gram positive, Gram negative bacterial strains and Candida fungus. Bioprocess Biosyst. Eng. 2016, 39, 575–584. [Google Scholar] [CrossRef] [PubMed]
- Alvarado, H.; Abrego, G.; Souto, E.B.; Garduño-Ramirez, M.; Clares, B.; García, M.; Calpena, A.C. Nanoemulsions for dermal controlled release of oleanolic and ursolic acids: In vitro, ex vivo and in vivo characterization. Colloids Surf. B Biointerfaces 2015, 130, 40–47. [Google Scholar] [CrossRef]
- Ghiulai, R.; Roşca, O.J.; Antal, D.S.; Mioc, M.; Mioc, A.; Racoviceanu, R.; Macaşoi, I.; Olariu, T.; Dehelean, C.; Creţu, O.M.; et al. Tetracyclic and pentacyclic triterpenes with high therapeutic efficiency in wound healing approaches. Molecules 2020, 25, 5557. [Google Scholar] [CrossRef] [PubMed]
- Mallavadhani, U.V.; Mahapatra, A.; Jamil, K.; Reddy, P.S. Antimicrobial activity of some pentacyclic triterpenes and their synthesized 3-O-lipophilic chains. Biol. Pharm. Bull. 2004, 27, 1576–1579. [Google Scholar] [CrossRef] [PubMed]
- Cunha, L.C.S.; Andrade e Silva, M.L.; Cardoso Furtado, N.A.; Vinhólis, A.H.; Martins, C.H.; da Silva Filho, A.A.; Cunha, W.R. Antibacterial activity of triterpene acids and semi-synthetic derivatives against oral pathogens. Z. Naturforsch. C. J. Biosci. 2007, 62, 668–672. [Google Scholar] [CrossRef] [PubMed]
- Horiuchi, K.; Shiota, S.; Hatano, T.; Yoshida, T.; Kuroda, T.; Tsuchiya, T. Antimicrobial activity of oleanolic acid from Salvia officinalis and related compounds on vancomycin-resistant enterococci (VRE). Biol. Pharm. Bull. 2007, 30, 1147–1149. [Google Scholar] [CrossRef] [PubMed]
- Ahmad, M.; Ahmad, W.; Khan, S.; Zeeshan, M.; Obaidullah, A.J.; Nisar, M.; Shaheen, F.; Ahmad, M. New antibacterial pentacyclic triterpenes from Myricaria elegans Royle (tamariscineae). J. Enzyme Inhib. Med. Chem. 2008, 23, 1023–1027. [Google Scholar] [CrossRef] [PubMed]
- Fontanay, S.; Grare, M.; Mayer, J.; Finance, C.; Duval, R.E. Ursolic, oleanolic and betulinic acids: Antibacterial spectra and selectivity indexes. J. Ethnopharmacol. 2008, 120, 272–276. [Google Scholar] [CrossRef]
- da Silva Filho, A.A.; de Sousa, J.P.; Soares, S.; Furtado, N.A.; Andrade e Silva, M.L.; Cunha, W.R.; Gregório, L.E.; Nanayakkara, N.P.; Bastos, J.K. Antimicrobial activity of the extract and isolated compounds from Baccharis dracunculifolia D. C. (Asteraceae). Z. Naturforsch. C. J. Biosci. 2008, 63, 40–46. [Google Scholar] [CrossRef]
- Cunha, W.R.; de Matos, G.X.; Souza, M.G.; Tozatti, M.G.; Andrade e Silva, M.L.; Martins, C.H.; da Silva, R.; Da Silva Filho, A.A. Evaluation of the antibacterial activity of the methylene chloride extract of Miconia ligustroides, isolated triterpene acids, and ursolic acid derivatives. Pharm. Biol. 2010, 48, 166–169. [Google Scholar] [CrossRef]
- Kurek, A.; Grudniak, A.M.; Szwed, M.; Klicka, A.; Samluk, L.; Wolska, K.I.; Janiszowska, W.; Popowska, M. Oleanolic acid and ursolic acid affect peptidoglycan metabolism in Listeria monocytogenes. Antonie Van Leeuwenhoek 2010, 97, 61–68. [Google Scholar] [CrossRef]
- Sultana, T.; Rashid, M.A.; Ali, M.A.; Mahmood, S.F. Hepatoprotective and antibacterial activity of ursolic acid extracted from Hedyotis corymbosa L. Bangladesh J. Sci. Ind. Res. 2010, 45, 27–34. [Google Scholar] [CrossRef]
- Acebey-Castellon, I.L.; Voutquenne-Nazabadioko, L.; Doan Thi Mai, H.; Roseau, N.; Bouthagane, N.; Muhammad, D.; Le Magrex Debar, E.; Gangloff, S.C.; Litaudon, M.; Sevenet, T.; et al. Triterpenoid saponins from Symplocos lancifolia. J. Nat. Prod. 2011, 74, 163–168. [Google Scholar] [CrossRef]
- Filocamo, A.; Bisignano, C.; D’Arrigo, M.; Ginestra, G.; Mandalari, G.; Galati, E.M. Norfloxacin and ursolic acid: In vitro association and postantibiotic effect against Staphylococcus aureus. Lett. Appl. Microbiol. 2011, 53, 193–197. [Google Scholar] [CrossRef]
- Kim, M.J.; Kim, C.S.; Park, J.Y.; Lim, Y.K.; Park, S.N.; Ahn, S.J.; Jin, D.C.; Kim, T.H.; Kook, J.K. Antimicrobial effects of ursolic acid against mutans Streptococci isolated from Koreans. Int. J. Oral Biol. 2011, 36, 7–11. [Google Scholar]
- Moodley, R.; Chenia, H.; Jonnalagadda, S.B.; Koorbanally, N. Antibacterial and anti-adhesion activity of the pentacyclic triterpenoids isolated from the leaves and edible fruits of Carissa macrocarpa. J. Med. Plant. Res. 2011, 5, 4851–4858. [Google Scholar]
- Kim, S.G.; Kim, M.J.; Jin, D.; Park, S.N.; Cho, E.; Freire, M.O.; Jang, S.J.; Park, Y.J.; Kook, J.K. Antimicrobial effect of ursolic acid and oleanolic acid against methicillin-resistant Staphylococcus aureus. Korean J. Microbiol. 2012, 48, 212–215. [Google Scholar] [CrossRef]
- Kurek, A.; Nadkowska, P.; Pliszka, S.; Wolska, K.I. Modulation of antibiotic resistance in bacterial pathogens by oleanolic acid and ursolic acid. Phytomedicine 2012, 19, 515–519. [Google Scholar] [CrossRef]
- Wong, K.C.; Hag Ali, D.M.; Boey, P.L. Chemical constituents and antibacterial activity of Melastoma malabathricum L. Nat. Prod. Res. 2012, 26, 609–618. [Google Scholar] [CrossRef]
- Wojnicz, D.; Kicia, M.; Tichaczek-Goska, D. Effect of asiatic and ursolic acids on morphology, hydrophobicity and adhesion of UPECs to uroepithelial cells. Folia Microbiol. 2013, 58, 245–252. [Google Scholar]
- Wojnicz, D.; Tichaczek-Goska, D.; Kicia, M. Effect of asiatic and ursolic acids on growth and virulence factors of uropathogenic Escherichia coli strains. Turk. J. Biol. 2013, 37, 556–564. [Google Scholar] [CrossRef]
- Do Nascimento, P.G.G.; Lemos, T.L.G.; Bizerra, A.M.C.; Arriaga, Â.M.; Ferreira, D.A.; Santiago, G.M.; Braz-Filho, R.; Costa, J.G. Antibacterial and antioxidant activities of ursolic acid and derivatives. Molecules 2014, 19, 1317–1327. [Google Scholar] [CrossRef] [PubMed]
- Dwivedi, G.R.; Maurya, A.; Yadav, D.K.; Khan, F.; Darokar, M.P.; Srivastava, S.K. Drug resistance reversal potential of ursolic acid derivatives against nalidixic acid- and multidrug-resistant Escherichia coli. Chem. Biol. Drug. Des. 2015, 86, 272–283. [Google Scholar] [CrossRef]
- Park, S.N.; Ahn, S.J.; Kook, J.K. Oleanolic acid and ursolic acid inhibit peptidoglycan biosynthesis in Streptococcus mutans UA159. Braz. J. Microbiol. 2015, 46, 613–617. [Google Scholar] [CrossRef]
- Wang, C.M.; Chen, H.T.; Wu, Z.Y.; Jhan, Y.L.; Shyu, C.L.; Chou, C.H. Antibacterial and synergistic activity of pentacyclic triterpenoids isolated from Alstonia scholaris. Molecules 2016, 21, 139. [Google Scholar] [CrossRef]
- Wang, C.M.; Jhan, Y.L.; Tsai, S.J.; Chou, C.H. The pleiotropic antibacterial mechanisms of ursolic acid against methicillin-resistant Staphylococcus aureus (MRSA). Molecules 2016, 21, 884. [Google Scholar] [CrossRef]
- Catteau, L.; Reichmann, N.T.; Olson, J.; Pinho, M.G.; Nizet, V.; Van Bambeke, F.; Quetin-Leclercq, J. Synergy between ursolic and oleanolic acids from Vitellaria paradoxa leaf extract and β-lactams against methicillin-resistant Staphylococcus aureus: In vitro and in vivo activity and underlying mechanisms. Molecules 2017, 22, 2245. [Google Scholar] [CrossRef]
- Oloyede, H.O.B.; Ajiboye, H.O.; Salawu, M.O.; Ajiboye, T.O. Influence of oxidative stress on the antibacterial activity of betulin, betulinic acid and ursolic acid. Microb. Pathog. 2017, 111, 338–344. [Google Scholar] [CrossRef]
- Wojnicz, D.; Tichaczek-Goska, D.; Korzekwa, K.; Kicia, M.; Hendrich, A. Anti-enterococcal activities of pentacyclic triterpenes. Adv. Clin. Exp. Med. 2017, 26, 483–490. [Google Scholar] [CrossRef]
- Sundaramoorthy, N.S.; Mohan, H.M.; Subramaniam, S.; Raman, T.; Selva Ganesan, S.; Sivasubamanian, A.; Nagarajan, S. Ursolic acid inhibits colistin efflux and curtails colistin resistant Enterobacteriaceae. AMB Express 2019, 9, 27. [Google Scholar] [CrossRef]
- Ren, D.; Zuo, R.; Gonzalez Barrios, A.F.; Bedzyk, L.A.; Eldridge, G.R.; Pasmore, M.E.; Wood, T.K. Differential gene expression for investigation of Escherichia coli biofilm inhibition by plant extract ursolic acid. Appl. Environ. Microbiol. 2005, 71, 4022–4034. [Google Scholar] [CrossRef]
- Kim, S.; Song, M.; Roh, B.D.; Park, S.H.; Park, J.W. Inhibition of Streptococcus mutans biofilm formation on composite resins containing ursolic acid. Restor. Dent. Endod. 2013, 38, 65–72. [Google Scholar] [CrossRef]
- Zhou, L.; Ding, Y.; Chen, W.; Zhang, P.; Chen, Y.; Lv, X. The in vitro study of ursolic acid and oleanolic acid inhibiting cariogenic microorganisms as well as biofilm. Oral. Dis. 2013, 19, 494–500. [Google Scholar] [CrossRef]
- Kurek, A.; Markowska, K.; Grudniak, A.M.; Janiszowska, W.; Wolska, K.I. The effect of oleanolic and ursolic acids on the hemolytic properties and biofilm formation of Listeria monocytogenes. Pol. J. Microbiol. 2014, 63, 21–25. [Google Scholar] [CrossRef]
- Micota, B.; Sadowska, B.; Podsędek, A.; Redzynia, M.; Różalska, B. Leonurus cardiaca L. herb—A derived extract and an ursolic acid as the factors affecting the adhesion capacity of Staphylococcus aureus in the context of infective endocarditis. Acta Biochim. Pol. 2014, 61, 385–388. [Google Scholar] [CrossRef]
- Qin, N.; Tan, X.; Jiao, Y.; Liu, L.; Zhao, W.; Yang, S.; Jia, A. RNA-Seq-based transcriptome analysis of methicillin-resistant Staphylococcus aureus biofilm inhibition by ursolic acid and resveratrol. Sci. Rep. 2014, 4, 5467. [Google Scholar] [CrossRef]
- Zou, Y.; Lee, Y.; Huh, J.; Park, J.W. Synergistic effect of xylitol and ursolic acid combination on oral biofilms. Restor. Dent. Endod. 2014, 39, 288–295. [Google Scholar] [CrossRef]
- Gilabert, M.; Marcinkevicius, K.; Andujar, S.; Schiavone, M.; Arena, M.E.; Bardón, A. Sesqui- and triterpenoids from the liverwort Lepidozia chordulifera inhibitors of bacterial biofilm and elastase activity of human pathogenic bacteria. Phytomedicine 2015, 22, 77–85. [Google Scholar] [CrossRef]
- Lou, Z.; Tang, Y.; Song, X.; Wang, H. Metabolomics-based screening of biofilm-inhibitory compounds against Pseudomonas aeruginosa from burdock leaf. Molecules 2015, 20, 16266–16277. [Google Scholar] [CrossRef]
- Tan, X.; Qin, N.; Wu, C.; Sheng, J.; Yang, R.; Zheng, B.; Ma, Z.; Liu, L.; Peng, X.; Jia, A. Transcriptome analysis of the biofilm formed by methicillin-susceptible Staphylococcus aureus. Sci. Rep. 2015, 5, 11997. [Google Scholar] [CrossRef]
- Wojnicz, D.; Tichaczek-Goska, D.; Kicia, M. Pentacyclic triterpenes combined with ciprofloxacin help to eradicate the biofilm formed in vitro by Escherichia coli. Indian J. Med. Res. 2015, 141, 343–353. [Google Scholar] [CrossRef]
- Chung, S.H.; Cho, S.; Kim, K.; Lim, B.S.; Ahn, S.J. Antimicrobial and physical characteristics of orthodontic primers containing antimicrobial agents. Angle Orthod. 2017, 87, 307–312. [Google Scholar] [CrossRef]
- Ray, C.; Shenoy, A.T.; Orihuela, C.J.; González-Juarbe, N. Killing of Serratia marcescens biofilms with chloramphenicol. Ann. Clin. Microbiol. Antimicrob. 2017, 16, 19. [Google Scholar] [CrossRef]
- Jyothi, J.S.; Putty, K.; Reddy, Y.N.; Dhanalakshmi, K.; Umair, M.A.H. Antagonistic effect of ursolic acid on Staphylococcal biofilms. Vet. World 2018, 11, 1440–1444. [Google Scholar] [CrossRef]
- Silva, G.N.S.D.; Primon-Barros, M.; Macedo, A.J.; Gnoatto, S.C.B. Triterpene derivatives as relevant scaffold for new antibiofilm drugs. Biomolecules 2019, 9, 58. [Google Scholar] [CrossRef]
- Ahmed, N.; Khalid, H.; Mushtaq, M.; Basha, S.; Rabaan, A.A.; Garout, M.; Halwani, M.A.; Al Mutair, A.; Alhumaid, S.; Al Alawi, Z.; et al. The molecular characterization of virulence determinants and antibiotic resistance patterns in human bacterial uropathogens. Antibiotics 2022, 11, 516. [Google Scholar] [CrossRef]
- Lyu, X.; Wang, L.; Shui, Y.; Jiang, Q.; Chen, L.; Yang, W.; He, X.; Zeng, J.; Li, Y. Ursolic acid inhibits multi-species biofilms developed by Streptococcus mutans, Streptococcus sanguinis, and Streptococcus gordonii. Arch. Oral Biol. 2021, 125, 105107. [Google Scholar] [CrossRef]
- Di Bonaventura, G.; Spedicato, I.; D’Antonio, D.; Robuffo, I.; Piccolomini, R. Biofilm formation by Stenotrophomonas maltophilia modulation by quinolones, trimethoprim, sulfamethoxazole, and ceftazidime. Antimicrob. Agents Chemother. 2004, 48, 151–160. [Google Scholar] [CrossRef]
- O’Toole, G.A.; Kolter, R. Flagellar and twitching motility are necessary for Pseudomonas aeruginosa biofilm development. Mol. Microbiol. 1998, 30, 295–304. [Google Scholar] [CrossRef] [PubMed]
- Stepanović, S.; Vuković, D.; Hola, V.; Di Bonaventura, G.; Djukić, S.; Cirković, I.; Ruzicka, F. Quantification of biofilm in microtiter plates: Overview of testing conditions and practical recommendations for assessment of biofilm production by staphylococci. APMIS 2007, 115, 891–899. [Google Scholar] [CrossRef] [PubMed]
- Singh, A.K.; Prakash, P.; Achra, A.; Singh, G.P.; Das, A.; Singh, R.K. Standardization and classification of in vitro biofilm formation by clinical isolates of Staphylococcus aureus. J. Global. Infect. Dis. 2017, 9, 93–101. [Google Scholar]
- Kim, S.; Kim, M.J.; Kang, H.Y.; Seol, S.Y.; Cho, D.T.; Kim, J. A simple colorimetric method for testing antimicrobial susceptibility of biofilmed bacteria. J. Microbiol. 2010, 48, 709–711. [Google Scholar] [CrossRef]
- Haney, E.F.; Trimble, M.J.; Cheng, J.T.; Vallé, Q.; Hancock, R.E.W. Critical assessment of methods to quantify biofilm growth and evaluate antibiofilm activity of host defence peptides. Biomolecules 2018, 8, 29. [Google Scholar] [CrossRef]
- Machado, I.; Lopes, S.P.; Sousa, A.M.; Pereira, M.O. Adaptive response of single and binary Pseudomonas aeruginosa and Escherichia coli biofilms to benzalkonium chloride. J. Basic Microbiol. 2012, 52, 43–52. [Google Scholar] [CrossRef]
- Vanysacker, L.; Denis, C.; Declerck, P.; Piasecka, A.; Vankelecom, I.F. Microbial adhesion and biofilm formation on microfiltration membranes: A detailed characterization using model organisms with increasing complexity. Biomed. Res. Int. 2013, 2013, 470867. [Google Scholar] [CrossRef]
- Cerqueira, L.; Oliveira, J.A.; Nicolau, A.; Azevedo, N.F.; Vieira, M.J. Biofilm formation with mixed cultures of Pseudomonas aeruginosa/Escherichia coli on silicone using artificial urine to mimic urinary catheters. Biofouling 2013, 29, 829–840. [Google Scholar] [CrossRef]
- Kuznetsova, M.V.; Maslennikova, I.L.; Karpunina, T.I.; Nesterova, L.Y.; Demakov, V.A. Interactions of Pseudomonas aeruginosa in predominant biofilm or planktonic forms of existence in mixed culture with Escherichia coli in vitro. Can. J. Microbiol. 2013, 59, 604–610. [Google Scholar] [CrossRef]
- Oliveira, A.; Sousa, J.C.; Silva, A.C.; Melo, L.D.R.; Sillankorva, S. Chestnut honey and bacteriophage application to control Pseudomonas aeruginosa and Escherichia coli biofilms: Evaluation in an ex vivo wound model. Front. Microbiol. 2018, 9, 1725. [Google Scholar] [CrossRef]
- Solis-Velazquez, O.A.; Gutiérrez-Lomelí, M.; Guerreo-Medina, P.J.; Rosas-García, M.L.; Iñiguez-Moreno, M.; Avila-Novoa, M.G. Nosocomial pathogen biofilms on biomaterials: Different growth medium conditions and components of biofilms produced in vitro. J. Microbiol. Immunol. Infect. 2020, 54, 1038–1047. [Google Scholar] [CrossRef] [PubMed]
- Tashiro, Y.; Yawata, Y.; Toyofuku, M.; Uchiyama, H.; Nomura, N. Interspecies interaction between Pseudomonas aeruginosa and other microorganisms. Microbes Environ. 2013, 1, 13–24. [Google Scholar] [CrossRef]
- Pena, R.T.; Blasco, L.; Ambroa, A.; González-Pedrajo, B.; Fernández-García, L.; López, M.; Bleriot, I.; Bou, G.; García-Contreras, R.; Wood, T.K.; et al. Relationship between quorum sensing and secretion systems. Front. Microbiol. 2019, 10, 1100. [Google Scholar] [CrossRef]
- Mirani, Z.A.; Fatima, A.; Urooj, S.; Aziz, M.; Khan, M.N.; Abbas, T. Relationship of cell surface hydrophobicity with biofilm formation and growth rate: A study on Pseudomonas aeruginosa, Staphylococcus aureus and Escherichia coli. Iran J. Basic Med. Sci. 2018, 21, 760–769. [Google Scholar] [PubMed]
- Rahmani-Badi, A.; Sepehr, S.; Mohammadi, P.; Soudi, M.R.; Babaie-Naiej, H.; Fallahi, H. A combination of cis-2-decanoic acid and antibiotics eradicates pre-established catheter-associated biofilms. J. Med. Microbiol. 2014, 63, 1509–1516. [Google Scholar] [CrossRef]
- Davies, D.G.; Marques, C.N.H. A fatty acid messenger is responsible for inducing dispersion in microbial biofilms. J. Bacteriol. 2009, 191, 1393–1403. [Google Scholar] [CrossRef] [PubMed]
- Tiwari, R.; Karthik, K.; Rana, R.; Malik, Y.S.; Dhama, K.; Joshi, S.K. Quorum sensing inhibitors/antagonists countering food spoilage bacteria-need molecular and pharmaceutical intervention for protecting current issues of food safety. Int. J. Pharmacol. 2016, 12, 262–271. [Google Scholar] [CrossRef]
- Cao, T.; Morales-Soto, N.; Jia, J.; Baig, N.F.; Dunham, S.J.B.; Ellis, J.; Sweedler, J.V.; Shrout, J.D.; Bohn, P.W. Spatiotemporal dynamics of molecular messaging in bacterial co-cultures studied by multimodal chemical imaging. Proc. SPIE Int. Soc. Opt. Eng. 2019, 10863, 108630A. [Google Scholar] [PubMed]
- Lopes, S.P.; Machado, I.; Pereira, M.O. Role of planktonic and sessile extracellular metabolic by-products on Pseudomonas aeruginosa and Escherichia coli intra and interspecies relationships. J. Ind. Microbiol. Biotechnol. 2011, 38, 133–140. [Google Scholar] [CrossRef] [PubMed]
- Bhattacharjee, A.; Khan, M.; Kleiman, M.; Hochbaum, A.I. Effects of growth surface topography on bacterial signaling in coculture biofilms. ACS Appl. Mater. Interfaces 2017, 9, 18531–18539. [Google Scholar] [CrossRef]
- Chu, W.; Zere, T.R.; Weber, M.M.; Wood, T.K.; Whiteley, M.; Hidalgo-Romano, B.; Valenzuela, E., Jr.; McLean, R.J. Indole production promotes Escherichia coli mixed-culture growth with Pseudomonas aeruginosa by inhibiting quorum signaling. Appl. Environ. Microbiol. 2012, 78, 411–419. [Google Scholar] [CrossRef]
- Chen, L.; Zou, Y.; She, P.; Wu, Y. Composition, function, and regulation of T6SS in Pseudomonas aeruginosa. Microbiol. Res. 2015, 172, 19–25. [Google Scholar] [CrossRef] [PubMed]
- Navarro-Garcia, F.; Ruiz-Perez, F.; Cataldi, Á.; Larzábal, M. Type VI secretion system in pathogenic Escherichia coli: Structure, role in virulence, and acquisition. Front. Microbiol. 2019, 10, 1965. [Google Scholar] [CrossRef] [PubMed]
- Basler, M.; Ho, B.T.; Mekalanos, J.J. Tit-for-tat. Type VI secretion system counter attack during bacterial cell–cell interactions. Cell 2013, 4, 884–894. [Google Scholar] [CrossRef] [PubMed]
- Soria-Bustos, J.; Ares, M.A.; Gómez-Aldapa, C.A.; González-y-Merchand, J.A.; Girón, J.A.; De la Cruz, M.A. Two Type VI secretion systems of Enterobacter cloacae are required for bacterial competition, cell adherence, and intestinal colonization. Front. Microbiol. 2020, 11, 560488. [Google Scholar] [CrossRef] [PubMed]
- Ma, J.; Pan, Z.; Huang, J.; Sun, M.; Lu, C.; Yao, H. The Hcp proteins fused with diverse extended-toxin domains represent a novel pattern of antibacterial effectors in type VI secretion systems. Virulence 2017, 8, 1189–1202. [Google Scholar] [CrossRef] [PubMed]
- Zong, B.; Zhang, Y.; Wang, X.; Liu, M.; Zhang, T.; Zhu, Y.; Zheng, Y.; Hu, L.; Li, P.; Chen, H.; et al. Characterization of multiple type-VI secretion system (T6SS) VgrG proteins in the pathogenicity and antibacterial activity of porcine extra-intestinal pathogenic Escherichia coli. Virulence 2019, 10, 118–132. [Google Scholar] [CrossRef]
- Wang, X.; Lünsdorf, H.; Ehrén, I.; Brauner, A.; Römling, U. Characteristics of biofilms from urinary tract catheters and presence of biofilm-related components in Escherichia coli. Curr. Microbiol. 2010, 60, 446–453. [Google Scholar] [CrossRef] [PubMed]
- Qian, W.; Fu, Y.; Liu, M.; Zhang, J.; Wang, W.; Li, J.; Zeng, Q.; Wang, T.; Li, Y. Mechanisms of action of luteolin against single- and dual-species of Escherichia coli and Enterobacter cloacae and its antibiofilm activities. Appl. Biochem. Biotechnol. 2021, 193, 1397–1414. [Google Scholar] [CrossRef] [PubMed]
- Culotti, A.; Packman, A.I. Pseudomonas aeruginosa promotes Escherichia coli biofilm formation in nutrient-limited medium. PLoS ONE 2014, 9, e107186. [Google Scholar] [CrossRef] [PubMed]
- Sheraton, M.V.; Yam, J.K.H.; Tan, C.H.; Oh, H.S.; Mancini, E.; Yang, L.; Rice, S.A.; Sloot, P.M.A. Mesoscopic energy minimization drives Pseudomonas aeruginosa biofilm morphologies and consequent stratification of antibiotic activity based on cell metabolism. Antimicrob. Agents Chemother. 2018, 62, e02544-17. [Google Scholar] [CrossRef] [PubMed]
- Tielen, P.; Rosenau, F.; Wilhelm, S.; Jaeger, K.E.; Flemming, H.C.; Wingender, J. Extracellular enzymes affect biofilm formation of mucoid Pseudomonas aeruginosa. Microbiology 2010, 156, 2239–2252. [Google Scholar] [CrossRef] [PubMed]
- Yu, H.; He, X.; Xie, W.; Xiong, J.; Sheng, H.; Guo, S.; Huang, C.; Zhang, D.; Zhang, K. Elastase LasB of Pseudomonas aeruginosa promotes biofilm formation partly through rhamnolipid mediated regulation. Can. J. Microbiol. 2014, 60, 227–235. [Google Scholar] [CrossRef] [PubMed]
- Schwering, M.; Song, J.; Louie, M.; Turner, R.J.; Ceri, H. Multi-species biofilms defined from drinking water microorganisms provide increased protection against chlorine disinfection. Biofouling 2013, 29, 917–928. [Google Scholar] [CrossRef]
- Nostro, A.; Scaffaro, R.; D’Arrigo, M.; Botta, L.; Filocamo, A.; Marino, A.; Bisignano, G. Development and characterization of essential oil component-based polymer films: A potential approach to reduce bacterial biofilm. Appl. Microbiol. Biotechnol. 2013, 97, 9515–9523. [Google Scholar] [CrossRef] [PubMed]
- Kong, K.F.; Schneper, L.; Mathee, K. Beta-lactam antibiotics: From antibiosis to resistance and bacteriology. APMIS 2010, 118, 1–36. [Google Scholar] [CrossRef]
- El-Hajj, Z.W.; Newman, E.B. How much territory can a single E. coli cell control? Front. Microbiol. 2015, 6, 309. [Google Scholar] [CrossRef] [PubMed]
- Mizushina, Y.; Iida, A.; Ohta, K.; Sugawara, F.; Sakaguchi, K. Novel triterpenoids inhibit both DNA polymerase and DNA topoisomerase. Biochem. J. 2000, 350, 757–763. [Google Scholar] [CrossRef]
- Justice, S.S.; Hunstad, D.A.; Seed, P.C.; Hultgren, S.J. Filamentation by Escherichia coli subverts innate defenses during urinary tract infection. Proc. Natl. Acad. Sci. USA 2006, 103, 19884–19889. [Google Scholar] [CrossRef] [PubMed]
- Cheng, G.; Hao, H.; Dai, M.; Liu, Z.; Yuan, Z. Antibacterial action of quinolones: From target to network. Eur. J. Med. Chem. 2013, 66, 555–562. [Google Scholar] [CrossRef] [PubMed]
- Cushnie, T.P.; O’Driscoll, N.H.; Lamb, A.J. Morphological and ultrastructural changes in bacterial cells as an indicator of antibacterial mechanism of action. Cell. Mol. Life Sci. 2016, 73, 4471–4492. [Google Scholar] [CrossRef] [PubMed]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sycz, Z.; Wojnicz, D.; Tichaczek-Goska, D. Does Secondary Plant Metabolite Ursolic Acid Exhibit Antibacterial Activity against Uropathogenic Escherichia coli Living in Single- and Multispecies Biofilms? Pharmaceutics 2022, 14, 1691. https://doi.org/10.3390/pharmaceutics14081691
Sycz Z, Wojnicz D, Tichaczek-Goska D. Does Secondary Plant Metabolite Ursolic Acid Exhibit Antibacterial Activity against Uropathogenic Escherichia coli Living in Single- and Multispecies Biofilms? Pharmaceutics. 2022; 14(8):1691. https://doi.org/10.3390/pharmaceutics14081691
Chicago/Turabian StyleSycz, Zuzanna, Dorota Wojnicz, and Dorota Tichaczek-Goska. 2022. "Does Secondary Plant Metabolite Ursolic Acid Exhibit Antibacterial Activity against Uropathogenic Escherichia coli Living in Single- and Multispecies Biofilms?" Pharmaceutics 14, no. 8: 1691. https://doi.org/10.3390/pharmaceutics14081691





