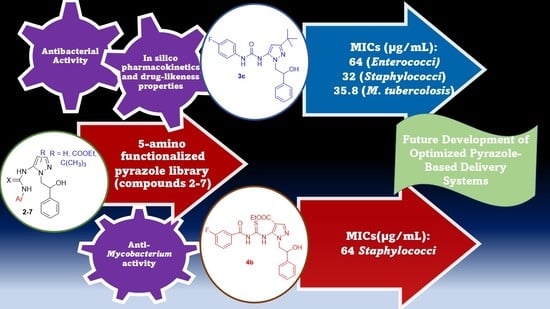
Microbiological Screening of 5-Functionalized Pyrazoles for the Future Development of Optimized Pyrazole-Based Delivery Systems
Abstract
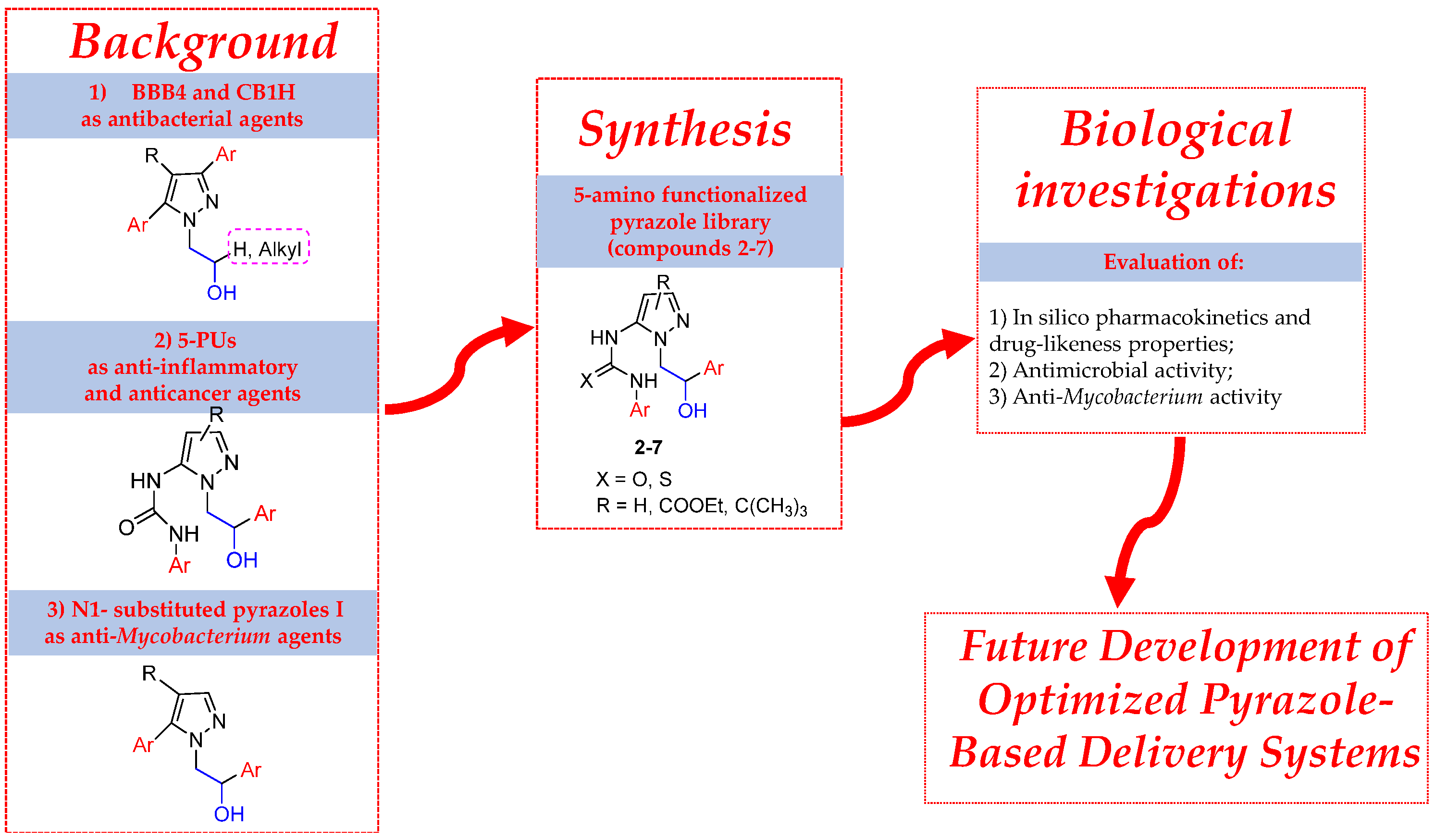
:1. Introduction
2. Results and Discussion
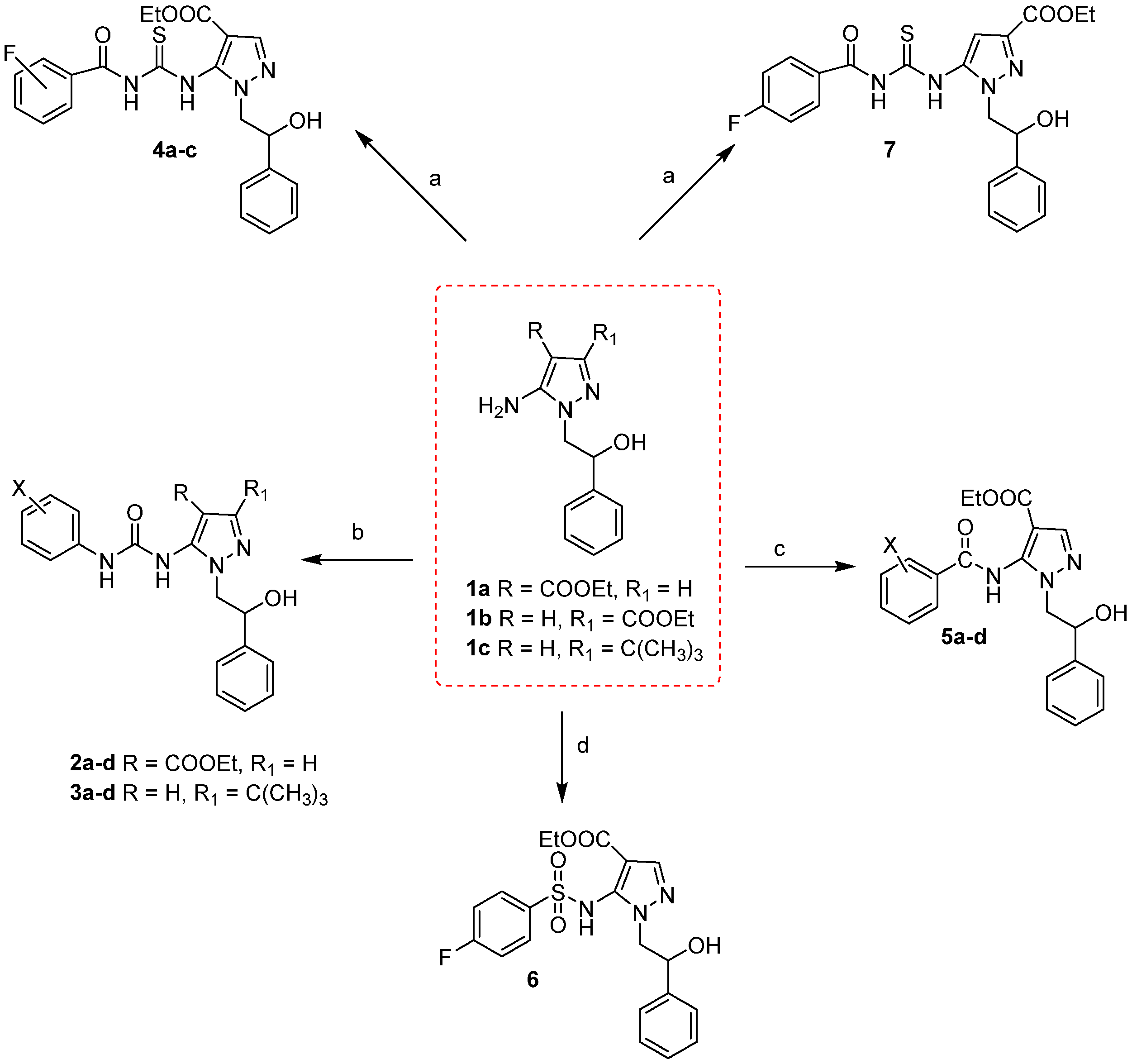
2.1. Chemistry
2.2. Pharmacokinetic Properties and Druglikeness Prediction
2.3. Antimicrobial Activity
2.4. Anti-Mycobacterium Activity
3. Materials and Methods
3.1. General Information
3.2. Synthesis of Compounds 2a-d and 3a-d
3.2.1. Synthesis of Compounds 4a–c
Synthesis of 2-Fluorobenzoyl isothiocyanate, 3-Fluorobenzoyl isothiocyanate and 4-Fluorobenzoyl isothiocyanate
3.2.2. Synthesis of Compounds 5a-d
3.2.3. Synthesis of Ethyl 5-((4-fluorophenyl)-sulfonamido)-1-(2-hydroxy-2-phenylethyl)-1H-pyrazole-4-carboxylate 6
3.2.4. Synthesis of Ethyl 5-(3-(4-fluorobenzoyl)-thioureido)-1-(2-hydroxy-2-phenylethyl)-1H-pyrazole-3-carboxylate 7
3.3. Biological Evaluation
3.3.1. Microbiology
Bacterial Species Considered in This Study
Determination of the Minimal Inhibitory Concentrations (MICs)
3.3.2. MABA and LORA Tests
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Patel, B.; Zunk, M.; Grant, G.; Rudrawar, S. Design, synthesis and bioactivity evaluation of novel pyrazole linked phenylthiazole derivatives in context of antibacterial activity. Bioorg. Med. Chem. Lett. 2021, 39, 127853. [Google Scholar] [CrossRef] [PubMed]
- Taher, A.T.; Sarg, M.T.M.; Ali, N.R.E.; Elnagdi, N.H. Design, synthesis, modeling studies and biological screening of novel pyrazole derivatives as potential analgesic and anti-inflammatory agents. Bioorg. Chem. 2019, 89, 103023. [Google Scholar] [CrossRef] [PubMed]
- Yu, B.; Zhou, S.; Cao, L.; Hao, Z.; Yang, D.; Guo, X.; Zhang, N.; Bakulev, V.A.; Fan, Z. Design, Synthesis, and Evaluation of the Antifungal Activity of Novel Pyrazole–Thiazole Carboxamides as Succinate Dehydrogenase Inhibitors. J. Agric. Food Chem. 2020, 68, 7093–7102. [Google Scholar] [CrossRef] [PubMed]
- Abdellatif, K.R.A.; El-Saadi, M.T.; Elzayat, S.G.; Amin, N.H. New substituted pyrazole derivatives targeting COXs as potential safe anti-inflammatory agents. Future Med. Chem. 2019, 11, 1871–1887. [Google Scholar] [CrossRef]
- Ebenezer, O.; Shapi, M.; Tuszynski, J.A. A Review of the Recent Development in the Synthesis and Biological Evaluations of Pyrazole Derivatives. Biomedicines 2022, 10, 1124. [Google Scholar] [CrossRef]
- Alfei, S.; Brullo, C.; Caviglia, D.; Zuccari, G. Preparation and Physicochemical Characterization of Water-Soluble Pyrazole-Based Nanoparticles by Dendrimer Encapsulation of an Insoluble Bioactive Pyrazole Derivative. Nanomaterials 2021, 11, 2662. [Google Scholar] [CrossRef]
- Brullo, C.; Rapetti, F.; Bruno, O. Pyrazolyl-Ureas as Interesting Scaffold in Medicinal Chemistry. Molecules 2020, 25, 3457. [Google Scholar] [CrossRef]
- Morretta, E.; Sidibè, A.; Spallarossa, A.; Petrella, A.; Meta, E.; Bruno, O.; Monti, M.C.; Brullo, C. Synthesis, functional proteomics and biological evaluation of new 5-pyrazolyl ureas as potential anti-angiogenic compounds. Eur. J. Med. Chem. 2021, 226, 113872. [Google Scholar] [CrossRef]
- Belvedere, R.; Morretta, E.; Novizio, N.; Morello, S.; Bruno, O.; Brullo, C.; Petrella, A. The pyrazolyl-urea GeGe3 inhibits the activity of ANXA1 in the angiogenesis induced by the pancreatic cancer derived EV. Biomolecules 2021, 11, 1758. [Google Scholar] [CrossRef]
- Signorello, M.G.; Rapetti, F.; Meta, E.; Sidibè, A.; Bruno, O.; Brullo, C. New Series of Pyrazoles and Imidazo-pyrazoles Targeting Different Cancer and Inflammation Pathways. Molecules 2021, 26, 5735. [Google Scholar] [CrossRef]
- Bruno, O.; Brullo, C.; Bondavalli, F.; Schenone, S.; Ranise, A.; Arduino, N.; Bertolotto, M.B.; Montecucco, F.; Ottonello, L.; Dallegri, F.; et al. Synthesis and biological evaluation of N-pyrazolyl-N′-alkyl/benzyl/phenylureas: A new class of potent inhibitors of interleukin 8-induced neutrophil chemotaxis. J. Med. Chem. 2007, 50, 3618–3626. [Google Scholar] [CrossRef] [PubMed]
- Meta, E.; Brullo, C.; Sidibè, A.; Imhof, B.A.; Bruno, O. Design, synthesis, and biological evaluation of new pyrazolyl-ureas and imidazopyrazolecarboxamides able to interfere with MAPK and PI3K upstream signalling involved in the angiogenesis. Eur. J. Med. Chem. 2017, 133, 24–35. [Google Scholar] [CrossRef] [Green Version]
- Meta, E.; Brullo, C.; Tonelli, M.; Franzblau, S.G.; Wang, Y.; Ma, R.; Baojie, W.; Orena, B.S.; Pasca, M.R.; Bruno, O. Pyrazole and imidazo[1,2-b]pyrazole derivatives as new potential anti-tuberculosis agents. Med. Chem. 2019, 15, 17–27. [Google Scholar] [CrossRef] [PubMed]
- Alfei, S.; Zuccari, G.; Caviglia, D.; Brullo, C. Synthesis and Characterization of Pyrazole-Enriched Cationic Nanoparticles as New Promising Antibacterial Agent by Mutual Cooperation. Nanomaterials 2022, 12, 1215. [Google Scholar] [CrossRef] [PubMed]
- Alfei, S.; Brullo, C.; Caviglia, D.; Piatti, G.; Zorzoli, A.; Marimpietri, D.; Zuccari, G.; Schito, A.M. Pyrazole-Based Water-Soluble Dendrimer Nanoparticles as a Potential New Agent against Staphylococci. Biomedicines 2022, 10, 17. [Google Scholar] [CrossRef]
- Schito, A.M.; Caviglia, D.; Brullo, C.; Zorzoli, A.; Marimpietri, D.; Alfei, S. Enhanced Antibacterial Activity of a Cationic Macromolecule by its Complexation with a Weakly Active Pyrazole Derivative. Biomedicines 2022, 10, 1607. [Google Scholar] [CrossRef]
- Bondavalli, F.; Botta, M.; Bruno, O.; Ciacci, A.; Corelli, F.; Fossa, P.; Lucacchini, A.; Manetti, F.; Martini, C.; Menozzi, G.; et al. Synthesis, Molecular Modeling Studies, and Pharmacological Activity of Selective A1 Receptor Antagonists. J. Med. Chem. 2002, 45, 4875–4887. [Google Scholar] [CrossRef]
- Lewis, R.T.; Bode, C.M.; Choquette, D.M.; Potashman, M.; Romero, K.; Stellwagen, J.C.; Teffera, Y.; Moore, E.; Whittington, D.A.; Chen, H.; et al. The discovery and optimization of a novel class of potent, selective, and orally bioavailable anaplastic lymphoma kinase (ALK) inhibitors with potential utility for the treatment of cancer. J. Med. Chem. 2012, 55, 6523–6540. [Google Scholar] [CrossRef]
- Daina, A.; Michielin, O.; Zoete, V. SwissADME: A free web tool to evaluate pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules. Sci. Rep. 2017, 7, 42717. [Google Scholar] [CrossRef] [Green Version]
- Brenk, R.; Schipani, A.; James, D.; Krasowski, A.; Gilbert, I.H.; Frearson, J.; Wyatt, P.G. Lessons learnt from assembling screening libraries for drug discovery for neglected diseases. ChemMedChem 2008, 3, 435–444. [Google Scholar] [CrossRef]
- Delaney, J.S. ESOL: Estimating Aqueous Solubility Directly from Molecular Structure. J. Chem. Inf. Model. 2004, 44, 1000–1005. [Google Scholar] [CrossRef] [PubMed]
- Cho, S.; Lee, H.S.; Franzblau, S. Microplate Alamar Blue Assay (MABA) and Low Oxygen Recovery Assay (LORA) for mycobacterium tuberculosis. Methods Mol. Biol. 2015, 1285, 281–292. [Google Scholar] [CrossRef] [PubMed]
- Lilly Launches Open Innovation Drug Discovery Platform to Help Find Potential New Medicines Where Medical Need is Great. Available online: https://investor.lilly.com/news-releases/news-release-details/lilly-launches-open-innovation-drug-discovery-platform-help-find (accessed on 23 May 2022).
- Schito, A.M.; Piatti, G.; Caviglia, D.; Zuccari, G.; Alfei, S. Broad-Spectrum Bactericidal Activity of a Synthetic Random Copolymer Based on 2-Methoxy-6-(4-Vinylbenzyloxy)-Benzylammonium Hydrochloride. Int. J. Mol. Sci. 2021, 22, 5021. [Google Scholar] [CrossRef] [PubMed]
- Schito, A.M.; Piatti, G.; Caviglia, D.; Zuccari, G.; Zorzoli, A.; Marimpietri, D.; Alfei, S. Bactericidal Activity of Non-Cytotoxic Cationic Nanoparticles against Clinically and Environmentally Relevant Pseudomonas spp. Isolates. Pharmaceutics 2021, 13, 1411. [Google Scholar] [CrossRef]
- EUCAST. European Committee on Antimicrobial Susceptibility Testing. Available online: https://www.eucast.org/ast_of_bacteria (accessed on 23 May 2022).
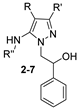
| Compd. | R | R’ | R’’ | M.p (°C) | Yield (%) |
|---|---|---|---|---|---|
| 2a | COOEt | H | 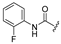 | [11] | [11] |
| 2b | COOEt | H | 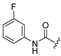 | [11] | [11] |
| 2c | COOEt | H |  | [11] | [11] |
| 2d | COOEt | H | 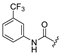 | [11] | [11] |
| 3a | H | C(CH3)3 | 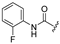 | [12] | [12] |
| 3b | H | C(CH3)3 |  | [12] | [12] |
| 3c | H | C(CH3)3 | 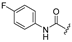 | [12] | [12] |
| 3d | H | C(CH3)3 |  | [12] | [12] |
| 4a | COOEt | H | 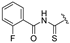 | 150–151 | 42 |
| 4b | COOEt | H | 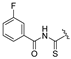 | 155–156 | 48 |
| 4c | COOEt | H | 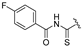 | 158–160 | 51 |
| 5a | COOEt | H | 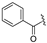 | 117–118 | 61 |
| 5b | COOEt | H | 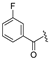 | 149–151 | 35 |
| 5c | COOEt | H |  | 98–100 | 30 |
| 5d | COOEt | H |  | Oil | 23 |
| 6 | COOEt | H |  | 180–181 | 34 |
| 7 | H | COOEt | 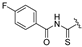 | 169–172 | 37 |
| 2a | 2b | 2c | 2d | 3a | 3b | 3c | 3d | |
|---|---|---|---|---|---|---|---|---|
| Physicochemical Prop. | ||||||||
| MW (g/mol) | 412.41 | 412.41 | 412.41 | 462.42 | 396.46 | 396.46 | 396.46 | 446.47 |
| Fraction Csp3 | 0.19 | 0.19 | 0.19 | 0.23 | 0.27 | 0.27 | 0.27 | 0.3 |
| Rotatable bonds | 10 | 10 | 10 | 11 | 8 | 8 | 8 | 9 |
| H-bond acceptors | 6 | 6 | 6 | 8 | 4 | 4 | 4 | 6 |
| H-bond donors | 3 | 3 | 3 | 3 | 3 | 3 | 3 | 3 |
| TPSA a (Å2) | 105.48 | 105.48 | 105.48 | 105.48 | 79.18 | 79.18 | 79.18 | 79.18 |
| Lipophilicity | ||||||||
| LogP b | 2.78 | 2.78 | 2.78 | 3.56 | 3.72 | 3.72 | 3.72 | 4.5 |
| Water solubility | ||||||||
| Solubility (mg/mL) c | 0.051 | 0.051 | 0.051 | 0.0115 | 0.0112 | 0.0112 | 0.0112 | 0.00256 |
| Solubility class | Soluble | Soluble | Soluble | Moderately soluble | Moderately soluble | Moderately soluble | Moderately soluble | Moderately soluble |
| Pharmacokinetics | ||||||||
| GI absorption | High | High | High | High | High | High | High | High |
| BBB permeant | No | No | No | No | No | No | No | No |
| Pgp substrate | Yes | Yes | Yes | No | No | No | Yes | No |
| CYP1A2 inhibitor | No | No | No | No | No | No | No | No |
| CYP2C19 inhibitor | No | Yes | Yes | Yes | Yes | Yes | Yes | Yes |
| CYP2C9 inhibitor | Yes | Yes | Yes | Yes | Yes | Yes | Yes | No |
| CYP2D6 inhibitor | No | No | No | No | No | No | No | No |
| CYP3A4 inhibitor | No | No | No | Yes | Yes | Yes | Yes | Yes |
| Druglikeness | ||||||||
| Lipinski violations | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Medicinal chemistry | ||||||||
| PAINS alerts | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Brenk alerts | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 4a | 4b | 4c | 5a | 5b | 5c | 5d | 6 | 7 | |
|---|---|---|---|---|---|---|---|---|---|
| Physicochemical Properties | |||||||||
| MW (g/mol) | 456.49 | 456.49 | 456.49 | 379.41 | 397.40 | 397.40 | 447.41 | 433.45 | 456.49 |
| Fraction Csp3 | 0.18 | 0.18 | 0.18 | 0.19 | 0.19 | 0.19 | 0.23 | 0.20 | 0.18 |
| Rotatable bonds | 11 | 11 | 11 | 9 | 9 | 9 | 10 | 9 | 11 |
| H-bond acceptors | 6 | 6 | 6 | 5 | 6 | 6 | 8 | 7 | 6 |
| H-bond donors | 3 | 3 | 3 | 2 | 2 | 2 | 2 | 2 | 3 |
| TPSA a (Å2) | 137.57 | 137.57 | 137.57 | 93.45 | 93.45 | 93.45 | 93.45 | 118.9 | 137.57 |
| Lipophilicity | |||||||||
| LogP b | 3.26 | 3.26 | 3.26 | 2.95 | 3.05 | 3.05 | 3.83 | 2.74 | 3.59 |
| Water solubility | |||||||||
| Solubility (mg/mL) c | 0.0185 | 0.0185 | 0.0185 | 0.0471 | 0.0342 | 0.0342 | 0.00778 | 0.0361 | 0.0115 |
| Solubility class | Moderately soluble | Moderately soluble | Moderately soluble | Soluble | Moderately soluble | Moderately soluble | Moderately soluble | Moderately soluble | Moderately soluble |
| Pharmacokinetics | |||||||||
| GI absorption | Low | Low | Low | High | High | High | High | High | Low |
| BBB permeant | No | No | No | No | No | No | No | No | No |
| Pgp substrate | No | No | No | No | No | No | No | No | No |
| CYP1A2 inhibitor | No | No | No | No | Yes | Yes | No | No | No |
| CYP2C19 inhibitor | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes |
| CYP2C9 inhibitor | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes |
| CYP2D6 inhibitor | Yes | No | Yes | Yes | Yes | Yes | Yes | No | No |
| CYP3A4 inhibitor | Yes | Yes | Yes | No | No | No | Yes | Yes | Yes |
| Druglikeness | |||||||||
| Lipinski violations | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Medicinal chemistry | |||||||||
| PAINS alerts | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Brenk alerts | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 1 |
| Strains | MIC (µg/mL) | |||||
|---|---|---|---|---|---|---|
| 3a | 3b | 3c | 4a | 4b | 4c | |
| Enterococcus genus | ||||||
| E. faecalis 365 * | >128 | >128 | 128 | >128 | >128 | >128 |
| E. faecalis 450 * | 128 | 128 | 64 | >128 | >128 | >128 |
| E. faecalis 451 * | >128 | 128 | 64 | >128 | >128 | >128 |
| E. faecium 182 * | 128 | 64 | 64 | >128 | >128 | >128 |
| E. faecium 300 * | >128 | >128 | 64 | >128 | >128 | >128 |
| E. faecium 364 * | >128 | >128 | 64 | >128 | >128 | >128 |
| E. durans 103 | 64 | >128 | 64 | >128 | >128 | >128 |
| E. gallinarum 150 * | 32 | >128 | 128 | >128 | >128 | >128 |
| E. casseliflavus 184 | 64 | 64 | 128 | 128 | >128 | >128 |
| Staphylococcus genus | ||||||
| S. aureus 18 ** | >128 | >128 | 64 | >128 | 64 | 128 |
| S. aureus 187 ** | >128 | 128 | 32 | 128 | 64 | 64 |
| S. aureus 195 ** | 128 | 128 | 32 | >128 | 128 | 128 |
| S. epidermidis 180 *** | 128 | 64 | 32 | 128 | 64 | 128 |
| S. epidermidis 181 *** | >128 | 128 | 64 | 128 | 64 | 128 |
| S. epidermidis 363 ** | 128 | 128 | 32 | 128 | 128 | 128 |
| S. saprophyticus 41 | >128 | >128 | 64 | >128 | >128 | >128 |
| S. warneri 74 | 64 | 128 | 64 | 128 | >128 | >128 |
| S. hominis 125 # | 32 | 32 | 64 | 128 | 128 | 128 |
| S. lugdunensis 129 | 32 | 32 | 32 | 64 | 32 | 32 |
| S. simulans 163 # | >128 | >128 | 128 | >128 | >128 | >128 |
| S. haemolyticus 193 # | >128 | >128 | 128 | >128 | >128 | >128 |
| MIC (µg/mL) | |||||
|---|---|---|---|---|---|
| Strains | 3c | 4b | Ampicillin | Ciprofloxacin | Oxacillin |
| E. faecalis 365 * | 128 | >128 | 128 | - | - |
| E. faecalis 450 * | 64 | >128 | 128 | - | - |
| E. faecalis 451 * | 64 | >128 | 128 | - | - |
| E. faecium 182 * | 64 | >128 | 128 | - | - |
| E. faecium 300 * | 64 | >128 | 128 | - | - |
| E. faecium 364 * | 64 | >128 | 128 | - | - |
| S. aureus 18 ** | 64 | 64 | - | 128 | 512 |
| S. aureus 187 ** | 32 | 64 | - | 128 | 512 |
| S. aureus 195 ** | 32 | 128 | - | 128 | 512 |
| S. epidermidis 180 *** | 32 | 64 | - | 64 | 256 |
| S. epidermidis 181 *** | 64 | 64 | - | 64 | 256 |
| S. epidermidis 363 ** | 32 | 64 | - | 64 | 256 |
| Entry | % inhib. at 50 µg/mL | MIC (µg/mL) |
|---|---|---|
| 3a | 30 | >50 |
| 3b | 55 | >50 |
| 3c | 99 | 35.8 |
| 3d | 59 | >50 |
| 4a | 100 | 39.3 |
| 4b | 75 | >50 |
| 4c | 88 | >50 |
| Compd. | Molecular Formula | Elemental Analysis | %S | |||
|---|---|---|---|---|---|---|
| Values | %C | %H | %N | |||
| 4a | C22H21N4O4SF | Calcd. | 57.88 | 4.64 | 12.27 | 7.02 |
| Found | 57.77 | 4.50 | 12.29 | 6.76 | ||
| 4b | C22H21N4O4SF | Calcd. | 57.88 | 4.64 | 12.27 | 7.02 |
| Found | 57.87 | 4.50 | 12.36 | 6.83 | ||
| 4c | C22H21N4O4SF | Calcd. | 57.88 | 4.64 | 12.27 | 7.02 |
| Found | 57.57 | 4.91 | 12.40 | 7.24 | ||
| 5a | C21H21N3O4 | Calcd. | 66.48 | 5.58 | 11.08 | // |
| Found | 66.16 | 5.80 | 11.09 | // | ||
| 5b | C21H20N3O4F | Calcd. | 63.47 | 5.07 | 10.57 | // |
| Found | 63.35 | 4.87 | 10.46 | // | ||
| 5c | C21H20N3O4F | Calcd. | 63.47 | 5.07 | 10.57 | // |
| Found | 63.47 | 5.32 | 10.57 | // | ||
| 5d | C22H20N3O4F3 | Calcd. | 59.06 | 4.51 | 9.39 | // |
| Found | 59.00 | 4.60 | 9.20 | // | ||
| 6 | C20H20N3O5SF | Calcd. | 55.42 | 4.65 | 9.69 | 7.40 |
| Found | 55.88 | 4.77 | 9.41 | 7.14 | ||
| 7 | C22H21N4O4SF | Calcd. | 57.88 | 4.64 | 12.27 | 7.02 |
| Found | 57.92 | 4.36 | 12.27 | 7.15 | ||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Brullo, C.; Caviglia, D.; Spallarossa, A.; Alfei, S.; Franzblau, S.G.; Tasso, B.; Schito, A.M. Microbiological Screening of 5-Functionalized Pyrazoles for the Future Development of Optimized Pyrazole-Based Delivery Systems. Pharmaceutics 2022, 14, 1770. https://doi.org/10.3390/pharmaceutics14091770
Brullo C, Caviglia D, Spallarossa A, Alfei S, Franzblau SG, Tasso B, Schito AM. Microbiological Screening of 5-Functionalized Pyrazoles for the Future Development of Optimized Pyrazole-Based Delivery Systems. Pharmaceutics. 2022; 14(9):1770. https://doi.org/10.3390/pharmaceutics14091770
Chicago/Turabian StyleBrullo, Chiara, Debora Caviglia, Andrea Spallarossa, Silvana Alfei, Scott G. Franzblau, Bruno Tasso, and Anna Maria Schito. 2022. "Microbiological Screening of 5-Functionalized Pyrazoles for the Future Development of Optimized Pyrazole-Based Delivery Systems" Pharmaceutics 14, no. 9: 1770. https://doi.org/10.3390/pharmaceutics14091770
APA StyleBrullo, C., Caviglia, D., Spallarossa, A., Alfei, S., Franzblau, S. G., Tasso, B., & Schito, A. M. (2022). Microbiological Screening of 5-Functionalized Pyrazoles for the Future Development of Optimized Pyrazole-Based Delivery Systems. Pharmaceutics, 14(9), 1770. https://doi.org/10.3390/pharmaceutics14091770