Influence of Lipid Composition of Cationic Liposomes 2X3-DOPE on mRNA Delivery into Eukaryotic Cells
Abstract
:1. Introduction
2. Materials and Methods
2.1. Preparation of Cationic Liposomes
2.2. In Vitro Transcription
2.3. Preparation of Complexes of Cationic Liposomes with mRNA
2.4. Size and Zeta Potential Measurement
2.5. Transmission Electron Microscopy (TEM)
2.6. Atomic Force Microscopy (AFM)
2.7. Gel Retardation Assay with Capillary Electrophoresis
2.8. Quantification of Encapsulation Efficiency
2.9. Cell Lines
2.10. Cell Transfection
2.11. Fluorescence Microscopy
2.12. Flow Cytometry Analysis
2.13. Luminescence Analysis
2.14. Cytotoxicity Studies
3. Results and Discussion
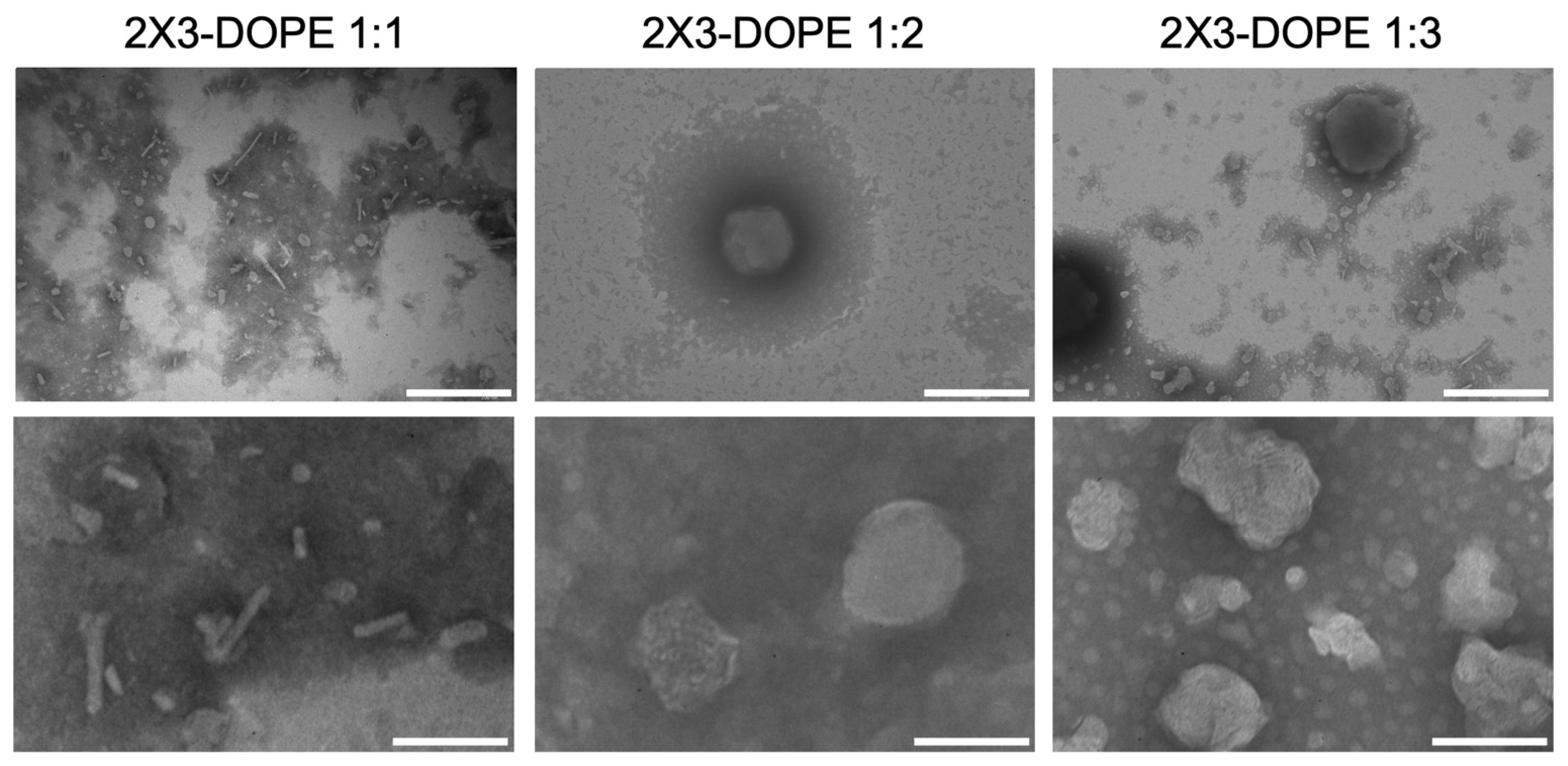
3.1. Preparation and Characterization of Liposomes: Effect of Cationic Lipid Composition on Lipoplex Formation with mRNA
3.2. Effect of Cationic Lipid Composition on mRNA Delivery into Eukaryotic Cells
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Schoenmaker, L.; Witzigmann, D.; Kulkarni, A.J.; Verbeke, R.; Kersten, G.; Jiskoot, W.; Crommelin, J.D. mRNA-lipid nanoparticle COVID-19 vaccines: Structure and stability. Int. J. Pharm. 2021, 601, 120586. [Google Scholar] [CrossRef] [PubMed]
- Kon, E.; Elia, U.; Peer, D. Principles for designing an optimal mRNA lipid nanoparticle vaccine. Curr. Opin. Biotechnol. 2022, 73, 329–336. [Google Scholar] [CrossRef] [PubMed]
- Paunovska, K.; Loughrey, D.; Dahlman, J.E. Drug delivery systems for RNA therapeutics. Nat. Rev. Genet. Nat. Rev. Genet. 2022, 23, 265–280. [Google Scholar] [CrossRef] [PubMed]
- Webb, C.; Ip, S.; Bathula, N.V.; Popova, P.; Soriano, S.K.; Ly, H.H.; Eryilmaz, B.; Huu, V.A.N.; Broadhead, R.; Rabel, M.; et al. Current status and future perspectives on mRNA drug manufacturing. Mol. Pharm. 2022, 19, 1047–1058. [Google Scholar] [CrossRef] [PubMed]
- Adams, D.; Gonzalez-Duarte, A.; O’Riordan, W.D.; Yang, C.C.; Ueda, M.; Kristen, A.V.; Tournev, I.; Tournev, H.H.; Tournev, T.; Tournev, J.L.; et al. Patisiran, an RNAi therapeutic, for hereditary transthyretin amyloidosis. N. Engl. J. Med. 2018, 379, 11–21. [Google Scholar] [CrossRef]
- Dammes, N.; Peer, D. Paving the road for RNA therapeutics. Trends Pharmacol. Sci. 2020, 41, 755–775. [Google Scholar] [CrossRef]
- Samaridou, E.; Heyes, J.; Lutwyche, P. Lipid nanoparticles for nucleic acid delivery: Current perspectives. Adv. Drug Deliv. Rev. 2020, 154–155, 37–63. [Google Scholar] [CrossRef]
- Gaspar, R.; Coelho, F.; Silva, B.F. Lipid-nucleic acid complexes: Physicochemical aspects and prospects for cancer treatment. Molecules 2020, 25, 5006. [Google Scholar] [CrossRef]
- Petukhov, I.A.; Maslov, M.A.; Morozova, N.G.; Serebrennikova, G.A. Synthesis of polycationic lipids based on cholesterol and spermine. Russ. Chem. Bull. 2010, 59, 260–268. [Google Scholar] [CrossRef]
- Maslov, M.A.; Kabilova, T.O.; Petukhov, I.A.; Morozova, N.G.; Serebrennikova, G.A.; Vlassov, V.V.; Zenkova, M.A. Novel cholesterol spermine conjugates provide efficient cellular delivery of plasmid DNA and small interfering RNA. J. Control. Release 2012, 160, 182–193. [Google Scholar] [CrossRef]
- Markov, O.O.; Mironova, N.L.; Maslov, M.A.; Petukhov, I.A.; Morozova, N.G.; Vlassov, V.V.; Zenkova, M.A. Novel cationic liposomes provide highly efficient delivery of DNA and RNA into dendritic cell progenitors and their immature offsets. J. Control. Release 2012, 160, 200–210. [Google Scholar] [CrossRef]
- Kabilova, T.; Shmendel, E.; Gladkikh, D.; Morozova, N.; Maslov, M.; Chernolovskaya, E.; Vlassov, V.; Zenkova, M. Novel PEGylated liposomes enhance immunostimulating activity of is RNA. Molecules 2018, 23, 3101. [Google Scholar] [CrossRef] [Green Version]
- Mikheev, A.A.; Shmendel, E.V.; Zhestovskaya, E.S.; Nazarov, G.V.; Maslov, M.A. Cationic liposomes as delivery systems for nucleic acids. Fine Chem. Technol. 2020, 15, 7–27. [Google Scholar] [CrossRef] [Green Version]
- Shmendel, E.; Kabilova, T.; Morozova, N.; Zenkova, M.; Maslov, M. Effects of spacers within a series of novel folate-containing lipoconjugates on the targeted delivery of nucleic acids. J. Drug Deliv. Sci. Technol. 2020, 57, 101609. [Google Scholar] [CrossRef]
- Gao, X.; Huang, L. Cationic liposome-mediated gene transfer. Gene Ther. 1995, 2, 710–722. [Google Scholar]
- Hattori, Y.; Shimizu, S.; Ozaki, K.I.; Onishi, H. Effect of cationic lipid type in folate-PEG-modified cationic liposomes on folate receptor-mediated siRNA transfection in tumor cells. Pharmaceutics 2019, 11, 181. [Google Scholar] [CrossRef] [Green Version]
- Mochizuki, S.; Kanegae, N.; Nishina, K.; Kamikawa, Y.; Koiwai, K.; Masunaga, H.; Sakurai, K. The role of the helper lipid dioleoylphosphatidylethanolamine (DOPE) for DNA transfection cooperating with a cationic lipid bearing ethylenediamine. Biochim. Biophys. Acta. 2013, 1828, 412–418. [Google Scholar] [CrossRef] [Green Version]
- Du, Z.; Munye, M.M.; Tagalakis, A.D.; Manunta, M.D.; Hart, S.L. The role of the helper lipid on the DNA transfection efficiency of lipopolyplex formulations. Sci. Rep. 2014, 4, 7107. [Google Scholar] [CrossRef] [Green Version]
- Farhood, H.; Serbina, N.; Huang, L. The role of dioleoyl phosphatidylethanolamine in cationic liposome mediated gene transfer. Biochim. Biophys. Acta 1995, 1235, 289–295. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Y.; Li, H.M.; Sun, J.; Gao, J.; Liu, W.; Li, B.; Gao, Y.; Chen, J.M. DC-Chol/DOPE cationic liposomes. A comparative study of the influence factors on plasmid pDNA and siRNA gene delivery. Int. J. Pharm. 2010, 390, 198–207. [Google Scholar] [CrossRef]
- Akhter, S.; Berchel, M.; Jaffrès, P.A.; Midoux, P.; Pichon, C. mRNA Lipoplexes with Cationic and Ionizable α-Amino-lipophosphonates: Membrane Fusion, Transfection, mRNA Translation and Conformation. Pharmaceutics 2022, 14, 581. [Google Scholar] [CrossRef] [PubMed]
- Tilcock, C.P. Lipid polymorphism. Chem. Phys. Lipids 1986, 40, 109–125. [Google Scholar] [CrossRef] [PubMed]
- Wasungu, L.; Hoekstra, D. Cationic lipids, lipoplexes and intracellular delivery of genes. J. Control. Release 2006, 116, 255–264. [Google Scholar] [CrossRef] [PubMed]
- Buck, J.; Grossen, P.; Cullis, P.R.; Huwyler, J.; Witzigmann, D. Lipid-Based DNA Therapeutics: Hallmarks of Non-Viral Gene Delivery. ACS Nano 2019, 13, 3754–3782. [Google Scholar] [CrossRef] [PubMed]
- Gottfried, L.F.; Dean, D.A. Extracellular and intracellular barriers to non-viral gene transfer. In Novel Gene Therapy Approaches; Wei, M., Good, D., Eds.; InTech: Rijeka, Croatia, 2013; pp. 75–88. [Google Scholar]
- Simonis, B.; Vignone, D.; Paz, O.G.; Donati, E.; Falchetti, M.L.; Bombelli, C.; Cellucci, A.; Auciello, G.; Fini, I.; Galantini, L.; et al. Transport of cationic liposomes in a human blood brain barrier model: Role of the stereochemistry of the gemini amphiphile on liposome biological features. J. Colloid Interface Sci. 2022, 627, 283–298. [Google Scholar] [CrossRef]
- Nečas, D.; Klapetek, P. Gwyddion: An open-source software for SPM data analysis. Open Phys. 2012, 10, 181–188. [Google Scholar] [CrossRef]
- Kabilova, T.O.; Sen’kova, A.V.; Nikolin, V.P.; Popova, N.A.; Zenkova, M.A.; Vlassov, V.V.; Chernolovskaya, E.L. Antitumor and antimetastatic effect of small immunostimulatory RNA against B16 melanoma in mice. PLoS ONE 2016, 11, e0150751. [Google Scholar] [CrossRef]
- Zuidam, N.J.; Barenholz, Y. Electrostatic and structural properties of complexes involving plasmid DNA and cationic lipids commonly used for gene delivery. BBA-Biomembranes 1998, 1368, 115–128. [Google Scholar] [CrossRef] [Green Version]
- Fujiwara, T.; Hasegawa, S.; Hirashima, N.; Nakanishi, M.; Ohwada, T. Gene transfection activities of amphiphilic steroid-polyamine conjugates. Biochim. Biophys. Acta 2000, 1468, 396–402. [Google Scholar] [CrossRef] [Green Version]
- Ramezani, M.; Khoshhamdam, M.; Dehshahri, A.; Malaekeh-Nikouei, B. The influence of size, lipid composition and bilayer fluidity of cationic liposomes on the transfection efficiency of nanolipoplexes. Colloids Surf. B Biointerfaces 2009, 72, 1–5. [Google Scholar] [CrossRef]
- Hallan, S.S.; Sguizzato, M.; Esposito, E.; Cortesi, R. Challenges in the physical characterization of lipid nanoparticles. Pharmaceutics 2021, 13, 549. [Google Scholar] [CrossRef]
- Angelov, B.; Garamus, V.M.; Drechsler, M.; Angelova, A. Structural analysis of nanoparticulate carriers for encapsulation of macromolecular drugs. J. Mol. Liq. 2017, 235, 83–89. [Google Scholar] [CrossRef]
- Mikheev, A.; Shmendel, E.; Nazarova, G.; Maslovb, M. Influence of Liposome Composition on Plasmid DNA Delivery to Eukaryotic Cells. Russ. J. Bioorganic Chem. 2021, 47, 1034–1042. [Google Scholar] [CrossRef]






| Sample | Size (nm) | PDI | ζ Potential (mV) |
|---|---|---|---|
| 2X3-DOPE 1:1 | 62.7 ± 0.1 | 0.23 ± 0.01 | 40.4 ±1.2 |
| 2X3-DOPE 1:2 | 95.6 ± 0.7 | 0.23 ± 0.01 | 51.7 ± 8.0 |
| 2X3-DOPE 1:3 | 70.5 ± 0.4 | 0.26 ± 0.00 | 58.1 ± 2.0 |
| 2X3-DOPE 1:1 | 2X3-DOPE 1:2 | 2X3-DOPE 1:3 | |
|---|---|---|---|
| Pure | 195 ± 183 | 147 ± 116 | 109 ± 62 |
| & mRNA-eGFP | 47 ± 24 | 53 ± 27 | 58 ± 30 |
| & mRNA-FLuc | 40 ± 17 | 48 ± 24 | 60 ± 35 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Vysochinskaya, V.; Shishlyannikov, S.; Zabrodskaya, Y.; Shmendel, E.; Klotchenko, S.; Dobrovolskaya, O.; Gavrilova, N.; Makarova, D.; Plotnikova, M.; Elpaeva, E.; et al. Influence of Lipid Composition of Cationic Liposomes 2X3-DOPE on mRNA Delivery into Eukaryotic Cells. Pharmaceutics 2023, 15, 8. https://doi.org/10.3390/pharmaceutics15010008
Vysochinskaya V, Shishlyannikov S, Zabrodskaya Y, Shmendel E, Klotchenko S, Dobrovolskaya O, Gavrilova N, Makarova D, Plotnikova M, Elpaeva E, et al. Influence of Lipid Composition of Cationic Liposomes 2X3-DOPE on mRNA Delivery into Eukaryotic Cells. Pharmaceutics. 2023; 15(1):8. https://doi.org/10.3390/pharmaceutics15010008
Chicago/Turabian StyleVysochinskaya, Vera, Sergey Shishlyannikov, Yana Zabrodskaya, Elena Shmendel, Sergey Klotchenko, Olga Dobrovolskaya, Nina Gavrilova, Darya Makarova, Marina Plotnikova, Ekaterina Elpaeva, and et al. 2023. "Influence of Lipid Composition of Cationic Liposomes 2X3-DOPE on mRNA Delivery into Eukaryotic Cells" Pharmaceutics 15, no. 1: 8. https://doi.org/10.3390/pharmaceutics15010008
APA StyleVysochinskaya, V., Shishlyannikov, S., Zabrodskaya, Y., Shmendel, E., Klotchenko, S., Dobrovolskaya, O., Gavrilova, N., Makarova, D., Plotnikova, M., Elpaeva, E., Gorshkov, A., Moshkoff, D., Maslov, M., & Vasin, A. (2023). Influence of Lipid Composition of Cationic Liposomes 2X3-DOPE on mRNA Delivery into Eukaryotic Cells. Pharmaceutics, 15(1), 8. https://doi.org/10.3390/pharmaceutics15010008






