Gliadin Nanoparticles Containing Doxorubicin Hydrochloride: Characterization and Cytotoxicity
Abstract
1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Preparation of GNPs
2.3. Physico-Chemical Characterization
2.4. Entrapment Efficiency (EE), Loading Capacity (LC) and Release Profiles
2.5. Cell Cultures and Cytotoxicity
2.6. Statistical Analysis
3. Results and Discussion
3.1. Characterization of Nanosystems
3.2. Retention Rate of DOX and Release Profiles
3.3. In Vitro Cytotoxic Effects of DOX-Loaded GNPs
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- van der Zanden, S.Y.; Qiao, X.; Neefjes, J. New insights into the activities and toxicities of the old anticancer drug doxorubicin. FEBS J. 2021, 288, 6095–6111. [Google Scholar] [CrossRef] [PubMed]
- Naderinezhad, S.; Amoabediny, G.; Haghiralsadat, F. Co-delivery of hydrophilic and hydrophobic anticancer drugs using biocompatible pH-sensitive lipid-based nano-carriers for multidrug-resistant cancers. RSC Adv. 2017, 7, 30008–30019. [Google Scholar] [CrossRef]
- World Health Organization. Model List of Essential Medicines. Available online: https://www.who.int/publications/i/item/WHO-MHP-HPS-EML-2021.02 (accessed on 28 January 2022).
- Yang, F.; Teves, S.S.; Kemp, C.J.; Henikoff, S. Doxorubicin, DNA torsion, and chromatin dynamics. Biochim. Biophys. Acta (BBA)-Rev. Cancer 2014, 1845, 84–89. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, Y.; Nagoshi, T.; Yoshii, A.; Oi, Y.; Takahashi, H.; Kimura, H.; Ito, K.; Kashiwagi, Y.; Tanaka, T.D.; Yoshimura, M. Xanthine oxidase inhibition attenuates doxorubicin-induced cardiotoxicity in mice. Free Radic. Biol. Med. 2021, 162, 298–308. [Google Scholar] [CrossRef]
- Sritharan, S.; Sivalingam, N. A comprehensive review on time-tested anticancer drug doxorubicin. Life Sci. 2021, 278, 119527. [Google Scholar] [CrossRef]
- Wu, B.B.; Leung, K.T.; Poon, E.N.-Y. Mitochondrial-Targeted Therapy for Doxorubicin-Induced Cardiotoxicity. Int. J. Mol. Sci. 2022, 23, 1912. [Google Scholar] [CrossRef]
- Sobot, D.; Mura, S.; Couvreur, P. How can nanomedicines overcome cellular-based anticancer drug resistance? J. Mater. Chem. B 2016, 4, 5078–5100. [Google Scholar] [CrossRef]
- Mohammad, I.S.; He, W.; Yin, L. Understanding of human ATP binding cassette superfamily and novel multidrug resistance modulators to overcome MDR. Biomed. Pharmacother. 2018, 100, 335–348. [Google Scholar] [CrossRef]
- Cagel, M.; Grotz, E.; Bernabeu, E.; Moretton, M.A.; Chiappetta, D.A. Doxorubicin: Nanotechnological overviews from bench to bedside. Drug Discov. Today 2017, 22, 270–281. [Google Scholar] [CrossRef]
- Gabizon, A.; Shmeeda, H.; Barenholz, Y. Pharmacokinetics of pegylated liposomal doxorubicin. Clin. Pharmacokinet. 2003, 42, 419–436. [Google Scholar] [CrossRef]
- Barenholz, Y.C. Doxil®—The first FDA-approved nano-drug: Lessons learned. J. Control. Release 2012, 160, 117–134. [Google Scholar] [CrossRef] [PubMed]
- Gonçalves, M.; Mignani, S.; Rodrigues, J.; Tomás, H. A glance over doxorubicin based-nanotherapeutics: From proof-of-concept studies to solutions in the market. J. Control. Release 2020, 317, 347–374. [Google Scholar] [CrossRef] [PubMed]
- Ruiz-Gatón, L.; Espuelas, S.; Huarte, J.; Larrañeta, E.; Martin-Arbella, N.; Irache, J.M. Nanoparticles from Gantrez® AN-poly (ethylene glycol) conjugates as carriers for oral delivery of docetaxel. Int. J. Pharm. 2019, 571, 118699. [Google Scholar] [CrossRef] [PubMed]
- Yu, W.; Liu, R.; Zhou, Y.; Gao, H. Size-tunable strategies for a tumor targeted drug delivery system. ACS Cent. Sci. 2020, 6, 100–116. [Google Scholar] [CrossRef]
- Ambrosio, N.; Voci, S.; Gagliardi, A.; Palma, E.; Fresta, M.; Cosco, D. Application of Biocompatible Drug Delivery Nanosystems for the Treatment of Naturally Occurring Cancer in Dogs. J. Funct. Biomater. 2022, 13, 116. [Google Scholar] [CrossRef]
- Akbal, Ö.; Erdal, E.; Vural, T.; Kavaz, D.; Denkbaş, E.B. Comparison of protein-and polysaccharide-based nanoparticles for cancer therapy: Synthesis, characterization, drug release, and interaction with a breast cancer cell line. Artif. Cells Nanomed. Biotechnol. 2017, 45, 193–203. [Google Scholar] [CrossRef]
- Iannone, M.; Mare, R.; Paolino, D.; Gagliardi, A.; Froiio, F.; Cosco, D.; Fresta, M. Characterization and in vitro anticancer properties of chitosan-microencapsulated flavan-3-ols-rich grape seed extracts. Int. J. Biol. Macromol. 2017, 104, 1039–1045. [Google Scholar] [CrossRef]
- Gou, Y.; Miao, D.; Zhou, M.; Wang, L.; Zhou, H.; Su, G. Bio-inspired protein-based nanoformulations for cancer theranostics. Front. Pharmacol. 2018, 9, 421. [Google Scholar] [CrossRef]
- Luis de Redín, I.; Expósito, F.; Agüeros, M.; Collantes, M.; Peñuelas, I.; Allemandi, D.; Llabot, J.M.; Calvo, A.; Irache, J.M. In vivo efficacy of bevacizumab-loaded albumin nanoparticles in the treatment of colorectal cancer. Drug Deliv. Transl. Res. 2020, 10, 635–645. [Google Scholar] [CrossRef]
- Liang, K.; Chen, H. Protein-based nanoplatforms for tumor imaging and therapy. Wiley Interdiscip. Rev. Nanomed. Nanobiotechnol. 2020, 12, e16162020. [Google Scholar] [CrossRef]
- Martínez-López, A.L.; Pangua, C.; Reboredo, C.; Campión, R.; Morales-Gracia, J.; Irache, J.M. Protein-based nanoparticles for drug delivery purposes. Int. J. Pharm. 2020, 581, 119289. [Google Scholar] [CrossRef] [PubMed]
- Gagliardi, A.; Voci, S.; Bonacci, S.; Iriti, G.; Procopio, A.; Fresta, M.; Cosco, D. SCLAREIN (SCLAREol contained in zeIN) nanoparticles: Development and characterization of an innovative natural nanoformulation. Int. J. Biol. Macromol. 2021, 193, 713–720. [Google Scholar] [CrossRef] [PubMed]
- Voci, S.; Gagliardi, A.; Molinaro, R.; Fresta, M.; Cosco, D. Recent Advances of Taxol-Loaded Biocompatible Nanocarriers Embedded in Natural Polymer-Based Hydrogels. Gels 2021, 7, 33. [Google Scholar] [CrossRef] [PubMed]
- Voci, S.; Fresta, M.; Cosco, D. Gliadins as versatile biomaterials for drug delivery applications. J. Control. Release 2021, 329, 385–400. [Google Scholar] [CrossRef]
- Mehanna, M.; Mneimneh, A. Updated but not outdated “Gliadin”: A plant protein in advanced pharmaceutical nanotechnologies. Int. J. Pharm. 2020, 587, 119672. [Google Scholar] [CrossRef]
- Ezpeleta, I.; Irache, J.M.; Stainmesse, S.; Chabenat, C.; Gueguen, J.; Popineau, Y.; Orecchioni, A.-M. Gliadin nanoparticles for the controlled release of all-trans-retinoic acid. Int. J. Pharm. 1996, 131, 191–200. [Google Scholar] [CrossRef]
- Gulfam, M.; Kim, J.; Lee, J.M.; Ku, B.; Chung, B.H.; Chung, B.G. Anticancer drug-loaded gliadin nanoparticles induce apoptosis in breast cancer cells. Langmuir 2012, 28, 8216–8223. [Google Scholar] [CrossRef]
- Sharma, K.; Deevenapalli, M.; Singh, D.; Chourasia, M.K.; Bathula, S.R. Preparation and characterization of paclitaxel-loaded gliadin nanoparticles. J. Biomater. Tissue Eng. 2014, 4, 399–404. [Google Scholar] [CrossRef]
- Khatik, R.; Dwivedi, P.; Junnuthula, V.R.; Sharma, K.; Chuttani, K.; Mishra, A.K.; Dwivedi, A.K. Potential in vitro and in vivo colon specific anticancer activity in a HCT-116 xenograft nude mice model: Targeted delivery using enteric coated folate modified nanoparticles. RSC Adv. 2015, 5, 16507–16520. [Google Scholar] [CrossRef]
- Banaee, F.; Poureini, F.; Mohammadi, M.; Najafpour, G.D.; Moghadamnia, A.A. Encapsulation of curcumin in gliadin-pectin in a core–shell nanostructure for efficient delivery of curcumin to cancer cells in vitro. Colloid Polym. Sci. 2022, 300, 1063–1073. [Google Scholar] [CrossRef]
- Abdelmoneem, M.A.; Mahmoud, M.; Zaky, A.; Helmy, M.W.; Sallam, M.; Fang, J.-Y.; Elkhodairy, K.A.; Elzoghby, A.O. Decorating protein nanospheres with lactoferrin enhances oral COX-2 inhibitor/herbal therapy of hepatocellular carcinoma. Nanomedicine 2018, 13, 2377–2395. [Google Scholar] [CrossRef] [PubMed]
- Xu, W.; Zheng, S.; Sun, H.; Li, Z.; Xi, R.; Luo, D.; Li, Y.; Shah, B.R. Green-step fabrication of gliadin/sodium caseinate nanogels for methotrexate release, cytotoxicity and cell phagocytosis. J. Drug Deliv. Sci. Technol. 2022, 67, 103028. [Google Scholar] [CrossRef]
- Voci, S.; Gagliardi, A.; Salvatici, M.C.; Fresta, M.; Cosco, D. Development of polyoxyethylene (2) oleyl ether-gliadin nanoparticles: Characterization and in vitro cytotoxicity. Eur. J. Pharm. Sci. 2021, 162, 105849. [Google Scholar] [CrossRef] [PubMed]
- Voci, S.; Gagliardi, A.; Fresta, M.; Cosco, D. Ascorbic acid-loaded gliadin nanoparticles as a novel nutraceutical formulation. Food Res. Int. 2022, 161, 111869. [Google Scholar] [CrossRef] [PubMed]
- Gagliardi, A.; Cosco, D.; Udongo, B.P.; Dini, L.; Viglietto, G.; Paolino, D. Design and Characterization of Glyceryl Monooleate-Nanostructures Containing Doxorubicin Hydrochloride. Pharmaceutics 2020, 12, 1017. [Google Scholar] [CrossRef] [PubMed]
- Gagliardi, A.; Voci, S.; Giuliano, E.; Salvatici, M.C.; Celano, M.; Fresta, M.; Cosco, D. Phospholipid/zein hybrid nanoparticles as promising carriers for the protection and delivery of all-trans retinoic acid. Mater. Sci. Eng. C 2021, 128, 112331. [Google Scholar] [CrossRef] [PubMed]
- Gagliardi, A.; Molinaro, R.; Fresta, M.; Duranti, A.; Cosco, D. α-Acylamino-β-lactone N-acylethanolamine-hydrolyzing acid amidase inhibitors encapsulated in PLGA nanoparticles: Improvement of the physical stability and protection of human cells from hydrogen peroxide-induced oxidative stress. Antioxidants 2022, 11, 686. [Google Scholar] [CrossRef] [PubMed]
- Yan, T.; Zhu, S.; Hui, W.; He, J.; Liu, Z.; Cheng, J. Chitosan based pH-responsive polymeric prodrug vector for enhanced tumor targeted co-delivery of doxorubicin and siRNA. Carbohydr. Polym. 2020, 250, 116781. [Google Scholar] [CrossRef]
- Akram, M.U.; Abbas, N.; Farman, M.; Manzoor, S.; Khan, M.I.; Osman, S.M.; Luque, R.; Shanableh, A. Tumor micro-environment sensitive release of doxorubicin through chitosan based polymeric nanoparticles: An in-vitro study. Chemosphere 2022, 313, 137332. [Google Scholar] [CrossRef]
- Cabeza, L.; Ortiz, R.; Prados, J.; Delgado, Á.V.; Martín-Villena, M.J.; Clares, B.; Perazzoli, G.; Entrena, J.M.; Melguizo, C.; Arias, J.L. Improved antitumor activity and reduced toxicity of doxorubicin encapsulated in poly (ε-caprolactone) nanoparticles in lung and breast cancer treatment: An in vitro and in vivo study. Eur. J. Pharm. Sci. 2017, 102, 24–34. [Google Scholar] [CrossRef]
- Hemaiswarya, S.; Doble, M. Combination of phenylpropanoids with 5-fluorouracil as anti-cancer agents against human cervical cancer (HeLa) cell line. Phytomedicine 2013, 20, 151–158. [Google Scholar] [CrossRef] [PubMed]
- Hasinoff, B.B.; Wu, X.; Yadav, A.A.; Patel, D.; Zhang, H.; Wang, D.-S.; Chen, Z.-S.; Yalowich, J.C. Cellular mechanisms of the cytotoxicity of the anticancer drug elesclomol and its complex with Cu (II). Biochem. Pharmacol. 2015, 93, 266–276. [Google Scholar] [CrossRef] [PubMed]
- Xu, W.; Sun, H.; Kang, M.; Zheng, S.; Ning, Y.; Jia, Y.; Hu, Y.; Luo, D.; Zhang, C. Ethanol-tolerant pickering emulsion stabilized by gliadin nanoparticles. LWT 2022, 162, 113440. [Google Scholar] [CrossRef]
- Duan, X.; Li, Y. Physicochemical characteristics of nanoparticles affect circulation, biodistribution, cellular internalization, and trafficking. Small 2013, 9, 1521–1532. [Google Scholar] [CrossRef]
- Yang, R.; An, Y.; Miao, F.; Li, M.; Liu, P.; Tang, Q. Preparation of folic acid-conjugated, doxorubicin-loaded, magnetic bovine serum albumin nanospheres and their antitumor effects in vitro and in vivo. Int. J. Nanomed. 2014, 9, 4231. [Google Scholar] [CrossRef]
- Chang, D.; Lim, M.; Goos, J.A.C.M.; Qiao, R.; Ng, Y.Y.; Mansfeld, F.M.; Jackson, M.; Davis, T.P.; Kavallaris, M. Biologically targeted magnetic hyperthermia: Potential and limitations. Front. Pharmacol. 2018, 9, 831. [Google Scholar] [CrossRef]
- Seynhaeve, A.L.B.; Amin, M.; Haemmerich, D.; Van Rhoon, G.C.; Ten Hagen, T.L.M. Hyperthermia and smart drug delivery systems for solid tumor therapy. Adv. Drug Deliv. Rev. 2020, 163, 125–144. [Google Scholar] [CrossRef]
- Qi, J.; Yao, P.; He, F.; Yu, C.; Huang, C. Nanoparticles with dextran/chitosan shell and BSA/chitosan core—Doxorubicin loading and delivery. Int. J. Pharm. 2010, 393, 177–185. [Google Scholar] [CrossRef]
- Yao, W.; Zha, Q.; Cheng, X.; Wang, X.; Wang, J.; Tang, R. Folic acid-conjugated soybean protein-based nanoparticles mediate efficient antitumor ability in vitro. J. Biomater. Appl. 2017, 31, 832–843. [Google Scholar] [CrossRef]
- Pandey, V.; Haider, T.; Chandak, A.R.; Chakraborty, A.; Banerjee, S.; Soni, V. Surface modified silk fibroin nanoparticles for improved delivery of doxorubicin: Development, characterization, in-vitro studies. Int. J. Biol. Macromol. 2020, 164, 2018–2027. [Google Scholar] [CrossRef]
- Aziz, A.; Sefidbakht, Y.; Rezaei, S.; Kouchakzadeh, H.; Uskoković, V. Doxorubicin-loaded, pH-sensitive Albumin Nanoparticles for Lung Cancer Cell Targeting. J. Pharm. Sci. 2022, 111, 1187–1196. [Google Scholar] [CrossRef]
- Ye, W.; Zhang, G.; Liu, X.; Ren, Q.; Huang, F.; Yan, Y. Fabrication of polysaccharide-stabilized zein nanoparticles by flash nanoprecipitation for doxorubicin sustained release. J. Drug Deliv. Sci. Technol. 2022, 70, 103183. [Google Scholar] [CrossRef]
- Kayani, Z.; Bordbar, A.-K.; Firuzi, O. Novel folic acid-conjugated doxorubicin loaded β-lactoglobulin nanoparticles induce apoptosis in breast cancer cells. Biomed. Pharmacother. 2018, 107, 945–956. [Google Scholar] [CrossRef]
- Wu, X.; Hu, Q.; Liang, X.; Fang, S. Fabrication of colloidal stable gliadin-casein nanoparticles for the encapsulation of natamycin: Molecular interactions and antifungal application on cherry tomato. Food Chem. 2022, 391, 133288. [Google Scholar] [CrossRef]
- Pandey, A.; Kulkarni, S.; Vincent, A.P.; Nannuri, S.H.; George, S.D.; Mutalik, S. Hyaluronic acid-drug conjugate modified core-shell MOFs as pH responsive nanoplatform for multimodal therapy of glioblastoma. Int. J. Pharm. 2020, 588, 119735. [Google Scholar] [CrossRef]
- Liu, B.; Sun, L.; Lu, X.; Yang, Y.; Peng, H.; Sun, Z.; Xu, J.; Chu, H. Real-time drug release monitoring from pH-responsive CuS-encapsulated metal–organic frameworks. RSC Adv. 2022, 12, 11119–11127. [Google Scholar] [CrossRef]
- Yang, Y.-Y.; Zhang, M.; Liu, Z.-P.; Wang, K.; Yu, D.-G. Meletin sustained-release gliadin nanoparticles prepared via solvent surface modification on blending electrospraying. Appl. Surf. Sci. 2018, 434, 1040–1047. [Google Scholar] [CrossRef]
- Wu, W.; Kong, X.; Zhang, C.; Hua, Y.; Chen, Y.; Li, X. Fabrication and characterization of resveratrol-loaded gliadin nanoparticles stabilized by gum Arabic and chitosan hydrochloride. LWT 2020, 129, 109532. [Google Scholar] [CrossRef]
- Kamaly, N.; Yameen, B.; Wu, J.; Farokhzad, O.C. Degradable controlled-release polymers and polymeric nanoparticles: Mechanisms of controlling drug release. Chem. Rev. 2016, 116, 2602–2663. [Google Scholar] [CrossRef]
- Lee, J.H.; Yeo, Y. Controlled drug release from pharmaceutical nanocarriers. Chem. Eng. Sci. 2015, 125, 75–84. [Google Scholar] [CrossRef]
- Shaikh, M.V.; Kala, M.; Nivsarkar, M. Formulation and optimization of doxorubicin loaded polymeric nanoparticles using Box-Behnken design: Ex-vivo stability and in-vitro activity. Eur. J. Pharm. Sci. 2017, 100, 262–272. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Wang, G.; Zhang, Y. Research on the biological activity and doxorubicin release behavior in vitro of mesoporous bioactive SiO2-CaO-P2O5 glass nanospheres. Appl. Surf. Sci. 2017, 419, 531–539. [Google Scholar] [CrossRef]
- Muniswamy, V.J.; Raval, N.; Gondaliya, P.; Tambe, V.; Kalia, K.; Tekade, R.K. ‘Dendrimer-Cationized-Albumin’encrusted polymeric nanoparticle improves BBB penetration and anticancer activity of doxorubicin. Int. J. Pharm. 2019, 555, 77–99. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Wu, H.; Dong, W.; Zhou, J.; Zhang, X.; Liu, L.; Zhang, X.; Cheng, H.; Guan, J.; Zhao, R. Molecular simulation approach to the rational design of self-assembled nanoparticles for enhanced peroral delivery of doxorubicin. Carbohydr. Polym. 2019, 218, 279–288. [Google Scholar] [CrossRef] [PubMed]
- Alibolandi, M.; Sadeghi, F.; Abnous, K.; Atyabi, F.; Ramezani, M.; Hadizadeh, F. The chemotherapeutic potential of doxorubicin-loaded PEG-b-PLGA nanopolymersomes in mouse breast cancer model. Eur. J. Pharm. Biopharm. 2015, 94, 521–531. [Google Scholar] [CrossRef]
- Zhang, L.; Chen, Z.; Wang, H.; Wu, S.; Zhao, K.; Sun, H.; Kong, D.; Wang, C.; Leng, X.; Zhu, D. Preparation and evaluation of PCL–PEG–PCL polymeric nanoparticles for doxorubicin delivery against breast cancer. Rsc Adv. 2016, 6, 54727–54737. [Google Scholar] [CrossRef]
- Tian, B.; Ding, Y.; Han, J.; Zhang, J.; Han, Y.; Han, J. N-Acetyl-D-glucosamine decorated polymeric nanoparticles for targeted delivery of doxorubicin: Synthesis, characterization and in vitro evaluation. Colloids Surf. B Biointerfaces 2015, 130, 246–254. [Google Scholar] [CrossRef]
- Iafisco, M.; Delgado-Lopez, J.M.; Varoni, E.M.; Tampieri, A.; Rimondini, L.; Gomez-Morales, J.; Prat, M. Cell surface receptor targeted biomimetic apatite nanocrystals for cancer therapy. Small 2013, 9, 3834–3844. [Google Scholar] [CrossRef]
- Wei, R.; Cheng, L.; Zheng, M.; Cheng, R.; Meng, F.; Deng, C.; Zhong, Z. Reduction-responsive disassemblable core-cross-linked micelles based on poly (ethylene glycol)-b-poly (N-2-hydroxypropyl methacrylamide)–lipoic acid conjugates for triggered intracellular anticancer drug release. Biomacromolecules 2012, 13, 2429–2438. [Google Scholar] [CrossRef]
- Lei, T.; Srinivasan, S.; Tang, Y.; Manchanda, R.; Nagesetti, A.; Fernandez-Fernandez, A.; McGoron, A.J. Comparing cellular uptake and cytotoxicity of targeted drug carriers in cancer cell lines with different drug resistance mechanisms. Nanomed. Nanotechnol. Biol. Med. 2011, 7, 324–332. [Google Scholar] [CrossRef]
- Dong, F.; Dong, X.; Zhou, L.; Xiao, H.; Ho, P.-Y.; Wong, M.-S.; Wang, Y. Doxorubicin-loaded biodegradable self-assembly zein nanoparticle and its anti-cancer effect: Preparation, in vitro evaluation, and cellular uptake. Colloids Surf. B Biointerfaces 2016, 140, 324–331. [Google Scholar] [CrossRef] [PubMed]
- Kilic, M.A.; Ozlu, E.; Calis, S. A novel protein-based anticancer drug encapsulating nanosphere: Apoferritin-doxorubicin complex. J. Biomed. Nanotechnol. 2012, 8, 508–514. [Google Scholar] [CrossRef] [PubMed]
- Vrignaud, S.; Benoit, J.-P.; Saulnier, P. Strategies for the nanoencapsulation of hydrophilic molecules in polymer-based nanoparticles. Biomaterials 2011, 32, 8593–8604. [Google Scholar] [CrossRef] [PubMed]

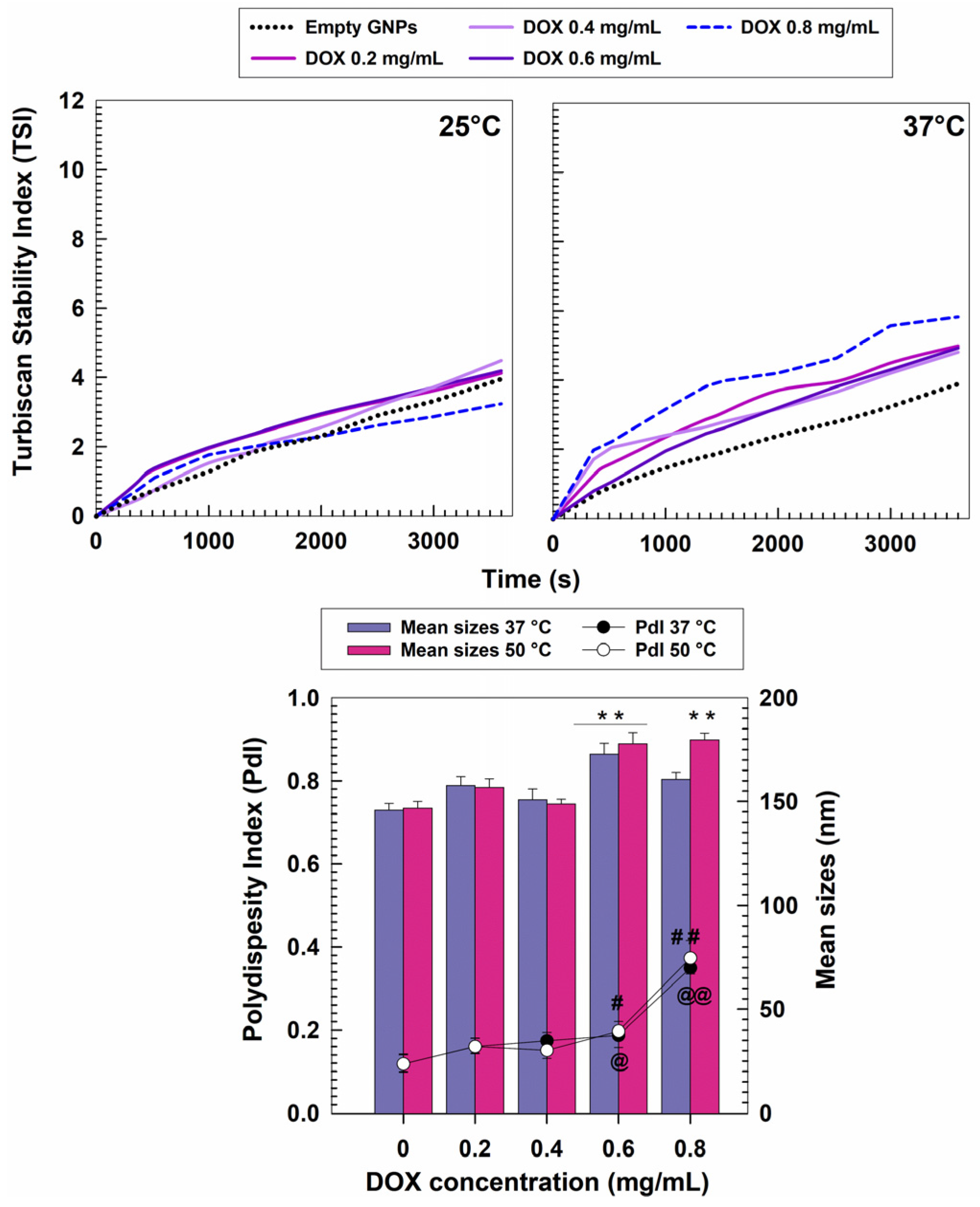
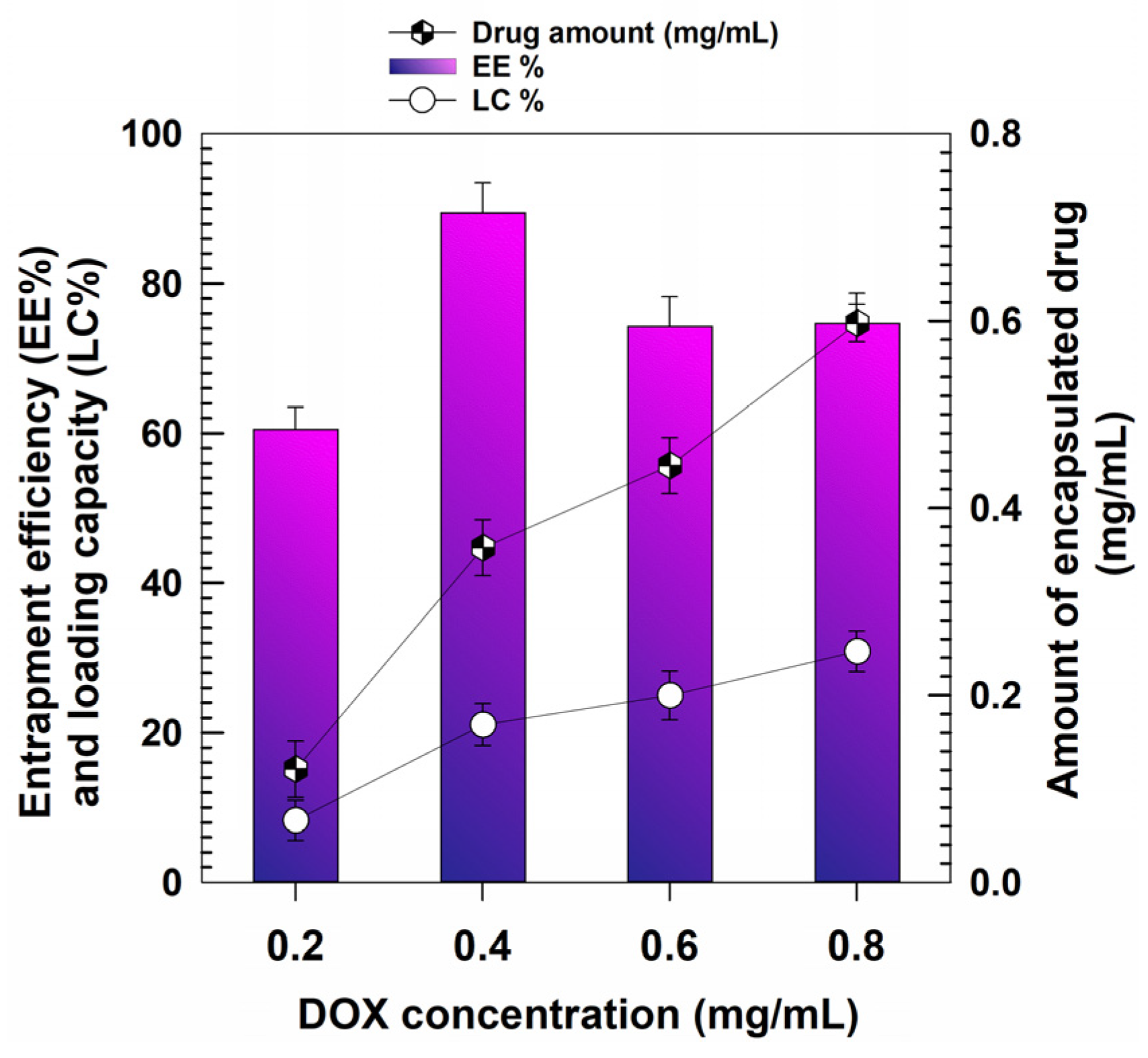
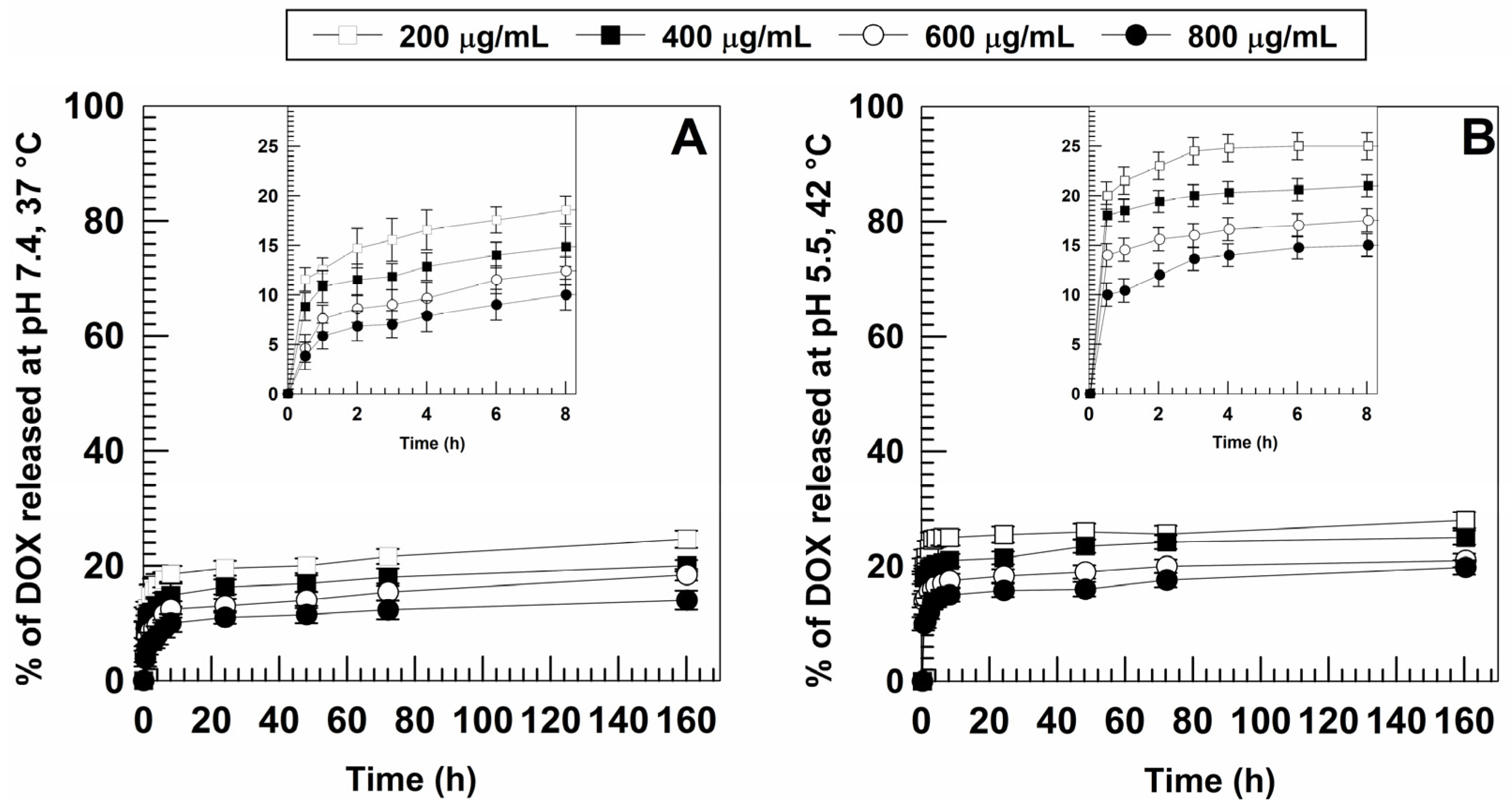
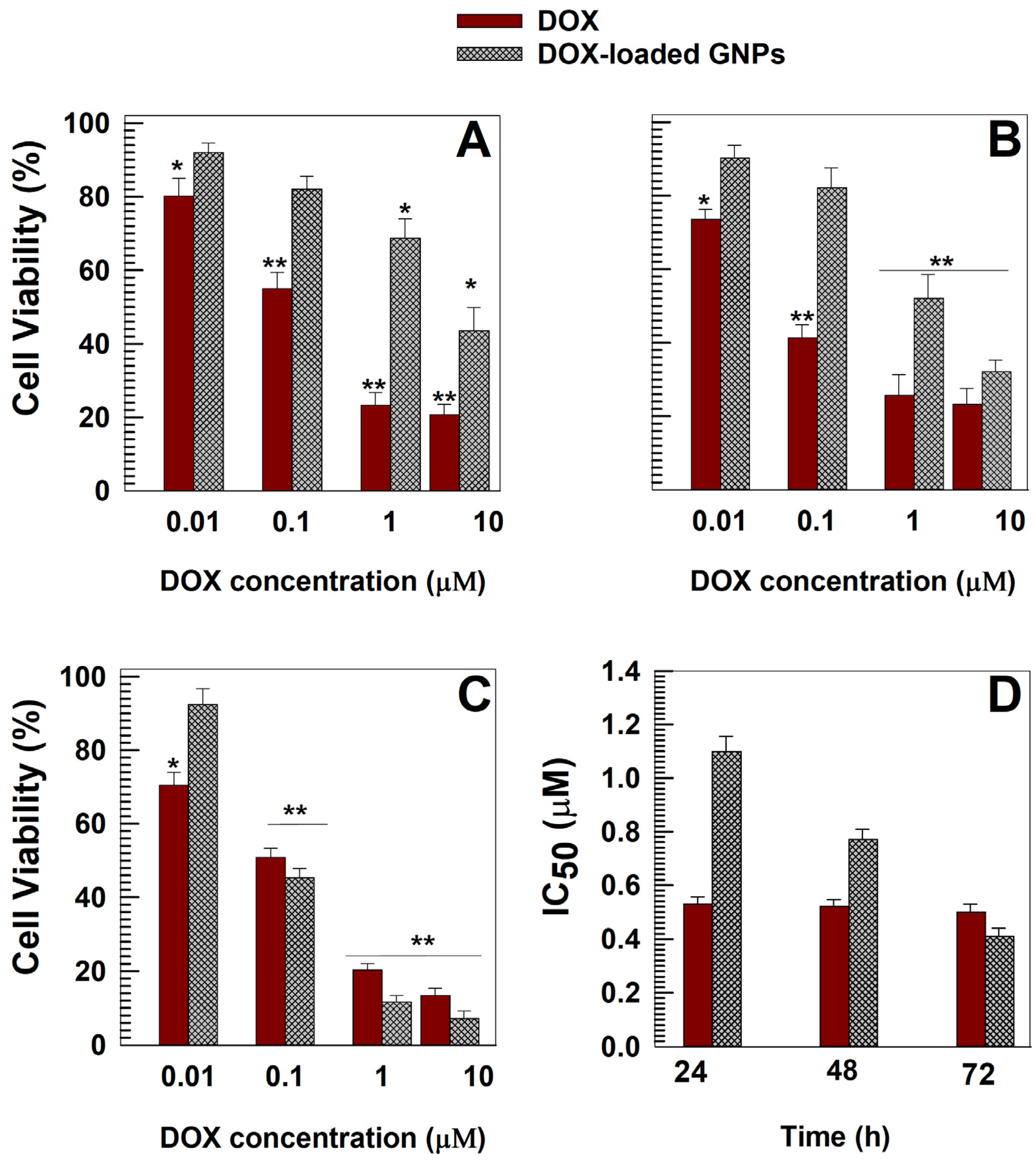
| DrugConcentration (mg/mL) | Temperature (°C) | Mean Sizes(nm) | PdI | Z-Potential(mV) |
|---|---|---|---|---|
| - | 25 | 143 ± 4 | 0.131 ± 0.004 | −28 ± 1 |
| 30 | 149 ± 2 | 0.131 ± 0.040 | −26 ± 1 | |
| 40 | 146 ± 3 | 0.120 ± 0.012 | −25 ± 1 | |
| 50 | 147 ± 3 | 0.120 ± 0.020 | −23 ± 1 | |
| 0.2 | 25 | 144 ± 2 | 0.125 ± 0.027 | −39 ± 1 |
| 30 | 156 ± 2 | 0.124 ± 0.031 | −49 ± 2 ** | |
| 40 | 158 ± 5 * | 0.162 ± 0.017 * | −44 ± 1 * | |
| 50 | 157 ± 4 * | 0.162 ± 0.048 * | −45 ± 1 * | |
| 0.4 | 25 | 141 ± 1 | 0.115 ± 0.010 | −50 ± 3 |
| 30 | 145 ± 1 | 0.161 ± 0.021 * | −49 ± 2 | |
| 40 | 151 ± 5 | 0.176 ± 0.013 * | −49 ± 3 | |
| 50 | 149 ± 2 | 0.153 ± 0.042 * | −43 ± 1 * | |
| 0.6 | 25 | 142 ± 3 | 0.160 ± 0.021 | −49 ± 2 |
| 30 | 160 ± 3 * | 0.197 ± 0.005 * | −42 ± 4 * | |
| 40 | 173 ± 5 * | 0.189 ± 0.031 * | −49 ± 2 | |
| 50 | 178 ± 2 * | 0.199 ± 0.007 ** | −44 ± 2 | |
| 0.8 | 25 | 173 ± 1 | 0.301 ± 0.026 | −35 ± 2 |
| 30 | 158 ± 2 * | 0.326 ± 0.004 * | −37 ± 4 | |
| 40 | 161 ± 3 * | 0.351 ± 0.007 * | −39 ± 3 | |
| 50 | 180 ± 3 | 0.355 ± 0.040 ** | −34 ± 1 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Voci, S.; Gagliardi, A.; Ambrosio, N.; Salvatici, M.C.; Fresta, M.; Cosco, D. Gliadin Nanoparticles Containing Doxorubicin Hydrochloride: Characterization and Cytotoxicity. Pharmaceutics 2023, 15, 180. https://doi.org/10.3390/pharmaceutics15010180
Voci S, Gagliardi A, Ambrosio N, Salvatici MC, Fresta M, Cosco D. Gliadin Nanoparticles Containing Doxorubicin Hydrochloride: Characterization and Cytotoxicity. Pharmaceutics. 2023; 15(1):180. https://doi.org/10.3390/pharmaceutics15010180
Chicago/Turabian StyleVoci, Silvia, Agnese Gagliardi, Nicola Ambrosio, Maria Cristina Salvatici, Massimo Fresta, and Donato Cosco. 2023. "Gliadin Nanoparticles Containing Doxorubicin Hydrochloride: Characterization and Cytotoxicity" Pharmaceutics 15, no. 1: 180. https://doi.org/10.3390/pharmaceutics15010180
APA StyleVoci, S., Gagliardi, A., Ambrosio, N., Salvatici, M. C., Fresta, M., & Cosco, D. (2023). Gliadin Nanoparticles Containing Doxorubicin Hydrochloride: Characterization and Cytotoxicity. Pharmaceutics, 15(1), 180. https://doi.org/10.3390/pharmaceutics15010180







