Chitosan Nanoparticles Loaded with Capparis cartilaginea Decne Extract: Insights into Characterization and Antigenotoxicity In Vivo
Abstract
1. Introduction
2. Materials and Methods
2.1. Materials
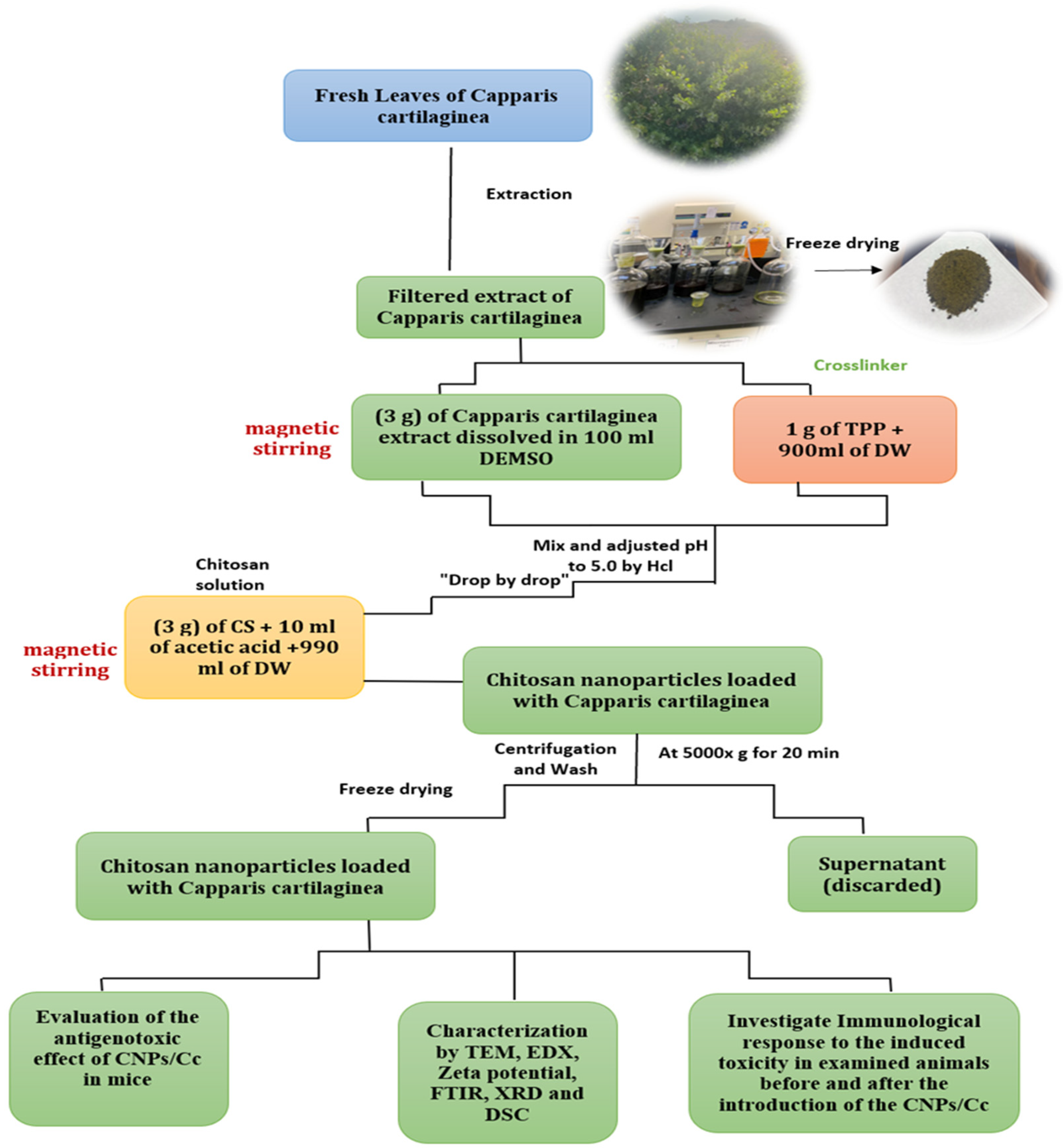
2.1.1. Preparation of C. cartilaginea Leaf Extract
2.1.2. C. cartilaginea Characterization
GC–MS Analysis
2.2. Synthesis and Optimization Process of CNPs/Cc
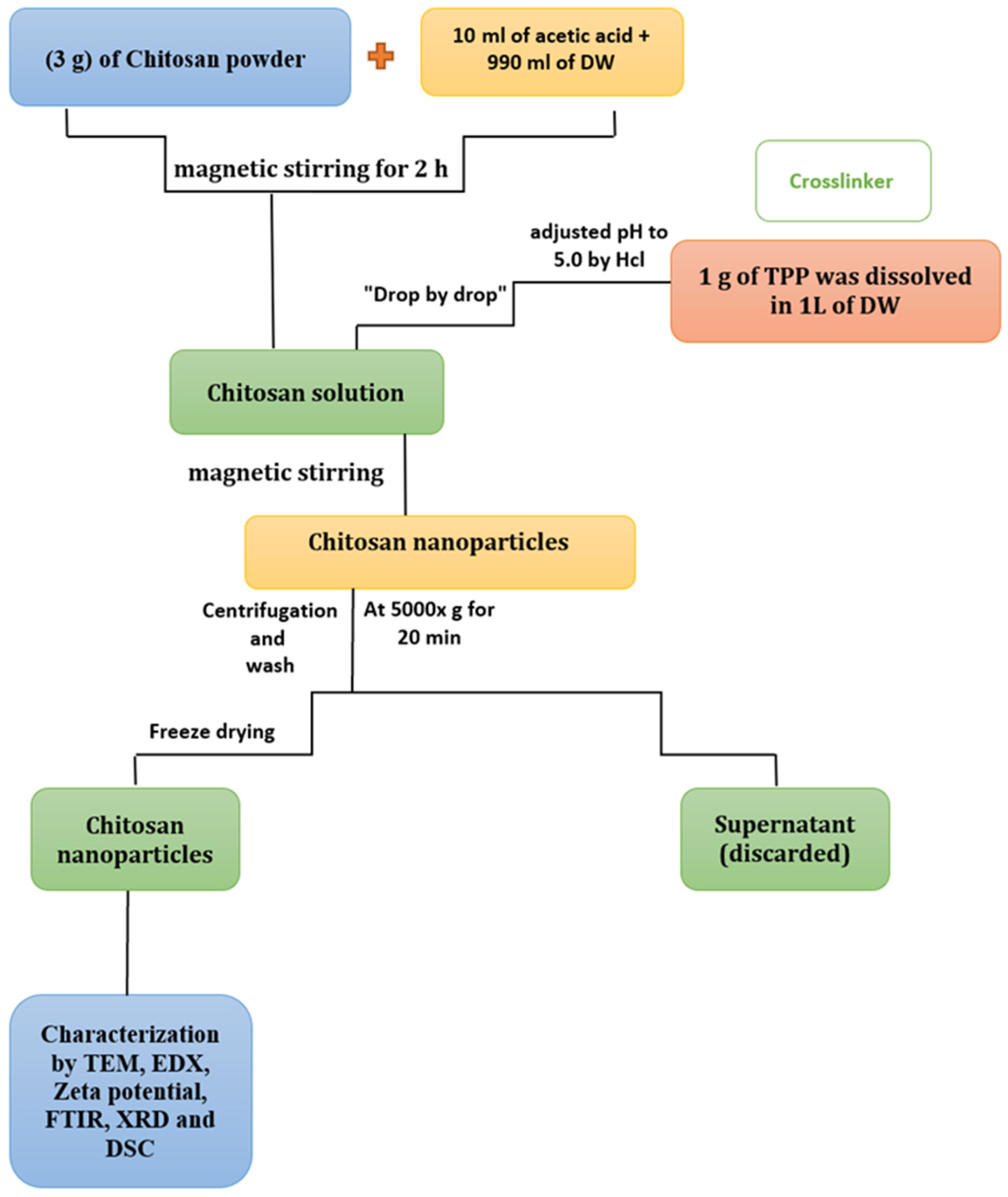
2.2.1. Synthesis of CNPs
2.2.2. Synthesis of CNPs/Cc
2.2.3. Characterization of CNPs and CNPs/Cc
2.3. In Vivo Antigenotoxicity
2.3.1. Experimental Animals
2.3.2. Experimental Design
2.3.3. Chromosome Abnormalities Assay
2.3.4. Sperm Morphology Assay
2.4. Statistical Analysis
3. Results and Discussion
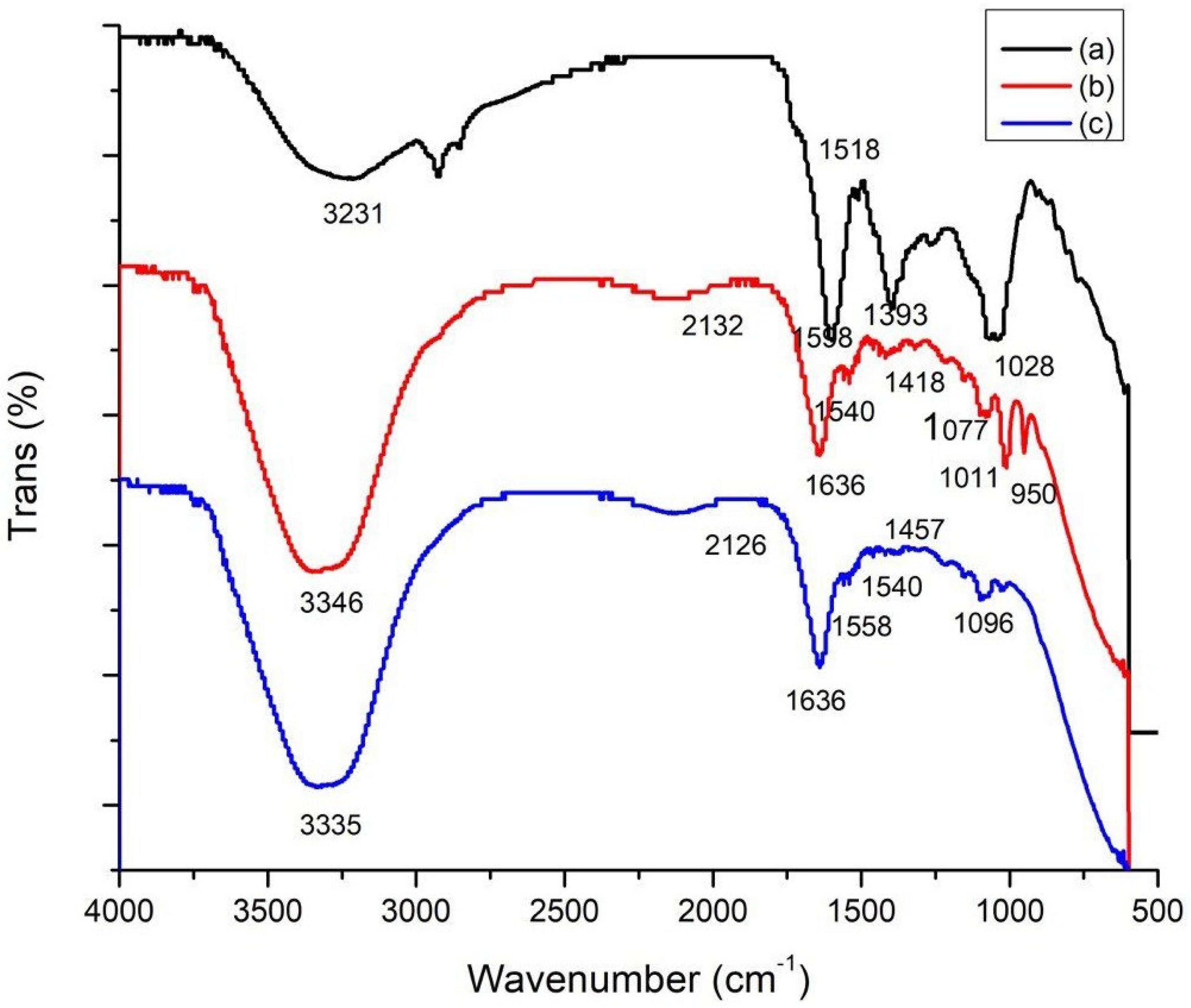
3.1. Fourier-Transform Infrared Spectroscopy (FTIR)
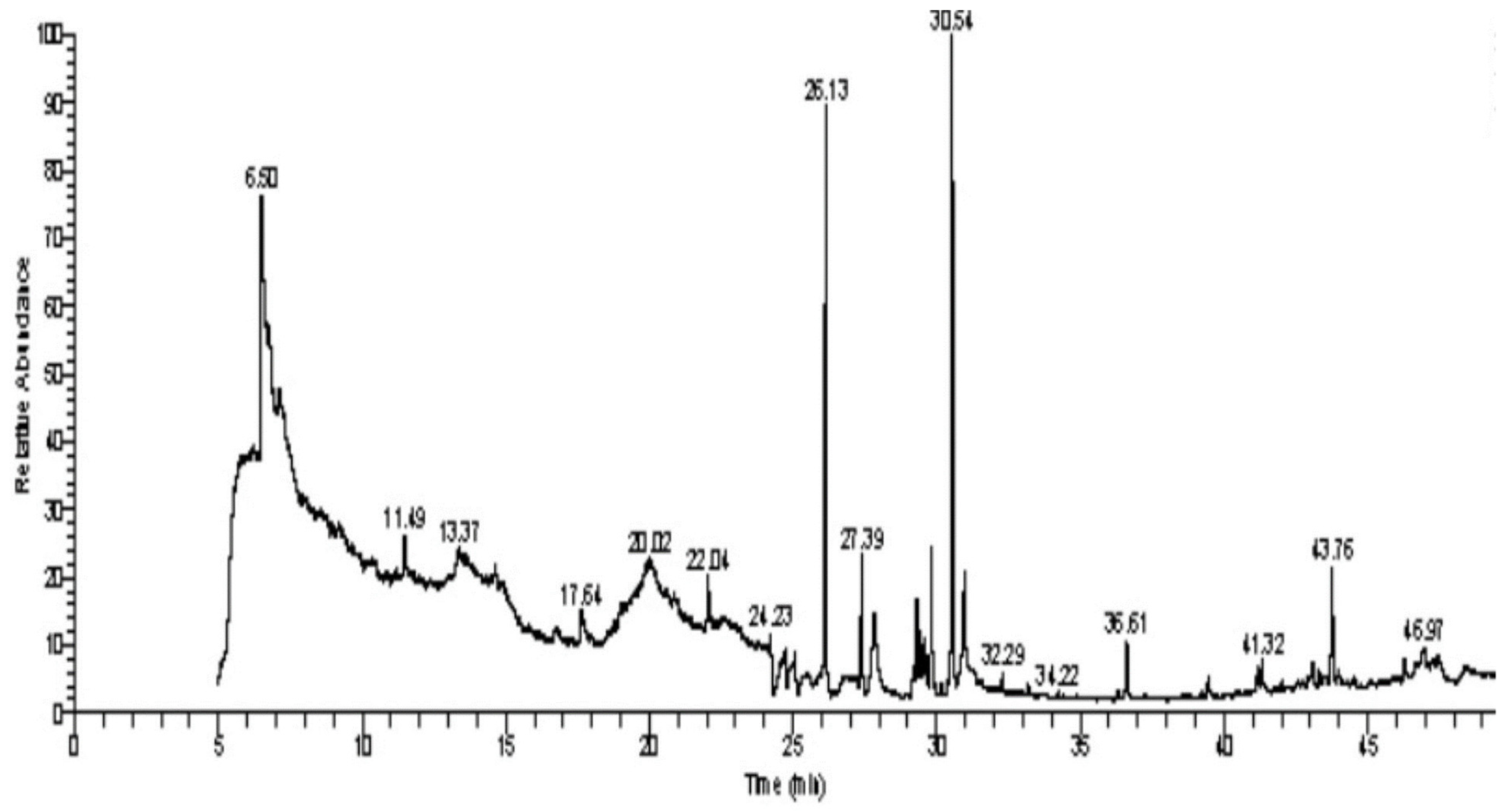
3.2. GC–MS Analysis of C. cartilaginea Extract
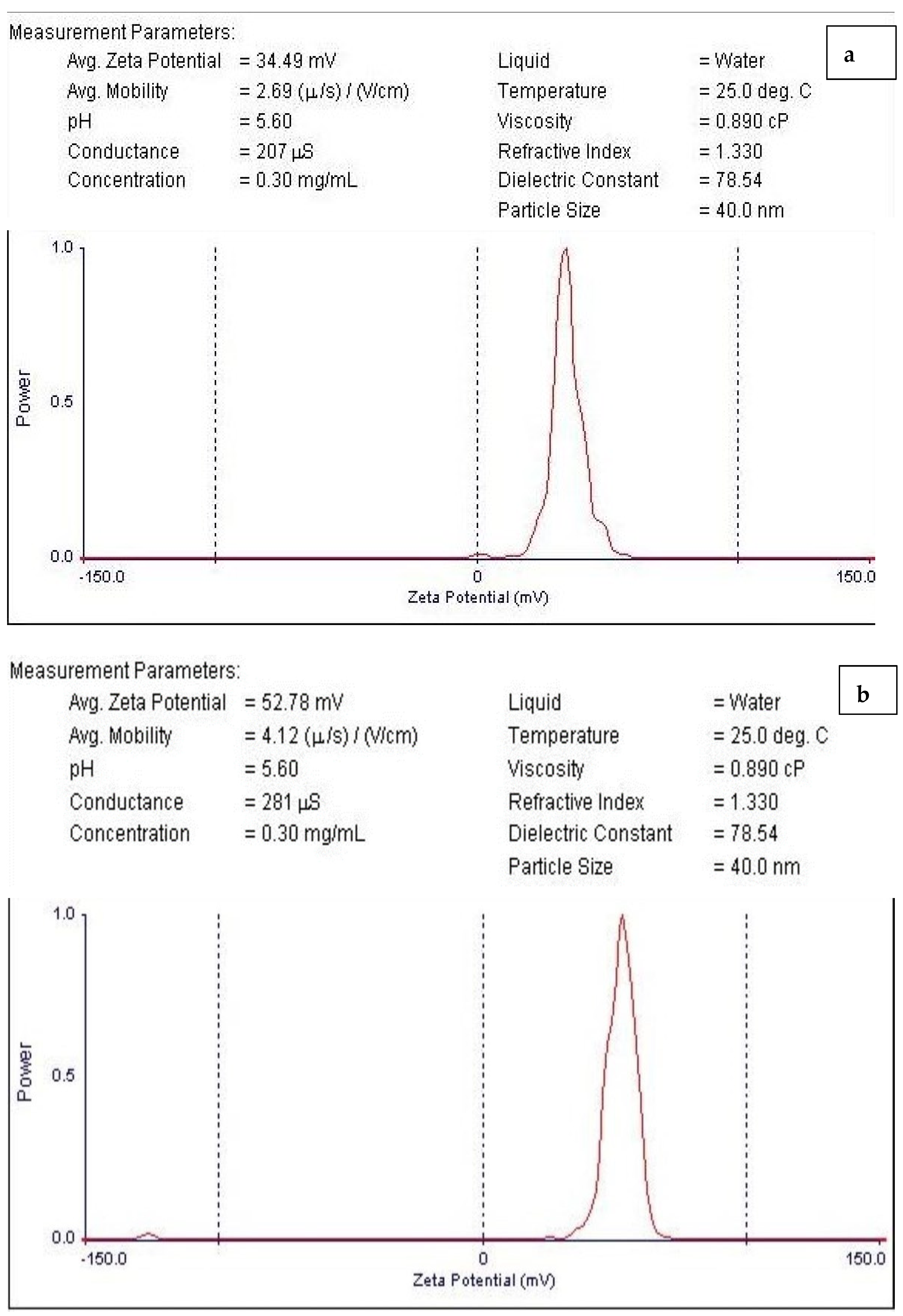
3.3. Zeta Potential Characterization
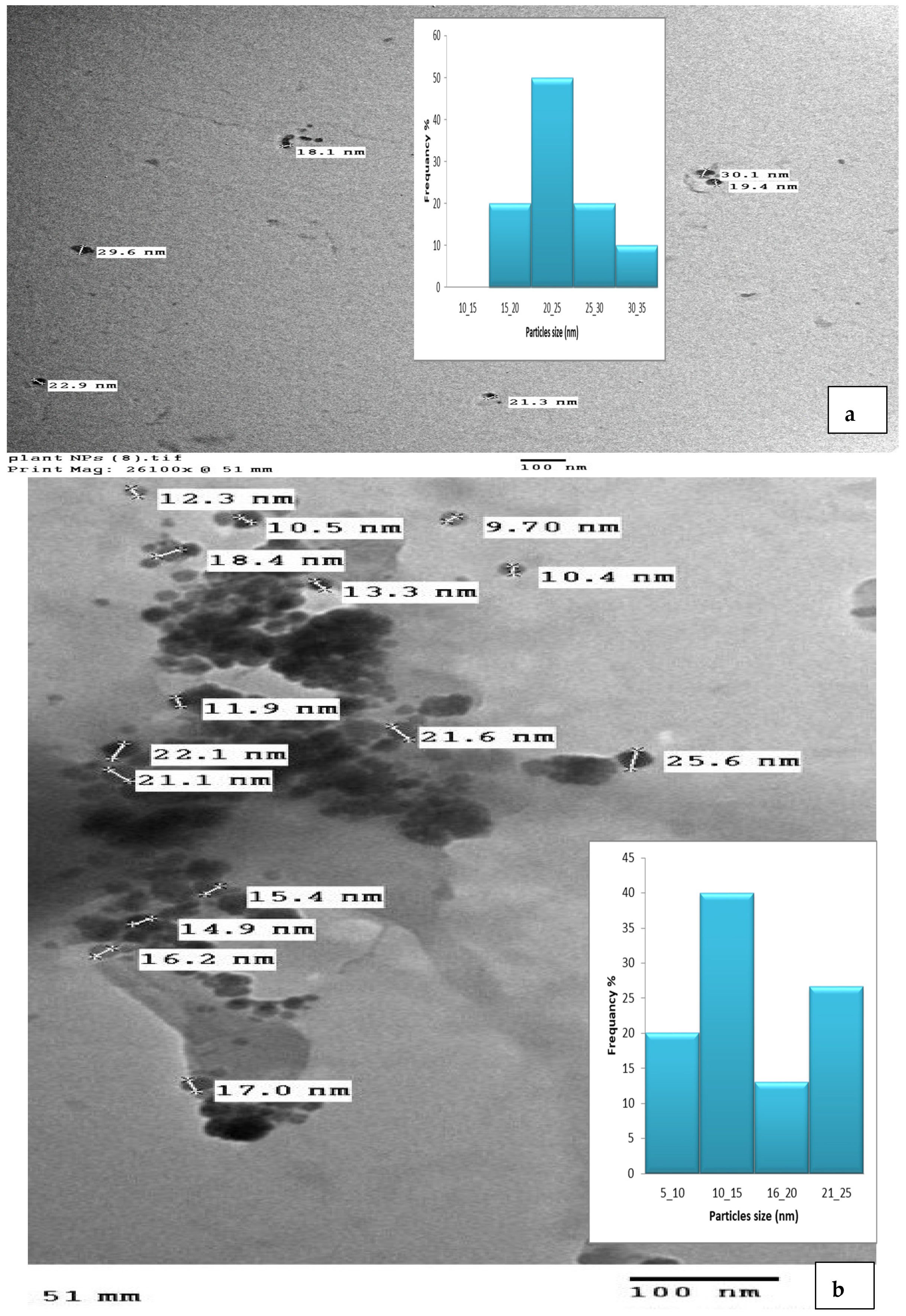
3.4. TEM Analysis
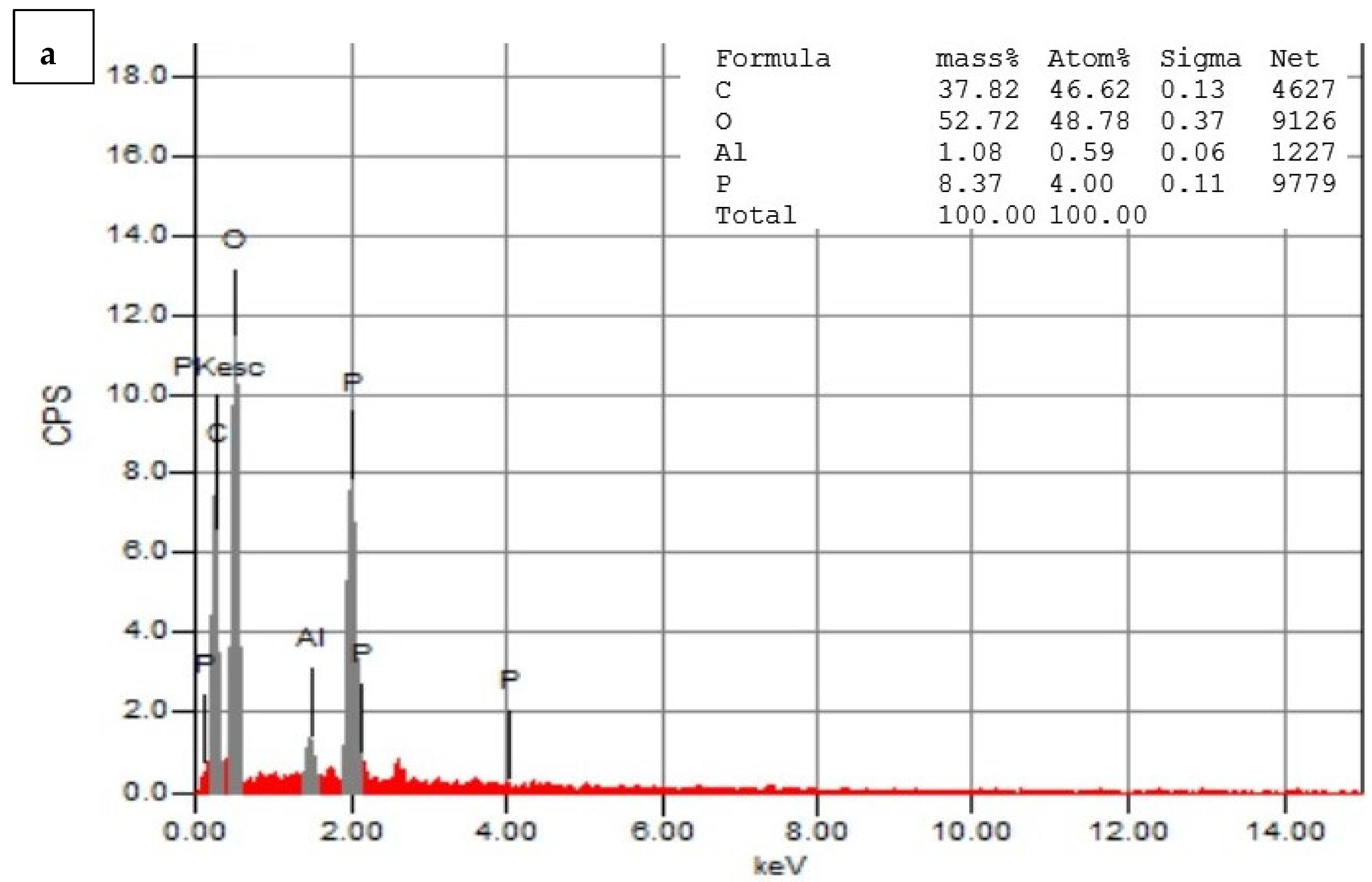
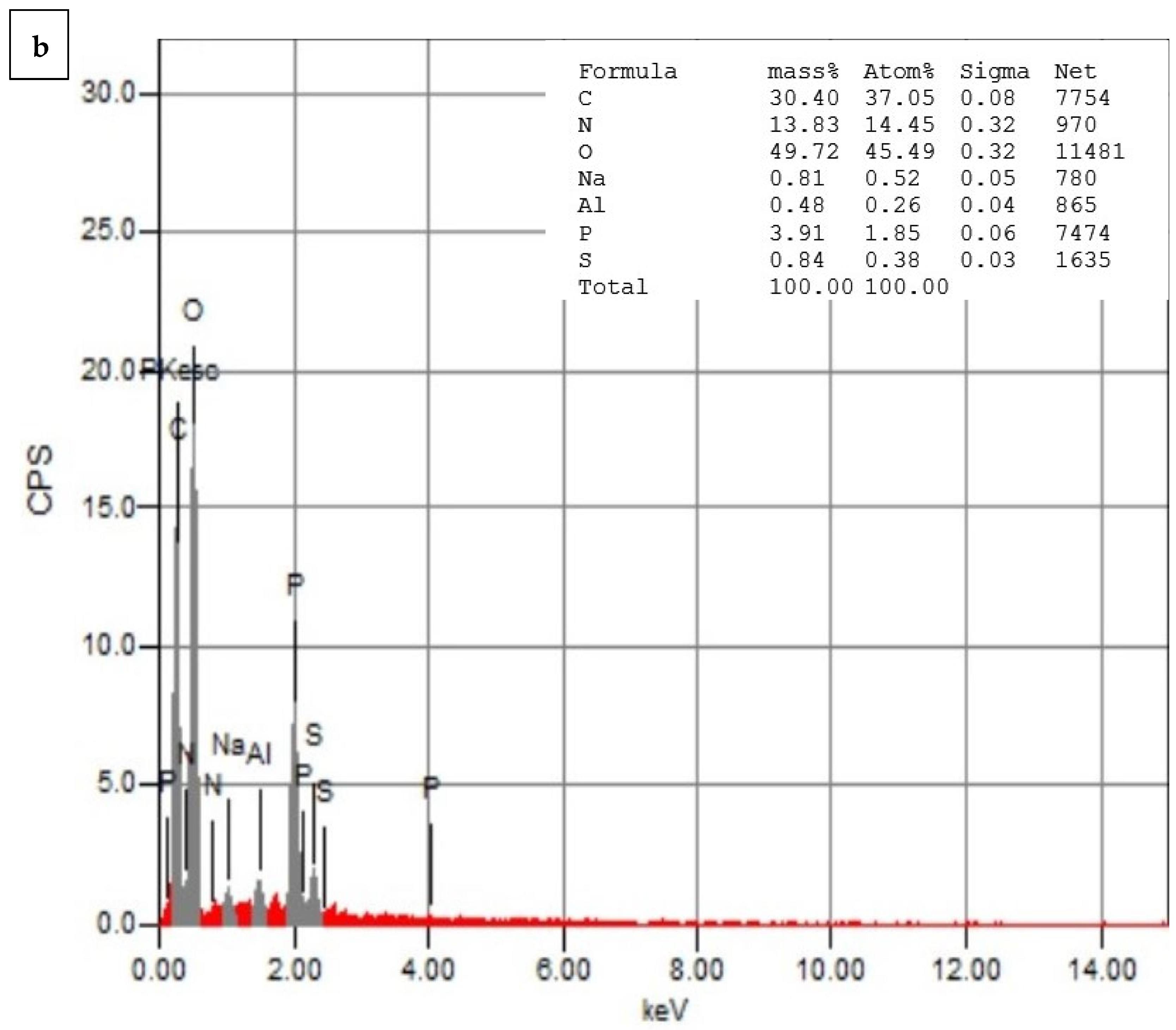
3.5. Energy-Dispersive Spectroscopy (EDS)
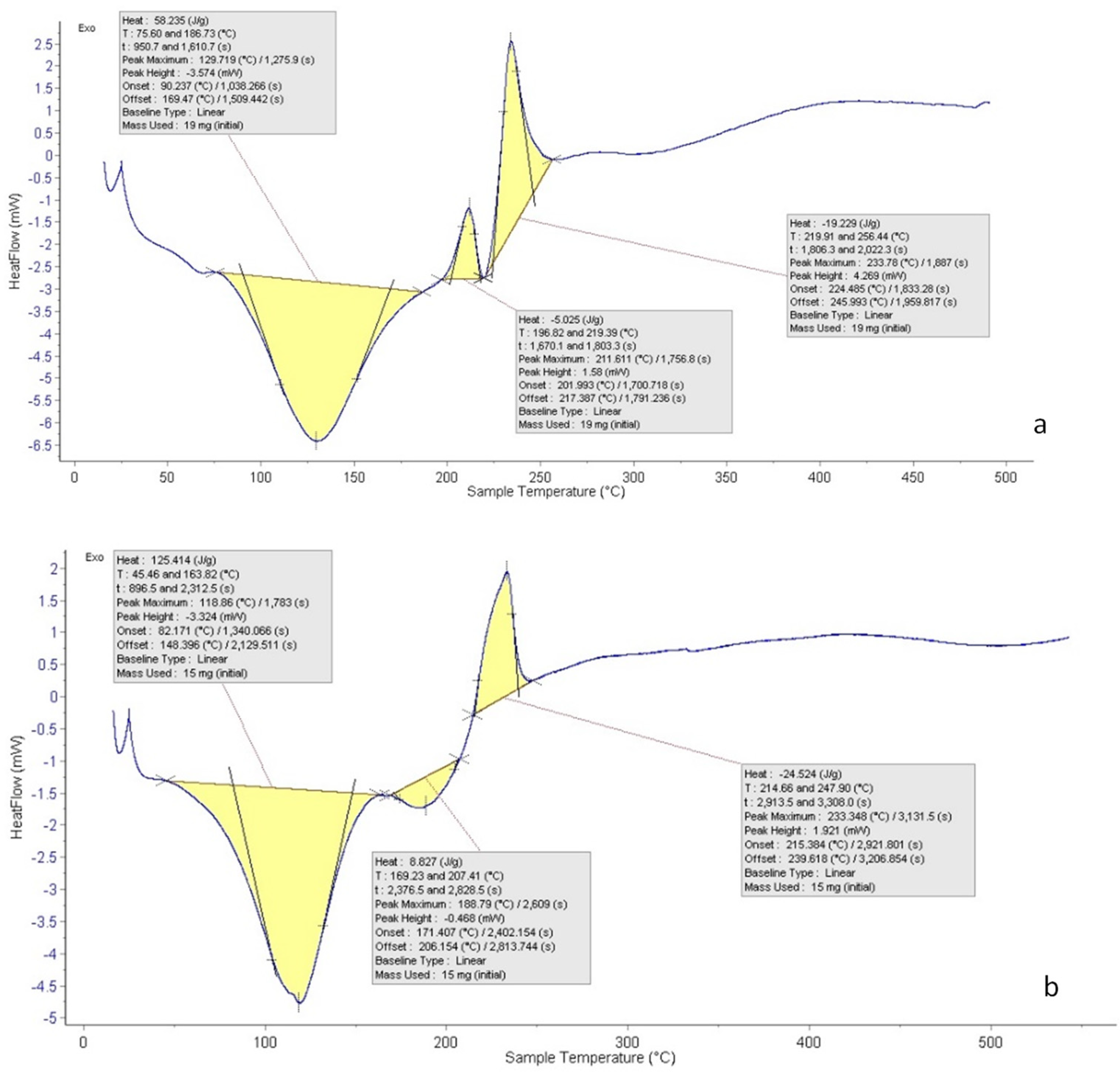
3.6. Differential Scanning Calorimetry (DSC)
3.7. Chromosomal Aberrations Observed in Bone Marrow Cells
3.8. Sperm Shape Abnormalities
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Barnes, J.L.; Zubair, M.; John, K.; Poirier, M.C.; Martin, F.L. Carcinogens and DNA damage. Biochem. Soc. Trans. 2018, 46, 1213–1224. [Google Scholar] [PubMed]
- Bray, F.; Laversanne, M.; Weiderpass, E.; Soerjomataram, I. The ever-increasing importance of cancer as a leading cause of premature death worldwide. Cancer 2021, 127, 3029–3030. [Google Scholar] [CrossRef] [PubMed]
- Basu, A.K. DNA damage, mutagenesis, and cancer. Int. J. Mol. Sci. 2018, 19, 970. [Google Scholar] [CrossRef] [PubMed]
- DeClercq, V.; Nearing, J.T.; Sweeney, E. Plant-based diets and cancer risk: What is the evidence? Curr. Nutr. Rep. 2022, 11, 354–369. [Google Scholar]
- Alshehri, K.M. Anticancer Plants Naturally Growing in Al-Baha Region, Saudi Arabia. Int. J. Pharm. Res. Allied Sci. 2020, 9, 92–101. [Google Scholar]
- Inocencio, C.; Rivera, D.; Obón, M.C.; Alcaraz, F.; Barreña, J.A. A systematic revision of capparis section Capparis (Capparaceae). Ann. Mo. Bot. Gard. 2006, 93, 122–149. [Google Scholar] [CrossRef]
- Eddouks, M.; Lemhadri, A.; Michel, J.B. Hypolipidemic activity of aqueous extract of Capparis spinosa L. in normal and diabetic rats. J. Ethnopharmacol. 2005, 98, 345–350. [Google Scholar] [CrossRef]
- Ghule, B.; Murugananthan, G.; Yeole, P. Analgesic and antipyretic effects of Capparis zeylanica leaves. Fitoterapia 2007, 78, 365–369. [Google Scholar] [CrossRef]
- Tlili, N.; Elfalleh, W.; Saadaoui, E.; Khaldi, A.; Triki, S.; Nasri, N. The caper (Capparis L.): Ethnopharmacology, phytochemical and pharmacological properties. Fitoterapia 2011, 82, 93–101. [Google Scholar]
- Peltonen, L.; Hirvonen, J. Drug nanocrystals–versatile option for formulation of poorly soluble materials. Int. J. Pharm. 2018, 537, 73–83. [Google Scholar]
- Aghili-Khorasani, H.; Makhzan, A. Source of Plant Research Institute for Islamic and complementary medicine Press. Iran Univ. Med. Sci. 2008, 729, 30. [Google Scholar]
- Soutter, W. Chitosan Nanoparticles—Properties and Applications, 2nd ed.; AZoNano: Sydney, Australia, 2013. [Google Scholar]
- Mohammed, M.A.; Syeda, J.T.; Wasan, K.M.; Wasan, E.K. An overview of chitosan nanoparticles and its application in non-parenteral drug delivery. Pharmaceutics 2017, 9, 53. [Google Scholar] [CrossRef]
- Sharifi-Rad, J.; Quispe, C.; Butnariu, M.; Rotariu, L.S.; Sytar, O.; Sestito, S.; Rapposelli, S.; Akram, M.; Iqbal, M.; Krishna, A.; et al. Chitosan nanoparticles as a promising tool in nanomedicine with particular emphasis on oncological treatment. Cancer Cell Int. 2021, 21, 318. [Google Scholar] [CrossRef]
- Jacinto, F.V.; Esteller, M. MGMT hypermethylation: A prognostic foe, a predictive friend. DNA Repair 2007, 6, 1155–1160. [Google Scholar] [CrossRef] [PubMed]
- Gerber, C.; Toelle, H.-G. What happened: The chemistry side of the incident with EMS contamination in Viracept tablets. Toxicol. Lett. 2009, 190, 248–253. [Google Scholar] [CrossRef] [PubMed]
- Flibotte, S.; Edgley, M.L.; Chaudhry, I.; Taylor, J.; Neil, S.E.; Rogula, A.; Zapf, R.; Hirst, M.; Butterfield, Y.; Jones, S.J.; et al. Whole-genome profiling of mutagenesis in Caenorhabditis elegans. Genetics 2010, 185, 431–441. [Google Scholar] [CrossRef] [PubMed]
- Anderson, P. Mutagenesis. Methods Cell Biol. 1995, 48, 31–58. [Google Scholar] [PubMed]
- Saito, T.; Ariizumi, T.; Okabe, Y.; Asamizu, E.; Hiwasa-Tanase, K.; Fukuda, N.; Mizoguchi, T.; Yamazaki, Y.; Aoki, K.; Ezura, H. TOMATOMA: A novel tomato mutant database distributing Micro-Tom mutant collections. Plant Cell Physiol. 2011, 52, 283–296. [Google Scholar] [CrossRef]
- Guruprasad, K.P.; Subramanian, A.; Singh, V.J.; Sharma, R.S.K.; Gopinath, P.M.; Sewram, V.; Satyamoorthy, K. Brahmarasayana protects against Ethyl methanesulfonate or Methyl methanesulfonate induced chromosomal aberrations in mouse bone marrow cells. BMC Complement. Altern. Med. 2012, 12, 113. [Google Scholar] [CrossRef]
- Kurniadi, A.S.; Irawati, F.; Putra, S.E.D.; Hardjo, P.H. Induction of Protocorm-Like Bodies (PLBs) Phalaenopsis spp. Hybrids Mutation through Ultraviolet Irradiation (UV254) and Ethyl Methane Sulfonate (EMS). Agriprima J. Appl. Agric. Sci. 2023, 7, 1–15. [Google Scholar] [CrossRef]
- Chaudhary, S.A. Flora of the Kingdom of Saudi Arabia, 2nd ed.; Ministry of Agriculture & Water: Riyadh, Saudi Arabia, 2001; pp. 455–457. [Google Scholar]
- Rautela, I.; Dheer, P.; Thapliyal, P.; Joshi, T.; Sharma, N.; Sharma, M.D. GC-MS analysis of plant leaf extract of Datura stramonium in different solvent system. Eur. J. Biomed. Pharm. Sci. 2018, 5, 236–245. [Google Scholar]
- Masarudin, M.J.; Cutts, S.M.; Evison, B.J.; Phillips, D.R.; Pigram, P.J. Factors determining the stability, size distribution, and cellular accumulation of small, monodisperse chitosan nanoparticles as candidate vectors for anticancer drug delivery: Application to the passive encapsulation of [14C]-doxorubicin. Nanotechnol. Sci. Appl. 2015, 8, 67–80. [Google Scholar] [CrossRef]
- Iglesias-Montes, M.L.; Soccio, M.; Siracusa, V.; Gazzano, M.; Lotti, N.; Cyras, V.P.; Manfredi, L.B. Chitin nanocomposite based on plasticized poly (lactic acid)/poly (3-hydroxybutyrate) (PLA/PHB) blends as fully biodegradable packaging materials. Polymers 2022, 14, 3177. [Google Scholar] [CrossRef] [PubMed]
- Moore, P.D.; Patlolla, A.K.; Tchounwou, P.B. Cytogenetic evaluation of malathion-induced toxicity in Sprague-Dawley rats. Mutat. Res./Genet. Toxicol. Environ. Mutagen. 2011, 725, 78–82. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Yosida, T.H.; Amano, K. Autosomal polymorphism in laboratory bred and wild Norway rats, Rattus norvegicus, found in Misima. Chromosoma 1965, 16, 658–667. [Google Scholar] [CrossRef] [PubMed]
- Rasgele, P.G. Abnormal sperm morphology in mouse germ cells after short-term exposures to acetamiprid, propineb, and their mixture. Arh. Hig. Rada Toksikol. 2014, 65, 47–55. [Google Scholar] [CrossRef]
- Tokarek, K.; Hueso, J.L.; Kuśtrowski, P.; Stochel, G.; Kyzioł, A. Green synthesis of chitosan-stabilized copper nanoparticles. Eur. J. Inorg. Chem. 2013, 2013, 4940–4947. [Google Scholar] [CrossRef]
- Wang, B.; Song, Y.; Zhang, X.; Chen, K.; Liu, M.; Hu, X.; He, L.; Huang, Q. Polymer derived SiBCN (O) ceramics with tunable element content. Ceram. Int. 2022, 48, 10280–10287. [Google Scholar] [CrossRef]
- Talari, A.C.S.; Martinez, M.A.G.; Movasaghi, Z.; Rehman, S.; Rehman, I.U. Advances in Fourier transform infrared (FTIR) spectroscopy of biological tissues. Appl. Spectrosc. Rev. 2017, 52, 456–506. [Google Scholar] [CrossRef]
- Umamaheswari, S.; Murali, M. FTIR spectroscopic study of fungal degradation of poly (ethylene terephthalate) and polystyrene foam. Chem. Eng. 2013, 64, 159. [Google Scholar]
- Trivedi, M.; Branton, A.; Trivedi, D.; Shettigar, H.; Bairwa, K.; Jana, S. Fourier transform infrared and ultraviolet-visible spectroscopic characterization of biofield treated salicylic acid and sparfloxacin. Nat. Prod. Chem. Res. 2015, 3, 1000186. [Google Scholar] [CrossRef]
- Shi, J.; Xing, D.; Lia, J. FTIR studies of the changes in wood chemistry from wood forming tissue under inclined treatment. Energy Procedia 2012, 16, 758–762. [Google Scholar] [CrossRef]
- Bianchi, C.L.; Stucchi, M.; Pirola, C.; Cerrato, G.; Sacchi, B.; Vitali, S.; Di Michele, A.; Capucci, V. Micro-sized TiO2 catalyst in powder form and as coating on porcelain grès tile for the photodegradation of phenol as model pollutant for water phase. Adv. Mater. Sci. 2017, 2, 1–6. [Google Scholar] [CrossRef][Green Version]
- Sahoo, S.; Chakraborti, C.K.; Mishra, S.C.; Nanda, U.; Naik, S. FTIR and XRD investigations of some fluoroquinolones. Int. J. Pharm. Pharm. Sci. 2011, 3, 165–170. [Google Scholar]
- Volkov, D.S.; Rogova, O.B.; Proskurnin, M.A. Organic matter and mineral composition of silicate soils: FTIR comparison study by photoacoustic, diffuse reflectance, and attenuated total reflection modalities. Agronomy 2021, 11, 1879. [Google Scholar] [CrossRef]
- Dutta, S.; Ray, S.; Nagarajan, K. Glutamic acid as anticancer agent: An overview. Saudi Pharm. J. 2013, 21, 337–343. [Google Scholar] [CrossRef] [PubMed]
- Aslam, S.; Wang, F.; Ahmad, M.; Zahoor, A.F.; Mansha, A.; Rasul, A.; Fu, L. Benzothiazine based acetohydrazides and acetamides as anticancer agents. Pak. J. Pharm. Sci. 2019, 32, 2795–2800. [Google Scholar]
- Maduabuchi, E.K.; Tobechukwu, O.P. Advanced phytochemistry and chemo-metric profiling of the bioactive medicinal components of n-hexane seed extract of Xylopia aethiopica using FTIR and GC-MS techniques. GSC Biol. Pharm. Sci. 2023, 22, 247–256. [Google Scholar] [CrossRef]
- Jan, N.; Andrabi, K.I.; John, R. Calendula officinalis—An important medicinal plant with potential biological properties. Proc. Indian Natl. Sci. Acad. 2017, 83, 769–787. [Google Scholar]
- Qanash, H.; Yahya, R.; Bakri, M.M.; Bazaid, A.S.; Qanash, S.; Shater, A.F.; TM, A. Anticancer, antioxidant, antiviral and antimicrobial activities of Kei Apple (Dovyalis caffra) fruit. Sci. Rep. 2022, 12, 5914. [Google Scholar] [CrossRef]
- Vijayabaskar, G.; Elango, V. Determination of phytocompounds in Withania somnifera and Smilax China using GC-MS technique. J. Pharmacogn. Phytochem. 2018, 7, 554–557. [Google Scholar]
- Sumathi, R. GC-MS Analysis of Methanol Extracts of Flowers of Allamanda neriifolia Hook. Indian J. Curr. Res. 2015, 2, 49–53. [Google Scholar]
- Mujeeb, F.; Bajpai, P.; Pathak, N. Phytochemical evaluation, antimicrobial activity, and determination of bioactive components from leaves of Aegle marmelos. BioMed Res. Int. 2014, 2014, 497606. [Google Scholar] [CrossRef] [PubMed]
- Awa, E.P.; Ibrahim, S.; Ameh, D.A. GC/MS analysis and antimicrobial activity of diethyl ether fraction of methanolic extract from the stem bark of Annona senegalensis Pers. Int. J. Pharm. Sci. Res. 2012, 3, 4213–4218. [Google Scholar]
- Belakhdar, G.; Benjouad, A.; Abdennebi, E.H. Determination of some bioactive chemical constituents from Thesium humile Vahl. J. Mater. Environ. Sci. 2015, 6, 2778–2783. [Google Scholar]
- Akpuaka, A.; Ekwenchi, M.M.; Dashak, D.A.; Dildar, A. Biological activities of characterized isolates of n-hexane extract of Azadirachta indica A. Juss (Neem) leaves. Nat. Sci. 2013, 11, 141–147. [Google Scholar]
- Brooks, J.D.; Milne, G.L.; Yin, H.; Sanchez, S.C.; Porter, N.A.; Morrow, J.D. Formation of highly reactive cyclopentenone isoprostane compounds (A3/J3-isoprostanes) in vivo from eicosapentaenoic acid. J. Biol. Chem. 2008, 283, 12043–12055. [Google Scholar] [CrossRef]
- Jiang, J.; Oberdörster, G.; Biswas, P. Characterization of size, surface charge, and agglomeration state of nanoparticle dispersions for toxicological studies. J. Nanoparticle Res. 2009, 11, 77–89. [Google Scholar] [CrossRef]
- Ali, S.W.; Joshi, M.; Rajendran, S. Synthesis and characterization of chitosan nanoparticles with enhanced antimicrobial activity. Int. J. Nanosci. 2011, 10, 979–984. [Google Scholar] [CrossRef]
- Santo Pereira, A.E.; Silva, P.M.; Oliveira, J.L.; Oliveira, H.C.; Fraceto, L.F. Chitosan nanoparticles as carrier systems for the plant growth hormone gibberellic acid. Colloids Surf. B. Biointerfaces 2017, 150, 141–152. [Google Scholar] [CrossRef]
- Hejjaji, E.M.; Smith, A.M.; Morris, G.A. Evaluation of the mucoadhesive properties of chitosan nanoparticles prepared using different chitosan to tripolyphosphate (CS: TPP) ratios. Int. J. Biol. Macromol. 2018, 120, 1610–1617. [Google Scholar] [CrossRef]
- Barrera-Necha, L.L.; Correa-Pacheco, Z.N.; Bautista-Baños, S.; Hernández-López, M.; Jiménez, J.E.M.; Mejía, A.F.M. Synthesis and characterization of chitosan nanoparticles loaded botanical extracts with antifungal activity on Colletotrichum gloeosporioides and Alternaria species. Adv. Microbiol. 2018, 8, 286. [Google Scholar] [CrossRef]
- Guo, H.; Li, F.; Qiu, H.; Liu, J.; Qin, S.; Hou, Y.; Wang, C. Preparation and characterization of chitosan nanoparticles for chemotherapy of melanoma through enhancing tumor penetration. Front. Pharmacol. 2020, 11, 317. [Google Scholar] [CrossRef] [PubMed]
- Elisa, F.R.; Taís, T.; de Barros, A.; Assis, O.B.G.; Junior, A.C.; Quiles, A.; Hernando, I.; Nicoletti, V.R. Chitosan and crosslinked chitosan nanoparticles: Synthesis, characterization and their role as Pickering emulsifiers. Carbohydr. Polym. 2020, 250, 116878. [Google Scholar]
- Müller, R.H.; Jacobs, C.; Kayser, O. Nanosuspensions as particulate drug formulations in therapy: Rationale for development and what we can expect for the future. Adv. Drug Deliv. Rev. 2001, 47, 3–19. [Google Scholar] [CrossRef]
- Kumar, A.; Dixit, C.K. Methods for Characterization of Nanoparticles. In Advances in Nanomedicine for the Delivery of Therapeutic Nucleic Acids; Elsevier: Amsterdam, The Netherlands, 2017; pp. 43–58. [Google Scholar]
- Huang, J.; Li, Q.; Sun, D.; Lu, Y.; Su, Y.; Yang, X.; Wang, H.; Wang, Y.; Shao, W.; He, N.; et al. Biosynthesis of silver and gold nanoparticles by novel sundried Cinnamomum camphora leaf. Nanotechnology 2007, 18, 105104. [Google Scholar] [CrossRef]
- Chandra Hembram, K.; Prabha, S.; Chandra, R.; Ahmed, B.; Nimesh, S. Advances in preparation and characterization of chitosan nanoparticles for therapeutics. Artif. Cells Nanomed. Biotechnol. 2016, 44, 305–314. [Google Scholar] [CrossRef]
- Rampino, A.; Borgogna, M.; Blasi, P.; Bellich, B.; Cesàro, A. Chitosan nanoparticles: Preparation, size evolution and stability. Int. J. Pharm. 2013, 455, 219–228. [Google Scholar] [CrossRef]
- Gokce, Y.; Cengiz, B.; Yildiz, N.; Calimli, A.; Aktas, Z. Ultrasonication of chitosan nanoparticle suspension: Influence on particle size. Colloids Surf. A Physicochem. Eng. Asp. 2014, 462, 75–81. [Google Scholar] [CrossRef]
- Khoerunnisa, F.; Nurhayati, M.; Dara, F.; Rizki, R.; Nasir, M.; Aziz, H.A.; Opaprakasit, P. Physicochemical properties of TPP-crosslinked chitosan nanoparticles as potential antibacterial agents. Fibers Polym. 2021, 22, 2954–2964. [Google Scholar] [CrossRef]
- Koukaras, E.N.; Papadimitriou, S.A.; Bikiaris, D.N.; Froudakis, G.E. Insight on the formation of chitosan nanoparticles through ionotropic gelation with tripolyphosphate. Mol. Pharm. 2012, 9, 2856–2862. [Google Scholar] [CrossRef]
- Yousefpour, P.; Atyabi, F.; Vasheghani-Farahani, E.; Movahedi, A.M.; Dinarvand, R. Targeted delivery of doxorubicin-utilizing chitosan nanoparticles surface-functionalized with anti-Her2 trastuzumab. Int. J. Nanomed. 2011, 6, 1977–1990. [Google Scholar] [CrossRef] [PubMed]
- Corazzari, I.; Nisticò, R.; Turci, F.; Faga, M.G.; Franzoso, F.; Tabasso, S.; Magnacca, G. Advanced physicochemical characterization of chitosan by means of TGA coupled on-line with FTIR and GCMS: Thermal degradation and water adsorption capacity. Polym. Degrad. Stab. 2015, 112, 1–9. [Google Scholar] [CrossRef]
- Kittur, F.; Prashanth, K.H.; Sankar, K.U.; Tharanathan, R. Characterization of chitin, chitosan and their carboxymethyl derivatives by differential scanning calorimetry. Carbohydr. Polym. 2002, 49, 185–193. [Google Scholar] [CrossRef]
- Norizadeh Tazehkand, M.; Topaktas, M.; Yilmaz, M.B.; Hajipour, O.; Valipour, E. Delineating the antigenotoxic and anticytotoxic potentials of 4-methylimidazole against ethyl methanesulfonate toxicity in bone marrow cell of Swiss albino mice. Bratisl. Med. J. 2016, 9248, 290–294. [Google Scholar] [CrossRef]
- Müller, L.; Gocke, E.; Lavé, T.; Pfister, T. Ethyl methanesulfonate toxicity in Viracept—A comprehensive human risk assessment based on threshold data for genotoxicity. Toxicol. Lett. 2009, 190, 317–329. [Google Scholar] [CrossRef] [PubMed]
- Zhu, X.; Huo, J.; Zeng, Z.; Liu, Y.; Li, R.; Chen, Y.; Zhang, L.; Chen, J. Determination of potential thresholds for N-ethyl-N-nitrosourea and ethyl methanesulfonate based on a multi-endpoint genotoxicity assessment platform in rats. Environ. Sci. Pollut. Res. 2022, 29, 85128–85142. [Google Scholar] [CrossRef]
- Yan, W.; Deng, X.W.; Yang, C.; Tang, X. The genome-wide EMS mutagenesis bias correlates with sequence context and chromatin structure in rice. Front. Plant Sci. 2021, 12, 579675. [Google Scholar] [CrossRef]
- Mohamed, N.R.; Badr, T.M.; Elnagar, M.R. Efficiency of curcumin and chitosan nanoparticles against toxicity of potassium dichromate in male mice. Int. J. Pharm. Pharm. Sci. 2021, 13, 22159. [Google Scholar] [CrossRef]
- Hamdan, D.I.; Tawfeek, N.; El-Shiekh, R.A.; Khalil, H.M.; Mahmoud, M.Y.; Bakr, A.F.; Zaafar, D.; Farrag, N.; Wink, M.; El-Shazly, A.M. Salix subserrata bark extract-loaded chitosan nanoparticles attenuate neurotoxicity induced by sodium arsenate in rats in relation with HPLC–PDA-ESI–MS/MS profile. AAPS PharmSciTech 2022, 24, 15. [Google Scholar] [CrossRef]
- Hamouda, R.A.; Salman, A.S.; Alharbi, A.A.; Alhasani, R.H.; Elshamy, M.M. Assessment of the Antigenotoxic Effects of Alginate and ZnO/Alginate–Nanocomposites Extracted from Brown Alga Fucus vesiculosus in Mice. Polymers 2021, 13, 3839. [Google Scholar] [CrossRef] [PubMed]
- Sinitsky, M.Y.; Kutikhin, A.G.; Tsepokina, A.V.; Shishkova, D.K.; Asanov, M.A.; Yuzhalin, A.E.; Minina, V.I.; Ponasenko, A.V. Mitomycin C induced genotoxic stress in endothelial cells is associated with differential expression of proinflammatory cytokines. Mutat. Res./Genet. Toxicol. Environ. Mutagen. 2020, 858, 503252. [Google Scholar] [CrossRef] [PubMed]
- Sharifi-Rad, M.; Anil Kumar, N.V.; Zucca, P.; Varoni, E.M.; Dini, L.; Panzarini, E.; Rajkovic, J.; Tsouh Fokou, P.V.; Azzini, E.; Peluso, I.; et al. Lifestyle, oxidative stress, and antioxidants: Back and forth in the pathophysiology of chronic diseases. Front. Physiol. 2020, 11, 694. [Google Scholar] [CrossRef]
- Moharram, B.A.; Al-Mahbashi, H.M.; Saif-Ali, R.; AliAqlan, F. Phytochemical, anti-inflammatory, antioxidant, cytotoxic and antibacterial study of Capparis cartilaginea decne from Yemen. Int. J. Pharm. Pharm. Sci. 2018, 10, 38. [Google Scholar] [CrossRef]
- He, J.R.; Zhu, J.J.; Yin, S.W.; Yang, X.Q. Bioaccessibility and intracellular antioxidant activity of phloretin embodied by gliadin/sodium carboxymethyl cellulose nanoparticles. Food Hydrocoll. 2022, 122, 107076. [Google Scholar] [CrossRef]
- Hu, Y.; Lin, Q.; Zhao, H.; Li, X.; Sang, S.; McClements, D.J.; Long, J.; Jin, Z.; Wang, J.; Qiu, C. Bioaccessibility and bioavailability of phytochemicals: Influencing factors, improvements, and evaluations. Food Hydrocoll. 2022, 108165. [Google Scholar] [CrossRef]
- Abdel-Wahhab, M.A.; Aljawish, A.; El-Nekeety, A.A.; Abdel-Aziem, S.H.; Hassan, N.S. Chitosan nanoparticles plus quercetin suppress the oxidative stress, modulate DNA fragmentation and gene expression in the kidney of rats fed ochratoxin A-contaminated diet. Food Chem. Toxicol. 2017, 99, 209–221. [Google Scholar] [CrossRef] [PubMed]
| Groups | Treatment and Doses | Treatment Day(s) |
|---|---|---|
| Control | Control (negative) | 1 |
| EMS: | intraperitoneal single injection with 240 mg/kg b.w | |
| CNPs: | oral administration with 350 mg/kg b.w. of chitosan nanoparticle | |
| HD of CNPs/Cc: | oral administration with 700 mg/kg b.w. of CNPs loaded with C. cartilaginea | |
| LD of CNPs/Cc: | oral administration with 350 mg/kg b.w. of CNPs loaded with C. cartilaginea | |
| CNPs + EMS | CNPs (350 mg/kg b.w.) + EMS (single injection with 240 mg/kg b.w) | |
| HD of CNPs/Cc +EMS | (single injection with 240 mg/kg b.w) | |
| LD of CNPs/Cc + EMS | (single injection with 240 mg/kg b.w) | |
| Control | Control (negative) | 7 |
| EMS: | intraperitoneal single injection with 240 mg/kg b.w, 24 h before the experiment. | |
| CNPs: | oral administration with 350 mg/kg b.w. of chitosan nanoparticle | |
| HD of CNPs/Cc: | oral administration with 700 mg/kg b.w. of CNPs loaded with C. cartilaginea | |
| LD of CNPs/Cc: | oral administration with 350 mg/kg b.w. of CNPs loaded with C. cartilaginea | |
| CNPs + EMS | CNPs (350 mg/kg b.w.) + EMS (single injection with 240 mg/kg b.w) | |
| HD of CNPs/Cc +EMS | (single injection with 240 mg/kg b.w) | |
| LD of CNPs/Cc + EMS | (single injection with 240 mg/kg b.w) |
| Wavenumber cm−1 | Plant | CNP/Cc | CNPs | Functional Groups | Ref. |
|---|---|---|---|---|---|
| 3346.88 | Nd | D | Nd | Hydroxyl groups | [29] |
| 3335.45 | Nd | Nd | D | Stretching N-H asymmetric | [30] |
| 3231.22 | D | Nd | Nd | O-H bond stretching | [31] |
| 2132.35 | Nd | D | −6 | Si–H stretching | [29] |
| 1636.33 | −38 | D | D | CHO stretching of carbonyl group | [30] |
| 1558.81 | Nd | Nd | D | C–C stretch aromatic rings (phenolic) | [32] |
| 1540.41 | −22 | D | D | Amide II | [30] |
| 1418.53 | −21 | D | +39 | Deformation C–H | [30] |
| 1267.27 | + | Nd | Nd | C-O stretching | [33] |
| 1077.43 | Nd | D | +19 | C=C bond | [34] |
| 1011.96 | +17 | D | Nd | C-F groups | [28] |
| 950.72 | Nd | D | Nd | Amines | [35] |
| 614.09 | D | Nd | +9 | C–S stretch | [36] |
| Biological Activity | Molecular Weight | Molecular Formula | Chemical Structure | Area % | Compound Name | RT |
|---|---|---|---|---|---|---|
| Antimicrobial, Anti-inflammatory, Antioxidant [37] | 152 | C9H12O2 | 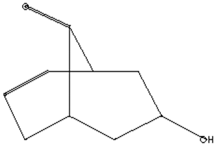 | 4.25 | 7-Hydroxy-bicyclo[3.3.1]n on-2-en-9-one | 5.53 |
| Antimicrobial, Anti-inflammatory, Antioxidant [37] | 152 | C9H12O2 | 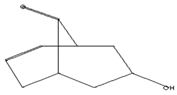 | 6.15 | 7-HYDROXY-BICYCLO[3.3.1]N ON-2-EN-9-ONE | 5.58 |
| Anticancer agent [38] | 129 | C5H7NO3 | 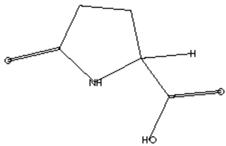 | 9.03 | D-Pyroglutamic acid | 6.50 |
| Acetohydrazides and acetamides possessed anticancer agents [39] | 196 | C10H16N2O2 | 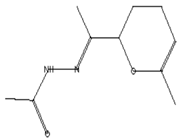 | 2.05 | Acethydrazide, n2-[1-(2,3-dihydro-6-methyl pyran-2-yl)ethylideno]- | 7.14 |
| Analgesic, digestive, and wound healing [40] Agonist activity at human TRPA1 channel expressed in HEK293 cell [41] | 150 | C10H14O | 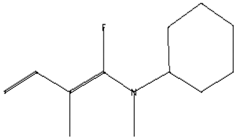 | 2.00 | 3,5-Heptadienal, 2-ethylidene-6-methyl- | 13.36 |
| The main compounds are found in Kei Apple fruits and have antioxidant and anticancer activities [42] | 157 | C2H7NO3S2 | 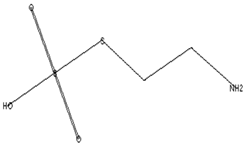 | 2.98 | 2-AMINOETHANETHIOL HYDROGEN SULFATE (ESTER) | 24.73 |
| Hepatoprotective Antiandrogenic, Antihistaminic, Anticoronary, Insectifuge, Anticancer [43] | 298 | C18H31ClO | 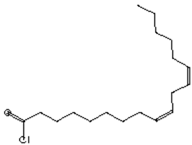 | 2.76 | 9,12-Octadecadienoyl chloride, (Z,Z)- | 25.07 |
| Anti-oxidant, decreases blood cholesterol, anti-inflammatory [44] | 270 | C17H34O2 |  | 11.79 | Hexadecanoic acid, methyl ester | 26.13 |
| Antimicrobial [45] | 284 | C18H36O2 |  | 3.09 | Hexadecanoic acid, ethyl ester | 27.39 |
| Antibacterial [46] | 282 | C18H34O2 | 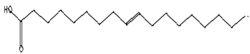 | 3.21 | 9-Octadecenoic acid (z)-, hexadecanoic acid | 27.84 |
| Antioxidant, anticancer [44,47] | 296 | C19H36O2 | 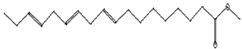 | 2.09 | 9-Octadecenoic acid (Z)-, methyl ester | 29.31 |
| Antimicrobial [48] | 298 | C19H38O2 | 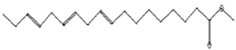 | 3.24 | Octadecanoic acid, methyl ester | 29.82 |
| Antibacterial, Anti-inflammatory, cancer preventive, hepatoprotective, nematicide, insectifuge [45] | 292 | C19H32O2 | 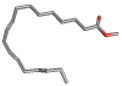 | 14.99 | 9,12,15-Octadecatrienoic acid, methyl ester | 30.54 |
| Antibacterial [46] | 282 | C18H34O2 | 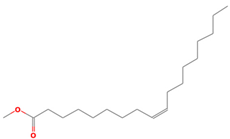 | 2.91 | 9-OCTADECENOIC ACID (Z)- 2-AMINOETHANETHIOL | 30.96 |
| Anti-atherosclerotic, anti-inflammatory, and anti-proliferative effects [49] | 302 | C20H30O2 |  21,736,059.79 | 3.22 | cis-5,8,11,14,17-Eicosapentaenoic acid | 43.76 |
| Treatment | No. of Metaphases with Aberrations | Chromosomal Aberrations | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Gap | Frag. and/or Break | Gap + (Frag. or Break) | Deletion | Ring | Total No. | Excluding Gaps Mean ± S.E. | Including Gaps Mean ± S.E. | Inhibition % | |
| One-day treatments | |||||||||
| Control | 2 | 4 | 0 | 0 | 0 | 6 | 0.8 ± 0.4 | 1.2 ± 0.4 | |
| EMS | 20 | 62 | 28 | 10 | 2 | 122 | 20.4 ± 0.7 a | 24.4 ± 0.4 a | |
| CNPs | 10 | 9 | 5 | 0 | 0 | 24 | 3.6 ± 0.48 | 4.8 ± 0.4 | |
| HD of CNPs/Cc | 12 | 5 | 2 | 2 | 0 | 21 | 4.6 ± 0.2 | 4.2 ± 0.2 | |
| LD of CNPs/Cc | 13 | 3 | 6 | 1 | 0 | 23 | 4.2 ± 0.3 | 4.106 ± 0.4 | |
| CNPs + EMS | 14 | 50 | 33 | 11 | 0 | 108 | 18.8 ± 0.37 | 21.6 ± 0.7 a | 11.4 |
| HD of CNPs/Cc + EMS | 11 | 61 | 15 | 9 | 1 | 97 | 17.2 ± 0.37 | 19.4 ± 0.6 a | 20.49 |
| LD of CNPs/Cc + EMS | 22 | 51 | 33 | 0 | 0 | 106 | 16.8 ± 0.6 a | 21.2 ± 0.6 a | 13.1 |
| Seven-day treatments | |||||||||
| Control | 2 | 4 | 0 | 0 | 0 | 6 | 0.8 ± 0.4 | 1.2 ± 0.1 | |
| EMS | 20 | 62 | 28 | 10 | 2 | 122 | 20.4 ± 0.7 a | 24.4 ± 0.4 a | |
| CNPs | 12 | 3 | 5 | 2 | 0 | 23 | 2.2 ± 0.2 | 4.6 ± 0.6 | |
| HD of CNPs/Cc | 12 | 5 | 3 | 0 | 0 | 20 | 1.6 ± 0.4 | 4.0 ± 0.1 | |
| LD of CNPs/Cc | 12 | 10 | 0 | 0 | 0 | 22 | 2.0 ± 0.4 | 4.4 ± 0.4 | |
| CNPs + EMS | 20 | 39 | 11 | 12 | 0 | 82 | 12.4 ± 0.6 ab | 16.4 ± 0.7 ab | 32.7 |
| HD of CNPs/Cc + EMS | 12 | 27 | 4 | 7 | 0 | 50 | 7.6 ± 0.5 ab | 10 ± 0.8 ab | 59.01 |
| LD of CNPs/Cc + EMS | 18 | 24 | 32 | 4 | 0 | 78 | 12 ± 0.7 ab | 15.6 ± 0.7 ab | 36.06 |
| Treatment and Doses (mg/kg b.wt.) | Sperm No. | No. of Sperm with Abnormalities in | Abnormal Sperm No. | Abnormal Sperm Mean % ± S.E. | Inhibition % | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Head | Tail | ||||||||||
| Amorphous | Tringle | Without Hook | Small | Big | Coiled | ||||||
| Control | 5000 | 29 | 36 | 28 | 0 | 0 | 6 | 99 | 1.98 ± 0.1 | ||
| EMS | 5000 | 259 | 183 | 47 | 3 | 1 | 229 | 722 | 14.4 ± 1.78 a | ||
| CNPs | 5000 | 31 | 25 | 6 | 0 | 0 | 24 | 86 | 1.72 ± 0.24 | ||
| HD of CNPs/Cc | 5000 | 25 | 32 | 5 | 0 | 0 | 21 | 83 | 1.66 ± 0.2 | ||
| LD of CNPs/Cc | 5000 | 32 | 21 | 7 | 0 | 0 | 35 | 95 | 1.9 ± 0.9 | ||
| CNPs + EMS | 5000 | 204 | 109 | 19 | 2 | 0 | 167 | 501 | 10.02 ± 0.98 ab | 30.6 | |
| HD of CNPs/Cc + EMS | 5000 | 158 | 93 | 23 | 7 | 2 | 80 | 363 | 7.26 ± 0.56 ab | 49.7 | |
| LD of CNPs/Cc + EMS | 5000 | 106 | 130 | 30 | 4 | 1 | 214 | 485 | 9.7 ± 0.22 ab | 32.8 | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Salman, A.S.; Alkhatib, S.N.; Ahmed, F.M.; Hamouda, R.A. Chitosan Nanoparticles Loaded with Capparis cartilaginea Decne Extract: Insights into Characterization and Antigenotoxicity In Vivo. Pharmaceutics 2023, 15, 2551. https://doi.org/10.3390/pharmaceutics15112551
Salman AS, Alkhatib SN, Ahmed FM, Hamouda RA. Chitosan Nanoparticles Loaded with Capparis cartilaginea Decne Extract: Insights into Characterization and Antigenotoxicity In Vivo. Pharmaceutics. 2023; 15(11):2551. https://doi.org/10.3390/pharmaceutics15112551
Chicago/Turabian StyleSalman, Asmaa S., Shaza N. Alkhatib, Fatimah M. Ahmed, and Ragaa A. Hamouda. 2023. "Chitosan Nanoparticles Loaded with Capparis cartilaginea Decne Extract: Insights into Characterization and Antigenotoxicity In Vivo" Pharmaceutics 15, no. 11: 2551. https://doi.org/10.3390/pharmaceutics15112551
APA StyleSalman, A. S., Alkhatib, S. N., Ahmed, F. M., & Hamouda, R. A. (2023). Chitosan Nanoparticles Loaded with Capparis cartilaginea Decne Extract: Insights into Characterization and Antigenotoxicity In Vivo. Pharmaceutics, 15(11), 2551. https://doi.org/10.3390/pharmaceutics15112551






