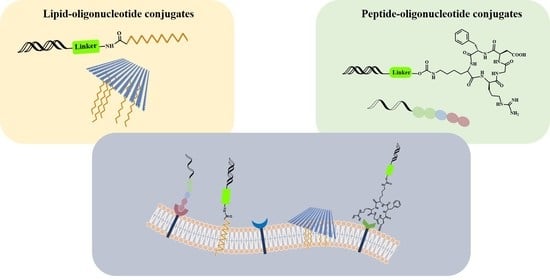
Lipid and Peptide-Oligonucleotide Conjugates for Therapeutic Purposes: From Simple Hybrids to Complex Multifunctional Assemblies
Abstract
1. Introduction
2. Results and Discussion
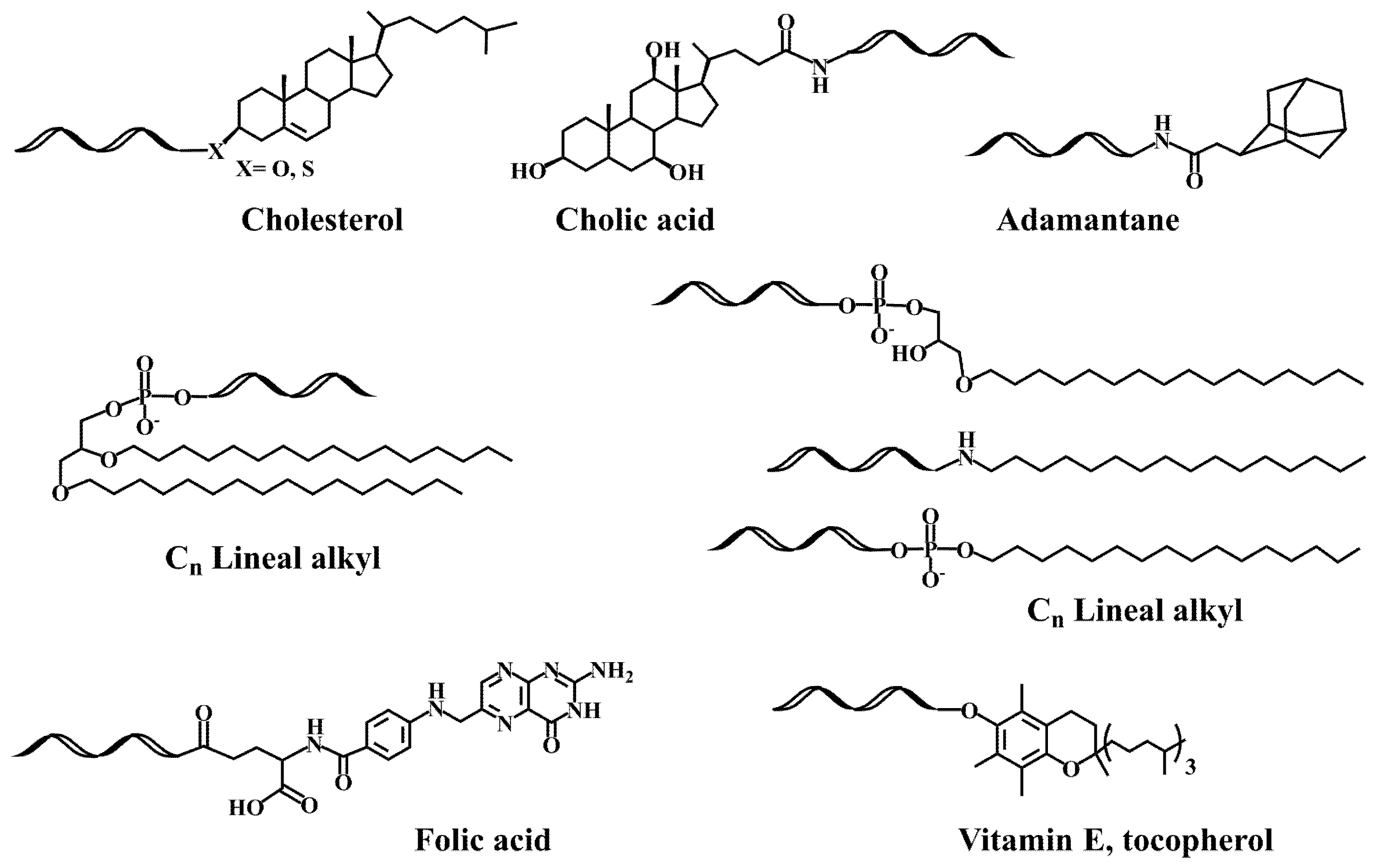
2.1. Early Developments in the Synthesis of Lipid-Oligonucleotide Conjugates
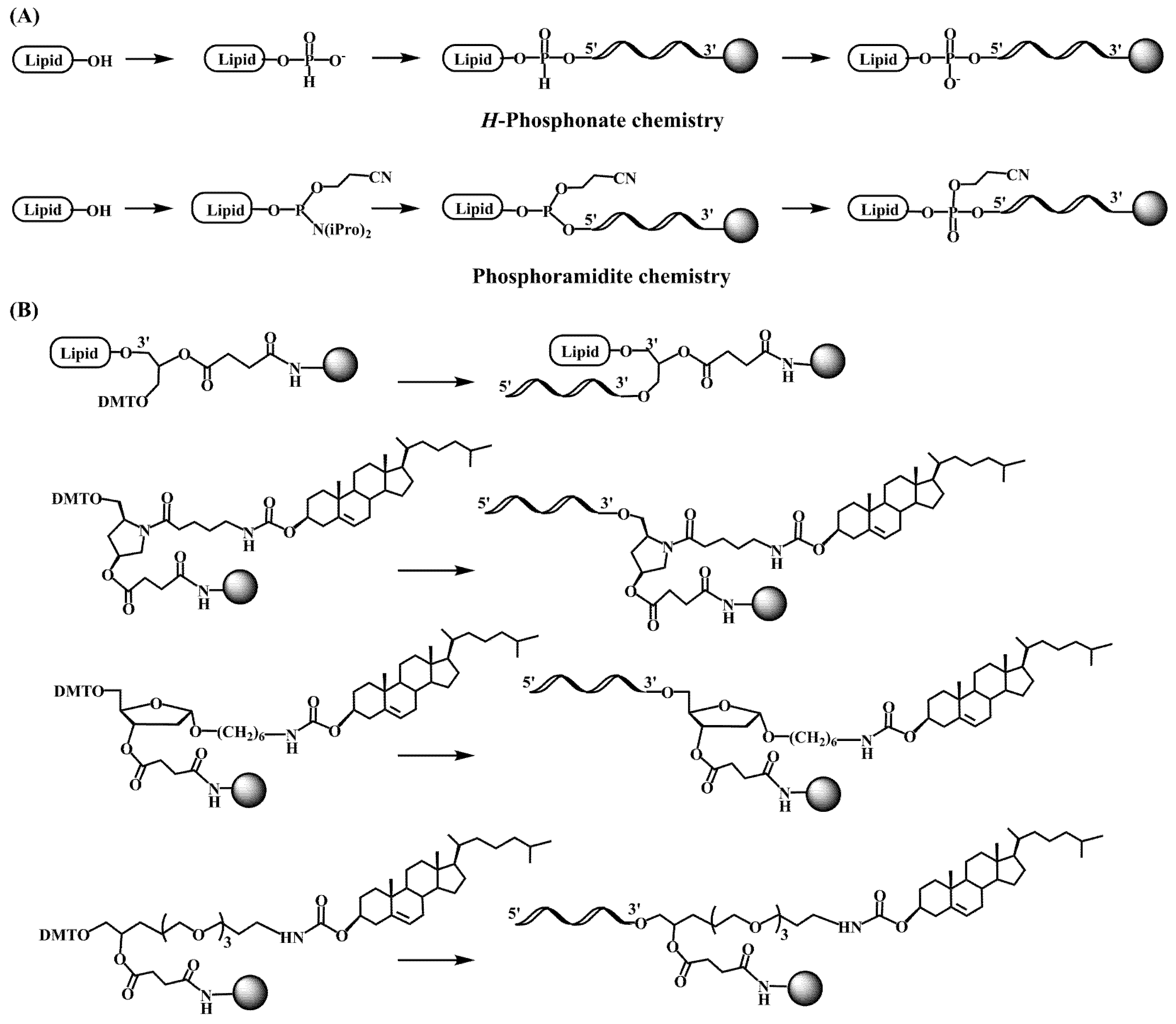
2.2. Lipid-Oligonucleotide Conjugates and the Development of RNA-Based Therapeutics
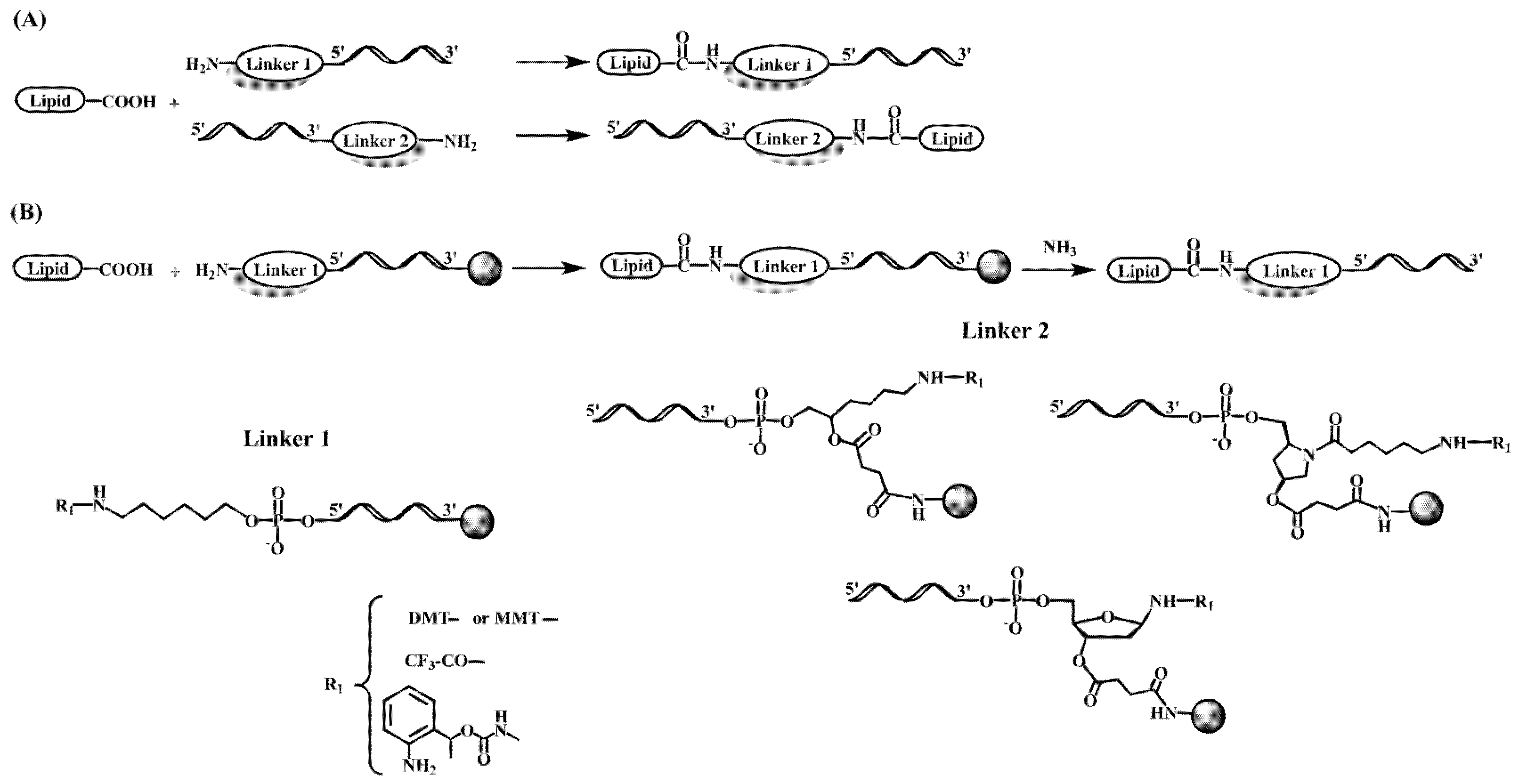
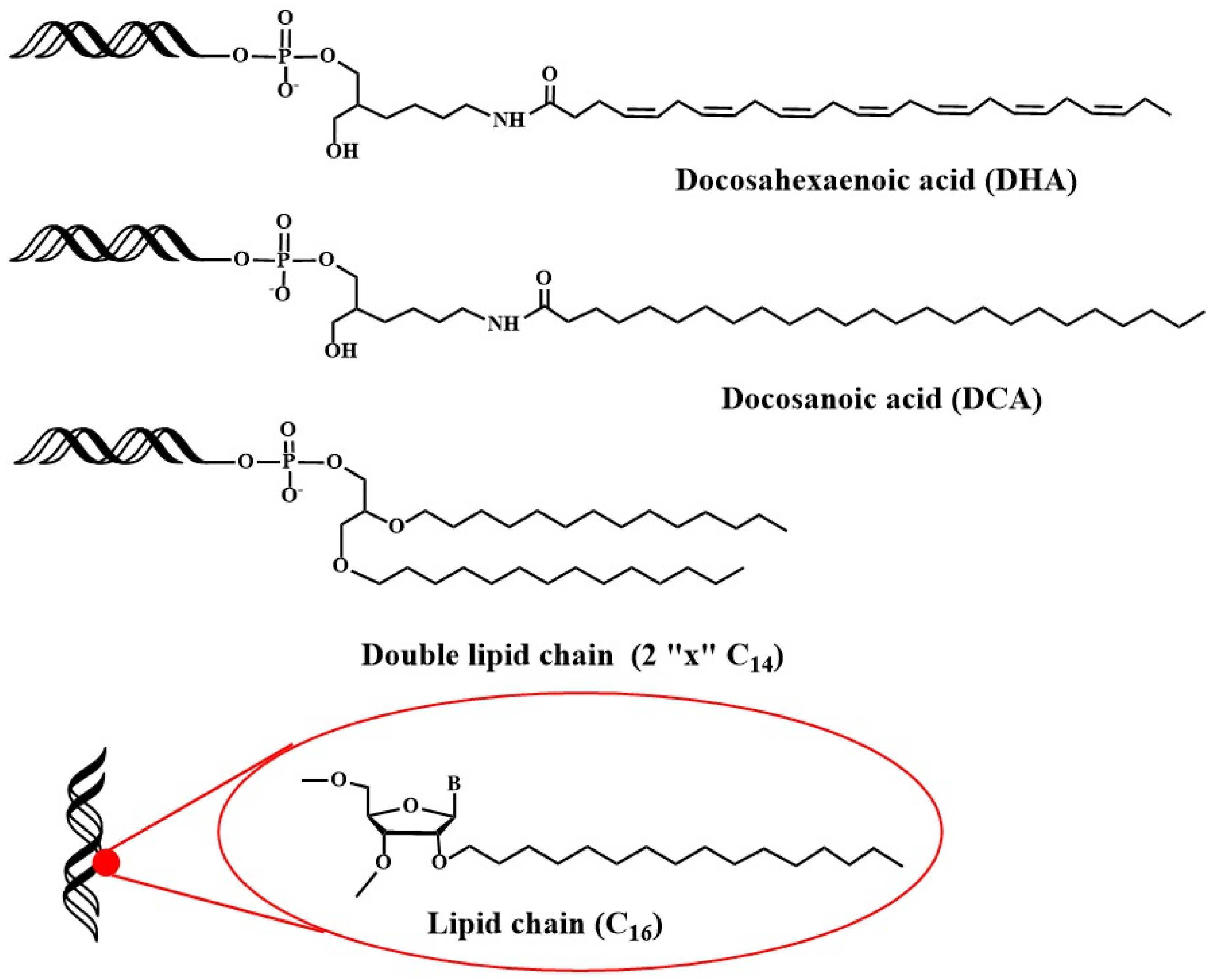
2.3. Early Developments in the Synthesis of Oligonucleotide-Peptide Conjugates

2.4. Peptide-Oligonucleotide Conjugates and the Development of RNA-Based Therapeutics
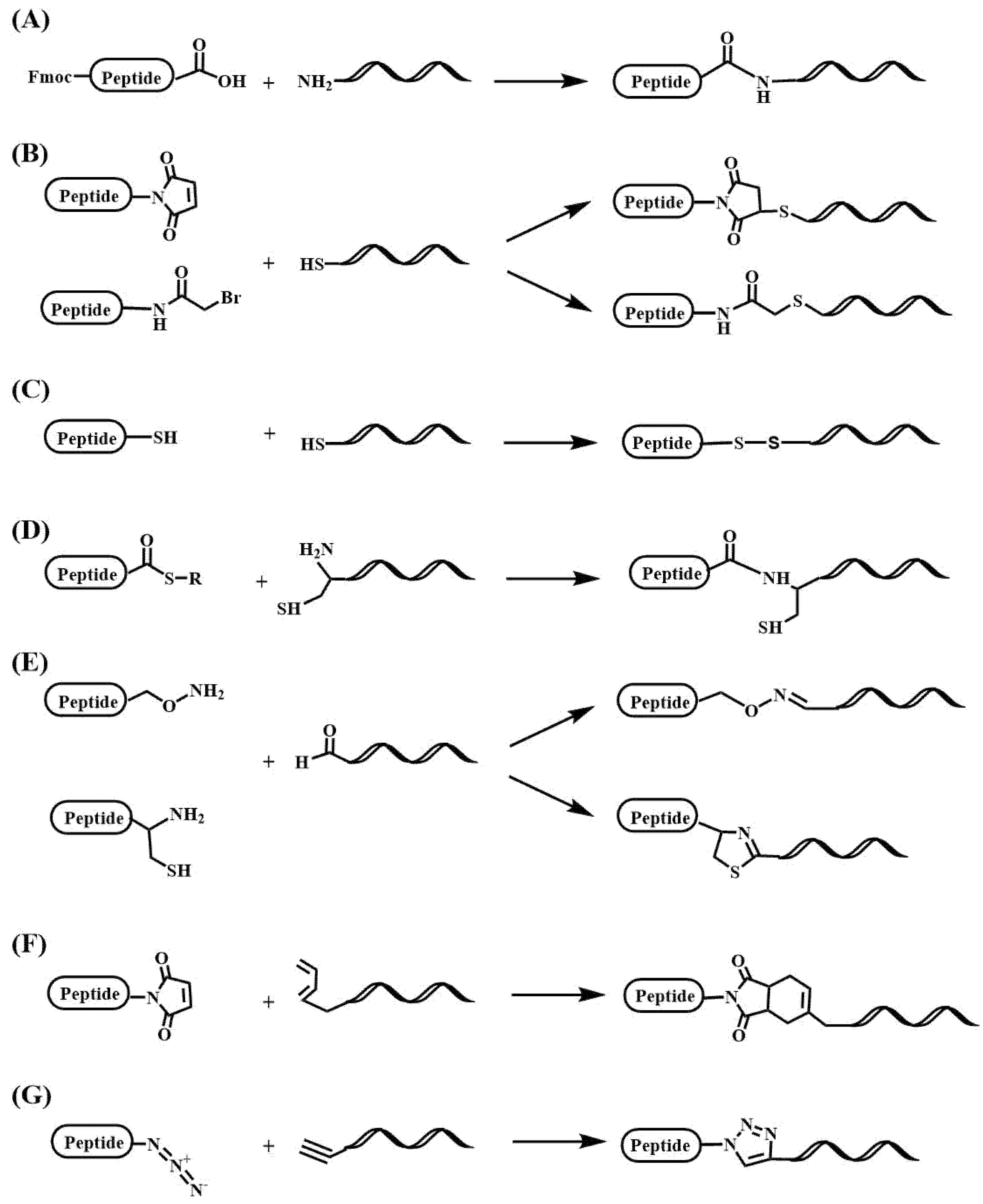
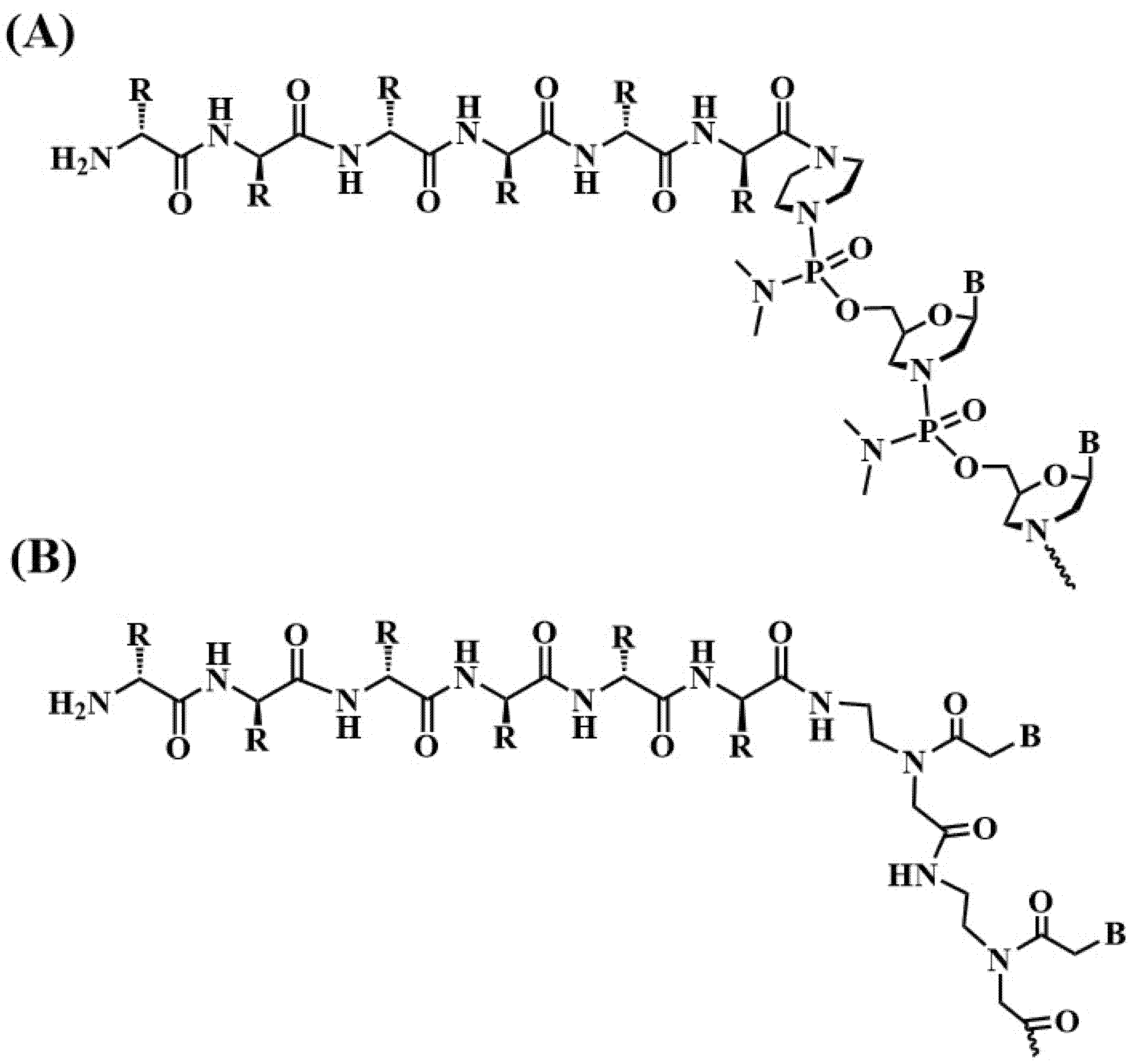
2.5. Multifunctional Conjugates

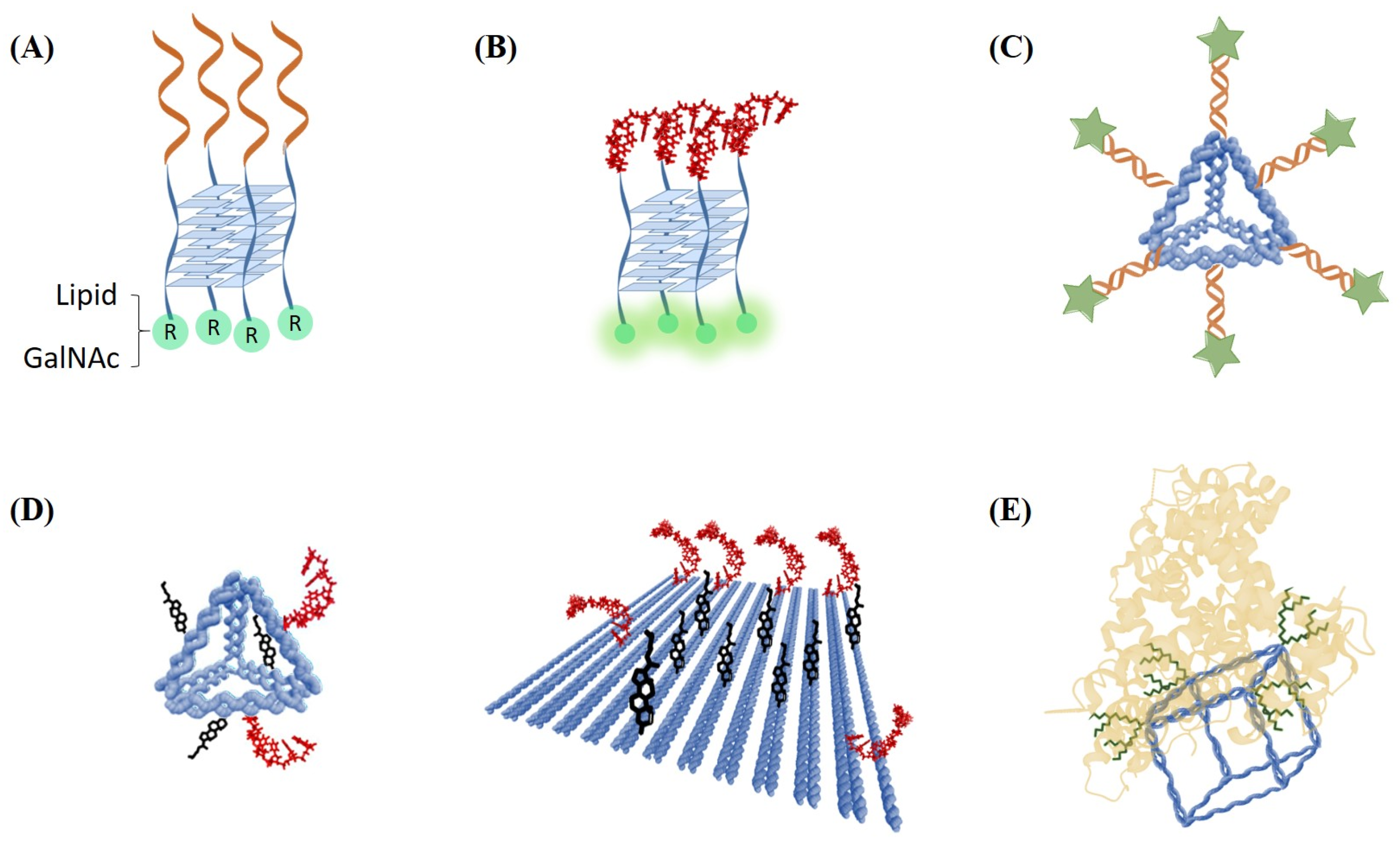
2.6. Oligonucleotide Conjugates Currently in Advanced Preclinical or Clinical Trials
3. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Belfort, R.; Goldstein, D.A.; Johnson, D.W.; Mansour, S.E.; Muccioli, C.; Sheppard, J.D.; Mora-Durate, J.; Chan, C.K.; Palestine, A.G.; Perez, J.E.; et al. A Randomized Controlled Clinical Trial of Intravitreous Fomivirsen for Treatment of Newly Diagnosed Peripheral Cytomegalovirus Retinitis in Patients with AIDS. Am. J. Ophthalmol. 2002, 133, 467–474. [Google Scholar]
- Finkel, R.S.; Mercuri, E.; Darras, B.T.; Connolly, A.M.; Kuntz, N.L.; Kirschner, J.; Chiriboga, C.A.; Saito, K.; Servais, L.; Tizzano, E. Nusinersen versus Sham Control in Infantile-Onset Spinal Muscular Atrophy. N. Engl. J. Med. 2017, 377, 1723–1732. [Google Scholar] [CrossRef] [PubMed]
- Charleston, J.S.; Schnell, F.J.; Dworzak, J.; Donoghue, C.; Lewis, S.; Chen, L.; Young, G.D.; Milici, A.J.; Voss, J.; DeAlwis, U. Eteplirsen Treatment for Duchenne Muscular Dystrophy: Exon Skipping and Dystrophin Production. Neurology 2018, 90, e2146–e2154. [Google Scholar] [CrossRef] [PubMed]
- Hu, B.; Zhong, L.; Weng, Y.; Peng, L.; Huang, Y.; Zhao, Y.; Liang, X.-J. Therapeutic SiRNA: State of the Art. Signal. Transduct. Target. Ther. 2020, 5, 101. [Google Scholar] [CrossRef] [PubMed]
- Crooke, S.T.; Liang, X.-H.; Baker, B.F.; Crooke, R.M. Antisense Technology: A Review. J. Biol. Chem. 2021, 296, 100416. [Google Scholar] [CrossRef]
- Mallikaratchy, P. Evolution of Complex Target SELEX to Identify Aptamers against Mammalian Cell-Surface Antigens. Molecules 2017, 22, 215. [Google Scholar] [CrossRef]
- Li, Z.; Rana, T.M. Therapeutic Targeting of MicroRNAs: Current Status and Future Challenges. Nat. Rev. Drug Discov. 2014, 13, 622–638. [Google Scholar] [CrossRef]
- Smith, C.I.E.; Zain, R. Therapeutic Oligonucleotides: State of the Art. Annu. Rev. Pharmacol. Toxicol. 2019, 59, 605–630. [Google Scholar] [CrossRef]
- Khvorova, A.; Watts, J.K. The Chemical Evolution of Oligonucleotide Therapies of Clinical Utility. Nat. Biotechnol. 2017, 35, 238–248. [Google Scholar] [CrossRef]
- Grijalvo, S.; Alagia, A.; Jorge, A.F.; Eritja, R. Covalent Strategies for Targeting Messenger and Non-Coding RNAs: An Updated Review on SiRNA, MiRNA and AntimiR Conjugates. Genes 2018, 9, 74. [Google Scholar] [CrossRef]
- Adams, D.; Gonzalez-Duarte, A.; O’Riordan, W.D.; Yang, C.-C.; Ueda, M.; Kristen, A.V.; Tournev, I.; Schmidt, H.H.; Coelho, T.; Berk, J.L. Patisiran, an RNAi Therapeutic, for Hereditary Transthyretin Amyloidosis. N. Engl. J. Med. 2018, 379, 11–21. [Google Scholar] [CrossRef] [PubMed]
- Nair, J.K.; Willoughby, J.L.S.; Chan, A.; Charisse, K.; Alam, M.R.; Wang, Q.; Hoekstra, M.; Kandasamy, P.; Kel’in, A.V.; Milstein, S. Multivalent N-Acetylgalactosamine-Conjugated SiRNA Localizes in Hepatocytes and Elicits Robust RNAi-Mediated Gene Silencing. J. Am. Chem. Soc. 2014, 136, 16958–16961. [Google Scholar] [CrossRef] [PubMed]
- Balwani, M.; Sardh, E.; Ventura, P.; Peiró, P.A.; Rees, D.C.; Stölzel, U.; Bissell, D.M.; Bonkovsky, H.L.; Windyga, J.; Anderson, K.E. Phase 3 Trial of RNAi Therapeutic Givosiran for Acute Intermittent Porphyria. N. Engl. J. Med. 2020, 382, 2289–2301. [Google Scholar] [CrossRef] [PubMed]
- Garrelfs, S.F.; Frishberg, Y.; Hulton, S.A.; Koren, M.J.; O’Riordan, W.D.; Cochat, P.; Deschênes, G.; Shasha-Lavsky, H.; Saland, J.M.; van’t Hoff, W.G. Lumasiran, an RNAi Therapeutic for Primary Hyperoxaluria Type 1. N. Engl. J. Med. 2021, 384, 1216–1226. [Google Scholar] [CrossRef] [PubMed]
- Ray, K.K.; Wright, R.S.; Kallend, D.; Koenig, W.; Leiter, L.A.; Raal, F.J.; Bisch, J.A.; Richardson, T.; Jaros, M.; Wijngaard, P.L.J. Two Phase 3 Trials of Inclisiran in Patients with Elevated LDL Cholesterol. N. Engl. J. Med. 2020, 382, 1507–1519. [Google Scholar] [CrossRef] [PubMed]
- Adams, D.; Tournev, I.L.; Taylor, M.S.; Coelho, T.; Planté-Bordeneuve, V.; Berk, J.L.; González-Duarte, A.; Gillmore, J.D.; Low, S.-C.; Sekijima, Y. Efficacy and Safety of Vutrisiran for Patients with Hereditary Transthyretin-Mediated Amyloidosis with Polyneuropathy: A Randomized Clinical Trial. Amyloid 2022, 1–9. [Google Scholar] [CrossRef]
- Kosmas, C.E.; Muñoz Estrella, A.; Sourlas, A.; Silverio, D.; Hilario, E.; Montan, P.D.; Guzman, E. Inclisiran: A New Promising Agent in the Management of Hypercholesterolemia. Diseases 2018, 6, 63. [Google Scholar] [CrossRef]
- Hawner, M.; Ducho, C. Cellular Targeting of Oligonucleotides by Conjugation with Small Molecules. Molecules 2020, 25, 5963. [Google Scholar] [CrossRef]
- Halloy, F.; Biscans, A.; Bujold, K.E.; Debacker, A.; Hill, A.C.; Lacroix, A.; Luige, O.; Strömberg, R.; Sundstrom, L.; Vogel, J. Innovative Developments and Emerging Technologies in RNA Therapeutics. RNA Biol. 2022, 19, 313–332. [Google Scholar] [CrossRef]
- Zamecnik, P.C.; Stephenson, M.L. Inhibition of Rous Sarcoma Virus Replication and Cell Transformation by a Specific Oligodeoxynucleotide. Proc. Natl. Acad. Sci. USA 1978, 75, 280–284. [Google Scholar] [CrossRef]
- Smith, C.C.; Aurelian, L.; Reddy, M.P.; Miller, P.S.; Ts’o, P.O. Antiviral Effect of an Oligo (Nucleoside Methylphosphonate) Complementary to the Splice Junction of Herpes Simplex Virus Type 1 Immediate Early Pre-MRNAs 4 and 5. Proc. Natl. Acad. Sci. USA 1986, 83, 2787–2791. [Google Scholar] [CrossRef] [PubMed]
- Letsinger, R.L.; Zhang, G.R.; Sun, D.K.; Ikeuchi, T.; Sarin, P.S. Cholesteryl-Conjugated Oligonucleotides: Synthesis, Properties, and Activity as Inhibitors of Replication of Human Immunodeficiency Virus in Cell Culture. Proc. Natl. Acad. Sci. USA 1989, 86, 6553–6556. [Google Scholar] [CrossRef] [PubMed]
- Stein, C.A.; Pal, R.; DeVico, A.L.; Hoke, G.; Mumbauer, S.; Kinstler, O.; Sarngadharan, M.G.; Letsinger, R.L. Mode of Action of 5’-Linked Cholesteryl Phosphorothioate Oligodeoxynucleotides in Inhibiting Syncytia Formation and Infection by HIV-1 and HIV-2 in Vitro. Biochemistry 1991, 30, 2439–2444. [Google Scholar] [CrossRef] [PubMed]
- Krieg, A.M.; ToNkINsoN, J.; MATsoN, S.; ZHAo, Q.; SAxoN, M.; Zhang, L.-M.; Bhanja, U.; Yakubov, L.; Stein, C.A. Modification of Antisense Phosphodiester Oligodeoxynucleotides by a 5’cholesteryl Moiety Increases Cellular Association and Improves Efficacy. Proc. Natl. Acad. Sci. USA 1993, 90, 1048–1052. [Google Scholar] [CrossRef] [PubMed]
- Shea, R.G.; Marsters, J.C.; Bischofberger, N. Synthesis, Hybridization Properties and Antiviral Activity of Lipid-Oligodeoxynucleotide Conjugates. Nucleic Acids Res. 1990, 18, 3777–3783. [Google Scholar] [CrossRef] [PubMed]
- Svinarchuk, F.P.; Konevetz, D.A.; Pliasunova, O.A.; Pokrovsky, A.G.; Vlassov, V. V Inhibition of HIV Proliferation in MT-4 Cells by Antisense Oligonucleotide Conjugated to Lipophilic Groups. Biochimie 1993, 75, 49–54. [Google Scholar] [CrossRef] [PubMed]
- Oberhauser, B.; Wagner, E. Effective Incorporation of 2’-O-Methyl-Oligoribonuclectides into Liposomes and Enhanced Cell Association through Modification with Thiocholesterol. Nucleic Acids Res. 1992, 20, 533–538. [Google Scholar] [CrossRef]
- Boutorine, A.S.; Kostina, E. V Reversible Covalent Attachment of Cholesterol to Oligodeoxyribonucleotides for Studies of the Mechanisms of Their Penetration into Eucaryotic Cells. Biochimie 1993, 75, 35–41. [Google Scholar] [CrossRef]
- Gryaznov, S.M.; Lloyd, D.H. Modulation of Oligonucleotide Duplex and Triplex Stability via Hydrophobic Interactions. Nucleic Acids Res. 1993, 21, 5909–5915. [Google Scholar] [CrossRef]
- Polushin, N.N.; Cohen, J.S. Antisense Pro-Drugs: 5’-Ester Oligodeoxynucleotides. Nucleic Acids Res. 1994, 22, 5492–5496. [Google Scholar] [CrossRef]
- MacKellar, C.; Graham, D.; Will, D.W.; Burgess, S.; Brown, T. Synthesis and Physical Properties of Anti-HIV Antisense Oligonucleotides Bearing Terminal Lipophilic Groups. Nucleic Acids Res. 1992, 20, 3411–3417. [Google Scholar] [CrossRef]
- Will, D.W.; Brown, T. Attachment of Vitamin E Derivatives to Oligonucleotides during Solid-Phase Synthesis. Tetrahedron Lett. 1992, 33, 2729–2732. [Google Scholar] [CrossRef]
- Vu, H.; Singh, P.; Lewis, L.; Zendegui, J.G.; Jayaraman, K. Synthesis of Cholesteryl Supports and Phosphoramidite for Automated DNA Synthesis of Triple-Helix Forming Oligonucleotides (TFOs). Nucleosides Nucleotides 1993, 12, 853–864. [Google Scholar] [CrossRef]
- Vu, H.; Hill, T.S.; Jayaraman, K. Synthesis and Properties of Cholesteryl-Modified Triple-Helix Forming Oligonucleotides Containing a Triglycyl Linker. Bioconjug. Chem. 1994, 5, 666–668. [Google Scholar] [CrossRef]
- Manoharan, M.; Johnson, L.K.; Bennett, C.F.; Vickers, T.A.; Ecker, D.J.; Cowsert, L.M.; Freier, S.M.; Cook, P.D. Cholic Acid-Oligonucleotide Conjugates for Antisense Applications. Bioorg. Med. Chem. Lett. 1994, 4, 1053–1060. [Google Scholar] [CrossRef]
- Manoharan, M.; Tivel, K.L.; Cook, P.D. Lipidic Nucleic Acids. Tetrahedron Lett. 1995, 36, 3651–3654. [Google Scholar] [CrossRef]
- Li, S.; Deshmukh, H.M.; Huang, L. Folate-Mediated Targeting of Antisense Oligodeoxynucleotides to Ovarian Cancer Cells. Pharm. Res. 1998, 15, 1540–1545. [Google Scholar] [CrossRef]
- Marasco, C.J., Jr.; Angelino, N.J.; Paul, B.; Dolnick, B.J. A Simplified Synthesis of Acridine and/or Lipid Containing Oligodeoxynucleotides. Tetrahedron Lett. 1994, 35, 3029–3032. [Google Scholar] [CrossRef]
- Grijalvo, S.; Ocampo, S.M.; Perales, J.C.; Eritja, R. Synthesis of Lipid–Oligonucleotide Conjugates for RNA Interference Studies. Chem. Biodivers. 2011, 8, 287–299. [Google Scholar] [CrossRef]
- Grijalvo, S.; Ocampo, S.M.; Perales, J.C.; Eritja, R. Synthesis of Oligonucleotides Carrying Amino Lipid Groups at the 3′-End for RNA Interference Studies. J. Org. Chem. 2010, 75, 6806–6813. [Google Scholar] [CrossRef]
- Manoharan, M. Oligonucleotide Conjugates as Potential Antisense Drugs with Improved Uptake, Biodistribution, Targeted Delivery, and Mechanism of Action. Antisense Nucleic Acid Drug Dev. 2002, 12, 103–128. [Google Scholar] [CrossRef] [PubMed]
- Crooke, S.T.; Graham, M.J.; Zuckerman, J.E.; Brooks, D.; Conklin, B.S.; Cummins, L.L.; Greig, M.J.; Guinosso, C.J.; Kornbrust, D.; Manoharan, M. Pharmacokinetic Properties of Several Novel Oligonucleotide Analogs in Mice. J. Pharmacol. Exp. Ther. 1996, 277, 923–937. [Google Scholar] [PubMed]
- De Smidt, P.C.; Le Doan, T.; de Falco, S.; van Berkel, T.J.C. Association of Antisense Oligonucleotides with Lipoproteins Prolongs the Plasma Half-Life and Modifies the Tissue Distribution. Nucleic Acids Res. 1991, 19, 4695–4700. [Google Scholar] [CrossRef] [PubMed]
- Wolfrum, C.; Shi, S.; Jayaprakash, K.N.; Jayaraman, M.; Wang, G.; Pandey, R.K.; Rajeev, K.G.; Nakayama, T.; Charrise, K.; Ndungo, E.M.; et al. Mechanisms and Optimization of in Vivo Delivery of Lipophilic SiRNAs. Nat. Biotechnol. 2007, 25, 1149–1157. [Google Scholar] [CrossRef]
- Jäschke, A.; Fürste, J.P.; Cech, D.; Erdmann, V.A. Automated Incorporation of Polyethylene Glycol into Synthetic Oligonucleotides. Tetrahedron Lett. 1993, 34, 301–304. [Google Scholar] [CrossRef]
- Jäschke, A.; Fürste, J.P.; Nordhoff, E.; Hillenkamp, F.; Cech, D.; Erdmann, V.A. Synthesis and Properties of Oligodeoxyribonucleotide—Polyethylene Glycol Conjugates. Nucleic Acids Res. 1994, 22, 4810–4817. [Google Scholar] [CrossRef]
- Bonora, G.M.; Ivanova, E.; Zarytova, V.; Burcovich, B.; Veronese, F.M. Synthesis and Characterization of High-Molecular Mass Polyethylene Glycol-Conjugated Oligonucleotides. Bioconjug. Chem. 1997, 8, 793–797. [Google Scholar] [CrossRef]
- Burcovich, B.; Veronese, F.M.; Zarytova, V.; Bonora, G.M. Branched Polyethylene Glycol (BPEG) Conjugated Antisense Oligonucleotides. Nucleosides Nucleotides 1998, 17, 1567–1570. [Google Scholar] [CrossRef]
- Gragoudas, E.S.; Adamis, A.P.; Cunningham Jr, E.T.; Feinsod, M.; Guyer, D.R. Pegaptanib for Neovascular Age-Related Macular Degeneration. N. Engl. J. Med. 2004, 351, 2805–2816. [Google Scholar] [CrossRef]
- Ng, E.W.M.; Shima, D.T.; Calias, P.; Cunningham, E.T.; Guyer, D.R.; Adamis, A.P. Pegaptanib, a Targeted Anti-VEGF Aptamer for Ocular Vascular Disease. Nat. Rev. Drug Discov. 2006, 5, 123–132. [Google Scholar] [CrossRef]
- Fire, A.; Xu, S.; Montgomery, M.K.; Kostas, S.A.; Driver, S.E.; Mello, C.C. Potent and Specific Genetic Interference by Double-Stranded RNA in Caenorhabditis Elegans. Nature 1998, 391, 806–811. [Google Scholar] [CrossRef] [PubMed]
- Elbashir, S.M.; Harborth, J.; Lendeckel, W.; Yalcin, A.; Weber, K.; Tuschl, T. Duplexes of 21-Nucleotide RNAs Mediate RNA Interference in Cultured Mammalian Cells. Nature 2001, 411, 494–498. [Google Scholar] [CrossRef]
- Dong, Y.; Siegwart, D.J.; Anderson, D.G. Strategies, Design, and Chemistry in SiRNA Delivery Systems. Adv. Drug Deliv. Rev. 2019, 144, 133–147. [Google Scholar] [CrossRef]
- Ha, M.; Kim, V.N. Regulation of MicroRNA Biogenesis. Nat. Rev. Mol. Cell Biol. 2014, 15, 509–524. [Google Scholar] [CrossRef]
- Soutscheck, J.; Akinc, A.; Bramlage, B.; Charisse, K.; Constien, R.; Donoghue, M.; Elbashir, S.; Geick, A.; Hadwiger, P.; Harborth, J.; et al. Therapeutic Silencing of an Endogenous Gene by Systemic Administration of Modified SiRNAs. Nature 2004, 432, 173–178. [Google Scholar] [CrossRef]
- Zimmermann, T.S.; Lee, A.C.; Akinc, A.; Bramlage, B.; Bumcrot, D.; Fedoruk, M.N.; Harborth, J.; Heyes, J.A.; Jeffs, L.B.; John, M.; et al. RNAi-Mediated Gene Silencing in Non-Human Primates. Nature 2006, 441, 111–114. [Google Scholar] [CrossRef]
- Heyes, J.; Palmer, L.; Bremner, K.; MacLachlan, I. Cationic Lipid Saturation Influences Intracellular Delivery of Encapsulated Nucleic Acids. J. Control. Release 2005, 107, 276–287. [Google Scholar] [CrossRef]
- Akinc, A.; Zumbuehl, A.; Goldberg, M.; Leshchiner, E.S.; Busini, V.; Hossain, N.; Bacallado, S.A.; Nguyen, D.N.; Fuller, J.; Alvarez, R. A Combinatorial Library of Lipid-like Materials for Delivery of RNAi Therapeutics. Nat. Biotechnol. 2008, 26, 561–569. [Google Scholar] [CrossRef]
- Semple, S.C.; Akinc, A.; Chen, J.; Sandhu, A.P.; Mui, B.L.; Cho, C.K.; Sah, D.W.Y.; Stebbing, D.; Crosley, E.J.; Yaworski, E. Rational Design of Cationic Lipids for SiRNA Delivery. Nat. Biotechnol. 2010, 28, 172–176. [Google Scholar] [CrossRef]
- Ueno, Y.; Kawada, K.; Naito, T.; Shibata, A.; Yoshikawa, K.; Kim, H.-S.; Wataya, Y.; Kitade, Y. Synthesis and Silencing Properties of SiRNAs Possessing Lipophilic Groups at Their 3′-Termini. Bioorg. Med. Chem. 2008, 16, 7698–7704. [Google Scholar] [CrossRef] [PubMed]
- Lorenz, C.; Hadwiger, P.; John, M.; Vornlocher, H.-P.; Unverzagt, C. Steroid and Lipid Conjugates of SiRNAs to Enhance Cellular Uptake and Gene Silencing in Liver Cells. Bioorg. Med. Chem. Lett. 2004, 14, 4975–4977. [Google Scholar] [CrossRef] [PubMed]
- Nishina, T.; Numata, J.; Nishina, K.; Yoshida-Tanaka, K.; Nitta, K.; Piao, W.; Iwata, R.; Ito, S.; Kuwahara, H.; Wada, T. Chimeric Antisense Oligonucleotide Conjugated to α-Tocopherol. Mol. Ther. Acids 2015, 4, e220. [Google Scholar] [CrossRef]
- Aviñó, A.; Ocampo, S.M.; Perales, J.C.; Eritja, R. Synthesis and in Vitro Inhibition Properties of SiRNA Conjugates Carrying Acridine and Quindoline Moieties. Chem. Biodivers. 2012, 9, 557–566. [Google Scholar] [CrossRef] [PubMed]
- Ugarte-Uribe, B.; Grijalvo, S.; Busto, J.V.; Martín, C.; Eritja, R.; Goñi, F.M.; Alkorta, I. Double-Tailed Lipid Modification as a Promising Candidate for Oligonucleotide Delivery in Mammalian Cells. Biochim. Biophys. Acta (BBA)-General Subj. 2013, 1830, 4872–4884. [Google Scholar] [CrossRef]
- Ugarte-Uribe, B.; Grijalvo, S.; Pertíñez, S.N.; Busto, J.V.; Martín, C.; Alagia, A.; Goñi, F.M.; Eritja, R.; Alkorta, I. Lipid-Modified Oligonucleotide Conjugates: Insights into Gene Silencing, Interaction with Model Membranes and Cellular Uptake Mechanisms. Bioorg. Med. Chem. 2017, 25, 175–186. [Google Scholar] [CrossRef] [PubMed]
- Ries, O.; Löffler, P.M.G.; Vogel, S. Convenient Synthesis and Application of Versatile Nucleic Acid Lipid Membrane Anchors in the Assembly and Fusion of Liposomes. Org. Biomol. Chem. 2015, 13, 9673–9680. [Google Scholar] [CrossRef]
- Liu, H.; Zhu, Z.; Kang, H.; Wu, Y.; Sefan, K.; Tan, W. DNA-based Micelles: Synthesis, Micellar Properties and Size-dependent Cell Permeability. Chem. Eur. J. 2010, 16, 3791–3797. [Google Scholar] [CrossRef]
- Patwa, A.; Gissot, A.; Bestel, I.; Barthélémy, P. Hybrid Lipid Oligonucleotide Conjugates: Synthesis, Self-Assemblies and Biomedical Applications. Chem. Soc. Rev. 2011, 40, 5844–5854. [Google Scholar] [CrossRef]
- Grijalvo, S.; Ocampo, S.M.; Perales, J.C.; Eritja, R. Preparation of Lipid-Conjugated SiRNA Oligonucleotides for Enhanced Gene Inhibition in Mammalian Cells. Methods Mol. Biol. 2021, 2282, 119–136. [Google Scholar]
- Campbell, A.; Morris, G.; Heller, J.P.; Langa, E.; Brindley, E.; Worm, J.; Jensen, M.A.; Miller, M.T.; Henshall, D.C.; Reschke, C.R. Antagomir-Mediated Suppression of MicroRNA-134 Reduces Kainic Acid-Induced Seizures in Immature Mice. Sci. Rep. 2021, 11, 340. [Google Scholar] [CrossRef]
- Haftmann, C.; Riedel, R.; Porstner, M.; Wittmann, J.; Chang, H.-D.; Radbruch, A.; Mashreghi, M.-F. Direct Uptake of Antagomirs and Efficient Knockdown of MiRNA in Primary B and T Lymphocytes. J. Immunol. Methods 2015, 426, 128–133. [Google Scholar] [CrossRef]
- Maschmeyer, P.; Petkau, G.; Siracusa, F.; Zimmermann, J.; Zügel, F.; Kühl, A.A.; Lehmann, K.; Schimmelpfennig, S.; Weber, M.; Haftmann, C. Selective Targeting of Pro-Inflammatory Th1 Cells by MicroRNA-148a-Specific Antagomirs in Vivo. J. Autoimmun. 2018, 89, 41–52. [Google Scholar] [CrossRef]
- Velu, C.S.; Grimes, H.L. Utilizing AntagomiR (Antisense MicroRNA) to Knock down MicroRNA in Murine Bone Marrow Cells. Methods Mol. Biol. 2012, 928, 185–195. [Google Scholar] [PubMed]
- Koutsoudakis, G.; Paris de León, A.; Herrera, C.; Dorner, M.; Pérez-Vilaró, G.; Lyonnais, S.; Grijalvo, S.; Eritja, R.; Meyerhans, A.; Mirambeau, G. Oligonucleotide-Lipid Conjugates Forming G-Quadruplex Structures Are Potent and Pangenotypic Hepatitis C Virus Entry Inhibitors in Vitro and Ex Vivo. Antimicrob. Agents Chemother. 2017, 61, e02354-16. [Google Scholar] [CrossRef] [PubMed]
- Musumeci, D.; Montesarchio, D. Synthesis of a Cholesteryl-HEG Phosphoramidite Derivative and Its Application to Lipid-Conjugates of the Anti-HIV 5’TGGGAG3’Hotoda’s Sequence. Molecules 2012, 17, 12378–12392. [Google Scholar] [CrossRef]
- Wolfe, J.L.; Goodchild, J. Modulation of Tetraplex Formation by Chemical Modifications of a G4-Containing Phosphorothioate Oligonucleotide. J. Am. Chem. Soc. 1996, 118, 6301–6302. [Google Scholar] [CrossRef]
- Godeau, G.; Staedel, C.; Barthéleémy, P. Lipid-Conjugated Oligonucleotides via “Click Chemistry” Efficiently Inhibit Hepatitis C Virus Translation. J. Med. Chem. 2008, 51, 4374–4376. [Google Scholar] [CrossRef] [PubMed]
- Terstappen, G.C.; Meyer, A.H.; Bell, R.D.; Zhang, W. Strategies for Delivering Therapeutics across the Blood–Brain Barrier. Nat. Rev. Drug Discov. 2021, 20, 362–383. [Google Scholar] [CrossRef]
- Alterman, J.F.; Hall, L.M.; Coles, A.H.; Hassler, M.R.; Didiot, M.-C.; Chase, K.; Abraham, J.; Sottosanti, E.; Johnson, E.; Sapp, E. Hydrophobically Modified SiRNAs Silence Huntingtin MRNA in Primary Neurons and Mouse Brain. Mol. Ther. Acids 2015, 4, e266. [Google Scholar] [CrossRef]
- Ly, S.; Navaroli, D.M.; Didiot, M.-C.; Cardia, J.; Pandarinathan, L.; Alterman, J.F.; Fogarty, K.; Standley, C.; Lifshitz, L.M.; Bellve, K.D. Visualization of Self-Delivering Hydrophobically Modified SiRNA Cellular Internalization. Nucleic Acids Res. 2017, 45, 15–25. [Google Scholar] [CrossRef]
- Nikan, M.; Osborn, M.F.; Coles, A.H.; Godinho, B.M.D.C.; Hall, L.M.; Haraszti, R.A.; Hassler, M.R.; Echeverria, D.; Aronin, N.; Khvorova, A. Docosahexaenoic Acid Conjugation Enhances Distribution and Safety of SiRNA upon Local Administration in Mouse Brain. Mol. Ther. Acids 2016, 5, e344. [Google Scholar] [CrossRef] [PubMed]
- Haraszti, R.A.; Miller, R.; Didiot, M.-C.; Biscans, A.; Alterman, J.F.; Hassler, M.R.; Roux, L.; Echeverria, D.; Sapp, E.; DiFiglia, M. Optimized Cholesterol-SiRNA Chemistry Improves Productive Loading onto Extracellular Vesicles. Mol. Ther. 2018, 26, 1973–1982. [Google Scholar] [CrossRef]
- Biscans, A.; Haraszti, R.A.; Echeverria, D.; Miller, R.; Didiot, M.-C.; Nikan, M.; Roux, L.; Aronin, N.; Khvorova, A. Hydrophobicity of Lipid-Conjugated SiRNAs Predicts Productive Loading to Small Extracellular Vesicles. Mol. Ther. 2018, 26, 1520–1528. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Gubu, A.; Xu, J.; Yan, N.; Su, W.; Feng, D.; Wang, Q.; Tang, X. Tetrazine-Induced Bioorthogonal Activation of Vitamin E-Modified SiRNA for Gene Silencing. Molecules 2022, 27, 4377. [Google Scholar] [CrossRef] [PubMed]
- Biscans, A.; Coles, A.; Haraszti, R.; Echeverria, D.; Hassler, M.; Osborn, M.; Khvorova, A. Diverse Lipid Conjugates for Functional Extra-Hepatic SiRNA Delivery in Vivo. Nucleic Acids Res. 2019, 47, 1082–1096. [Google Scholar] [CrossRef]
- Osborn, M.F.; Khvorova, A. Improving SiRNA Delivery in Vivo through Lipid Conjugation. Nucleic Acid Ther. 2018, 28, 128–136. [Google Scholar] [CrossRef]
- Biscans, A.; Caiazzi, J.; Davis, S.; McHugh, N.; Sousa, J.; Khvorova, A. The Chemical Structure and Phosphorothioate Content of Hydrophobically Modified SiRNAs Impact Extrahepatic Distribution and Efficacy. Nucleic Acids Res. 2020, 48, 7665–7680. [Google Scholar] [CrossRef]
- Ly, S.; Echeverria, D.; Sousa, J.; Khvorova, A. Single-Stranded Phosphorothioated Regions Enhance Cellular Uptake of Cholesterol-Conjugated SiRNA but Not Silencing Efficacy. Mol. Ther. Acids 2020, 21, 991–1005. [Google Scholar] [CrossRef]
- Biscans, A.; Coles, A.; Echeverria, D.; Khvorova, A. The Valency of Fatty Acid Conjugates Impacts SiRNA Pharmacokinetics, Distribution, and Efficacy in Vivo. J. Control. Release 2019, 302, 116–125. [Google Scholar] [CrossRef]
- Osborn, M.F.; Coles, A.H.; Biscans, A.; Haraszti, R.A.; Roux, L.; Davis, S.; Ly, S.; Echeverria, D.; Hassler, M.R.; Godinho, B.M.D.C. Hydrophobicity Drives the Systemic Distribution of Lipid-Conjugated SiRNAs via Lipid Transport Pathways. Nucleic Acids Res. 2019, 47, 1070–1081. [Google Scholar] [CrossRef]
- Hassler, M.R.; Turanov, A.A.; Alterman, J.F.; Haraszti, R.A.; Coles, A.H.; Osborn, M.F.; Echeverria, D.; Nikan, M.; Salomon, W.E.; Roux, L. Comparison of Partially and Fully Chemically-Modified SiRNA in Conjugate-Mediated Delivery in Vivo. Nucleic Acids Res. 2018, 46, 2185–2196. [Google Scholar] [CrossRef] [PubMed]
- Brown, K.M.; Nair, J.K.; Janas, M.M.; Anglero-Rodriguez, Y.I.; Dang, L.T.H.; Peng, H.; Theile, C.S.; Castellanos-Rizaldos, E.; Brown, C.; Foster, D. Expanding RNAi Therapeutics to Extrahepatic Tissues with Lipophilic Conjugates. Nat. Biotechnol. 2022, 40, 1500–1508. [Google Scholar] [CrossRef] [PubMed]
- Østergaard, M.E.; Jackson, M.; Low, A.; Chappell, A.E.; Lee, R.G.; Peralta, R.Q.; Yu, J.; Kinberger, G.A.; Dan, A.; Carty, R. Conjugation of Hydrophobic Moieties Enhances Potency of Antisense Oligonucleotides in the Muscle of Rodents and Non-Human Primates. Nucleic Acids Res. 2019, 47, 6045–6058. [Google Scholar] [CrossRef] [PubMed]
- Prakash, T.P.; Mullick, A.E.; Lee, R.G.; Yu, J.; Yeh, S.T.; Low, A.; Chappell, A.E.; Østergaard, M.E.; Murray, S.; Gaus, H.J. Fatty Acid Conjugation Enhances Potency of Antisense Oligonucleotides in Muscle. Nucleic Acids Res. 2019, 47, 6029–6044. [Google Scholar] [CrossRef]
- Wang, S.; Allen, N.; Prakash, T.P.; Liang, X.; Crooke, S.T. Lipid Conjugates Enhance Endosomal Release of Antisense Oligonucleotides Into Cells. Nucleic Acid Ther. 2019, 29, 245–255. [Google Scholar] [CrossRef]
- Relizani, K.; Echevarría, L.; Zarrouki, F.; Gastaldi, C.; Dambrune, C.; Aupy, P.; Haeberli, A.; Komisarski, M.; Tensorer, T.; Larcher, T. Palmitic Acid Conjugation Enhances Potency of Tricyclo-DNA Splice Switching Oligonucleotides. Nucleic Acids Res. 2022, 50, 17–34. [Google Scholar] [CrossRef]
- Marchesi, E.; Cortesi, R.; Preti, L.; Rimessi, P.; Sguizzato, M.; Bovolenta, M.; Perrone, D. Antisense Oligonucleotides Conjugated with Lipophilic Compounds: Synthesis and In Vitro Evaluation of Exon Skipping in Duchenne Muscular Dystrophy. Int. J. Mol. Sci. 2022, 23, 4270. [Google Scholar] [CrossRef]
- Anderson, E.J.; Rouphael, N.G.; Widge, A.T.; Jackson, L.A.; Roberts, P.C.; Makhene, M.; Chappell, J.D.; Denison, M.R.; Stevens, L.J.; Pruijssers, A.J. Safety and Immunogenicity of SARS-CoV-2 MRNA-1273 Vaccine in Older Adults. N. Engl. J. Med. 2020, 383, 2427–2438. [Google Scholar] [CrossRef]
- Polack, F.P.; Thomas, S.J.; Kitchin, N.; Absalon, J.; Gurtman, A.; Lockhart, S.; Perez, J.L.; Marc, G.P.; Moreira, E.D.; Zerbini, C. Safety and Efficacy of the BNT162b2 MRNA Covid-19 Vaccine. N. Engl. J. Med. 2020, 383, 2603–2615. [Google Scholar] [CrossRef]
- Hou, X.; Zaks, T.; Langer, R.; Dong, Y. Lipid Nanoparticles for MRNA Delivery. Nat. Rev. Mater. 2021, 37, 237–246. [Google Scholar] [CrossRef]
- Cheng, Q.; Wei, T.; Farbiak, L.; Johnson, L.T.; Dilliard, S.A.; Siegwart, D.J. Selective Organ Targeting (SORT) Nanoparticles for Tissue-Specific MRNA Delivery and CRISPR–Cas Gene Editing. Nat. Nanotechnol. 2020, 15, 313–320. [Google Scholar] [CrossRef]
- McClorey, G.; Banerjee, S. Cell-Penetrating Peptides to Enhance Delivery of Oligonucleotide-Based Therapeutics. Biomedicines 2018, 6, 51. [Google Scholar] [CrossRef] [PubMed]
- Klabenkova, K.; Fokina, A.; Stetsenko, D. Chemistry of Peptide-Oligonucleotide Conjugates: A Review. Molecules 2021, 26, 5420. [Google Scholar] [CrossRef] [PubMed]
- Lemaitre, M.; Bayard, B.; Lebleu, B. Specific Antiviral Activity of a Poly (L-Lysine)-Conjugated Oligodeoxyribonucleotide Sequence Complementary to Vesicular Stomatitis Virus N Protein MRNA Initiation Site. Proc. Natl. Acad. Sci. USA 1987, 84, 648–652. [Google Scholar] [CrossRef]
- Degols, G.; Leonetti, J.-P.; Gagnor, C.; Lemaitre, M.; Lebleu, B. Antiviral Activity and Possible Mechanisms of Action of Oligonucleotides-Poly (L-Lysine) Conjugates Targeted to Vesicular Stomatitis Virus MRNA and Genomic RNA. Nucleic Acids Res. 1989, 17, 9341–9394. [Google Scholar] [CrossRef] [PubMed]
- Arar, K.; Monsigny, M.; Mayer, R. Synthesis of Oligonucleotide-Peptide Conjugates Containing a KDEL Signal Sequence. Tetrahedron Lett. 1993, 34, 8087–8090. [Google Scholar] [CrossRef]
- Eritja, R.; Pons, A.; Escarcellar, M.; Giralt, E.; Albericio, F. Synthesis of Defined Peptide-Oligonucleotide Hybrids Containing a Nuclear Transport Signal Sequence. Tetrahedron 1991, 47, 4113–4120. [Google Scholar] [CrossRef]
- Gottschling, D.; Seliger, H.; Tarrasón, G.; Piulats, J.; Eritja, R. Synthesis of Oligodeoxynucleotides Containing N 4-Mercaptoethylcytosine and Their Use in the Preparation of Oligonucleotide–Peptide Conjugates Carrying c-Myc Tag-Sequence. Bioconjug. Chem. 1998, 9, 831–837. [Google Scholar] [CrossRef]
- Tung, C.-H.; Stein, S. Preparation and Applications of Peptide− Oligonucleotide Conjugates. Bioconjug. Chem. 2000, 11, 605–618. [Google Scholar] [CrossRef]
- Arar, K.; Aubertin, A.-M.; Roche, A.-C.; Monsigny, M.; Mayer, R. Synthesis and Antiviral Activity of Peptide-Oligonucleotide Conjugates Prepared by Using N. Alpha.-(Bromoacetyl) Peptides. Bioconjug. Chem. 1995, 6, 573–577. [Google Scholar] [CrossRef]
- Reed, M.W.; Fraga, D.; Schwartz, D.E.; Scholler, J.; Hinrichsen, R.D. Synthesis and Evaluation of Nuclear Targeting Peptide-Antisense Oligodeoxyribonucleotide Conjugates. Bioconjug. Chem. 1995, 6, 101–108. [Google Scholar] [CrossRef] [PubMed]
- Zanta, M.A.; Belguise-Valladier, P.; Behr, J.-P. Gene Delivery: A Single Nuclear Localization Signal Peptide Is Sufficient to Carry DNA to the Cell Nucleus. Proc. Natl. Acad. Sci. USA 1999, 96, 91–96. [Google Scholar] [CrossRef] [PubMed]
- de la Torre, B.G.; Aviñó, A.; Tarrason, G.; Piulats, J.; Albericio, F.; Eritja, R. Stepwise Solid-Phase Synthesis of Oligonucleotide-Peptide Hybrids. Tetrahedron Lett. 1994, 35, 2733–2736. [Google Scholar] [CrossRef]
- Truffert, J.-C.; Lorthioir, O.; Asseline, U.; Thuong, N.T.; Brack, A. On-Line Solid Phase Synthesis of Oligonucleotide-Peptide Hybrids Using Silica Supports. Tetrahedron Lett. 1994, 35, 2353–2356. [Google Scholar] [CrossRef]
- Haralambidis, J.; Duncan, L.; Tregear, G.W. The Solid Phase Synthesis of Oligonucleotides Containing a 3′-Peptide Moiety. Tetrahedron Lett. 1987, 28, 5199–5202. [Google Scholar] [CrossRef]
- De Napoli, L.; Messere, A.; Montesarchio, D.; Piccialli, G.; Benedetti, E.; Bucci, E.; Rossi, F. A New Solid-Phase Synthesis of Oligonucleotides 3′-Conjugated with Peptides. Bioorg. Med. Chem. 1999, 7, 395–400. [Google Scholar] [CrossRef] [PubMed]
- Aviñó, A.; Gómara, M.J.; Malakoutikhah, M.; Haro, I.; Eritja, R. Oligonucleotide-Peptide Conjugates: Solid-Phase Synthesis under Acidic Conditions and Use in ELISA Assays. Molecules 2012, 17, 13825–13843. [Google Scholar] [CrossRef]
- de la Torre, B.G.; Albericio, F.; Saison-Behmoaras, E.; Bachi, A.; Eritja, R. Synthesis and Binding Properties of Oligonucleotides Carrying Nuclear Localization Sequences. Bioconjug. Chem. 1999, 10, 1005–1012. [Google Scholar] [CrossRef]
- Antopolsky, M.; Azhayeva, E.; Tengvall, U.; Azhayev, A. Towards a General Method for the Stepwise Solid-Phase Synthesis of Peptide–Oligonucleotide Conjugates. Tetrahedron Lett. 2002, 43, 527–530. [Google Scholar] [CrossRef]
- Debéthune, L.; Marchán, V.; Fàbregas, G.; Pedroso, E.; Grandas, A. Towards Nucleopeptides Containing Any Trifunctional Amino Acid (II). Tetrahedron 2002, 58, 6965–6978. [Google Scholar] [CrossRef]
- Robles, J.; Beltrán, M.; Marchán, V.; Pérez, Y.; Travesset, I.; Pedroso, E.; Grandas, A. Towards Nucleopeptides Containing Any Trifunctional Amino Acid. Tetrahedron 1999, 55, 13251–13264. [Google Scholar] [CrossRef]
- Beltrán, M.; Pedroso, E.; Grandas, A. A Comparison of Histidine Protecting Groups in the Synthesis of Peptide-Oligonucleotide Conjugates. Tetrahedron Lett. 1998, 39, 4115–4118. [Google Scholar] [CrossRef]
- Peyrottes, S.; Mestre, B.; Burlina, F.; Gait, M.J. The Synthesis of Peptide-Oligonucleotide Conjugates by a Fragment Coupling Approach. Tetrahedron 1998, 54, 12513–12522. [Google Scholar] [CrossRef]
- Alam, M.R.; Ming, X.; Fisher, M.; Lackey, J.G.; Rajeev, K.G.; Manoharan, M.; Juliano, R.L. Multivalent Cyclic RGD Conjugates for Targeted Delivery of Small Interfering RNA. Bioconjug. Chem. 2011, 22, 1673–1681. [Google Scholar] [CrossRef]
- Derakhshankhah, H.; Jafari, S. Cell Penetrating Peptides: A Concise Review with Emphasis on Biomedical Applications. Biomed. Pharmacother. 2018, 108, 1090–1096. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.-P.; Zhang, L.-R.; Peng, Y.-F.; Wang, X.-B.; Wang, S.-Q.; Zhang, L.H. A Concise Method for the Preparation of Peptide and Arginine-Rich Peptide-Conjugated Antisense Oligonucleotide. Bioconjug. Chem. 2003, 14, 532–538. [Google Scholar] [CrossRef]
- Grijalvo, S.; Terrazas, M.; Aviñó, A.; Eritja, R. Stepwise Synthesis of Oligonucleotide–Peptide Conjugates Containing Guanidinium and Lipophilic Groups in Their 3′-Termini. Bioorg. Med. Chem. Lett. 2010, 20, 2144–2147. [Google Scholar] [CrossRef]
- Turner, J.J.; Arzumanov, A.A.; Gait, M.J. Synthesis, Cellular Uptake and HIV-1 Tat-Dependent Trans-Activation Inhibition Activity of Oligonucleotide Analogues Disulphide-Conjugated to Cell-Penetrating Peptides. Nucleic Acids Res. 2005, 33, 27–42. [Google Scholar] [CrossRef]
- Astriab-Fisher, A.; Sergueev, D.; Fisher, M.; Shaw, B.R.; Juliano, R.L. Conjugates of Antisense Oligonucleotides with the Tat and Antennapedia Cell-Penetrating Peptides: Effects on Cellular Uptake, Binding to Target Sequences, and Biologic Actions. Pharm. Res. 2002, 19, 744–754. [Google Scholar] [CrossRef]
- Gait, M.J. Peptide-Mediated Cellular Delivery of Antisense Oligonucleotides and Their Analogues. Cell. Mol. Life Sci. C 2003, 60, 844–853. [Google Scholar] [CrossRef]
- Langel, Ü. Cell-Penetrating Peptides and Transportan. Pharmaceutics 2021, 13, 987. [Google Scholar] [CrossRef] [PubMed]
- Wolf, Y.; Pritz, S.; Abes, S.; Bienert, M.; Lebleu, B.; Oehlke, J. Structural Requirements for Cellular Uptake and Antisense Activity of Peptide Nucleic Acids Conjugated with Various Peptides. Biochemistry 2006, 45, 14944–14954. [Google Scholar] [CrossRef]
- Pujals, S.; Giralt, E. Proline-Rich, Amphipathic Cell-Penetrating Peptides. Adv. Drug Deliv. Rev. 2008, 60, 473–484. [Google Scholar] [CrossRef] [PubMed]
- Grijalvo, S.; Eritja, R. Synthesis and in Vitro Inhibition Properties of Oligonucleotide Conjugates Carrying Amphipathic Proline-Rich Peptide Derivatives of the Sweet Arrow Peptide (SAP). Mol. Divers. 2012, 16, 307–317. [Google Scholar] [CrossRef]
- Rhee, M.; Davis, P. Mechanism of Uptake of C105Y, a Novel Cell-Penetrating Peptide. J. Biol. Chem. 2006, 281, 1233–1240. [Google Scholar] [CrossRef] [PubMed]
- Novopashina, D.; Vorobyeva, M.; Nazarov, A.; Davydova, A.; Danilin, N.; Koroleva, L.; Matveev, A.; Bardasheva, A.; Tikunova, N.; Kupryushkin, M. Novel Peptide Conjugates of Modified Oligonucleotides for Inhibition of Bacterial RNase P. Front. Pharmacol. 2019, 10, 813. [Google Scholar] [CrossRef]
- Antopolsky, M.; Azhayeva, E.; Tengvall, U.; Auriola, S.; Jääskeläinen, I.; Rönkkö, S.; Honkakoski, P.; Urtti, A.; Lönnberg, H.; Azhayev, A. Peptide− Oligonucleotide Phosphorothioate Conjugates with Membrane Translocation and Nuclear Localization Properties. Bioconjug. Chem. 1999, 10, 598–606. [Google Scholar] [CrossRef]
- Cooper, R.G.; Harbottle, R.P.; Schneider, H.; Coutelle, C.; Miller, A.D. Peptide Mini-vectors for Gene Delivery. Angew. Chemie Int. Ed. 1999, 38, 1949–1952. [Google Scholar] [CrossRef]
- Mier, W.; Eritja, R.; Mohammed, A.; Haberkorn, U.; Eisenhut, M. Preparation and Evaluation of Tumor-Targeting Peptide− Oligonucleotide Conjugates. Bioconjug. Chem. 2000, 11, 855–860. [Google Scholar] [CrossRef]
- Jirka, S.M.G.; Heemskerk, H.; Tanganyika-de Winter, C.L.; Muilwijk, D.; Pang, K.H.; de Visser, P.C.; Janson, A.; Karnaoukh, T.G.; Vermue, R.; ‘t Hoen, P.A.C. Peptide Conjugation of 2′-O-Methyl Phosphorothioate Antisense Oligonucleotides Enhances Cardiac Uptake and Exon Skipping in Mdx Mice. Nucleic Acid Ther. 2014, 24, 25–36. [Google Scholar] [CrossRef]
- Moschos, S.A.; Jones, S.W.; Perry, M.M.; Williams, A.E.; Erjefalt, J.S.; Turner, J.J.; Barnes, P.J.; Sproat, B.S.; Gait, M.J.; Lindsay, M.A. Lung Delivery Studies Using SiRNA Conjugated to TAT(48−60) and Penetratin Reveal Peptide Induced Reduction in Gene Expression and Induction of Innate Immunity. Bioconjug. Chem. 2007, 18, 1450–1459. [Google Scholar] [CrossRef] [PubMed]
- Kanazawa, T.; Morisaki, K.; Suzuki, S.; Takashima, Y. Prolongation of Life in Rats with Malignant Glioma by Intranasal SiRNA/Drug Codelivery to the Brain with Cell-Penetrating Peptide-Modified Micelles. Mol. Pharm. 2014, 11, 1471–1478. [Google Scholar] [CrossRef] [PubMed]
- Malhotra, M.; Tomaro-Duchesneau, C.; Prakash, S. Synthesis of TAT Peptide-Tagged PEGylated Chitosan Nanoparticles for SiRNA Delivery Targeting Neurodegenerative Diseases. Biomaterials 2013, 34, 1270–1280. [Google Scholar] [CrossRef] [PubMed]
- Abushahba, M.F.N.; Mohammad, H.; Thangamani, S.; Hussein, A.A.A.; Seleem, M.N. Impact of Different Cell Penetrating Peptides on the Efficacy of Antisense Therapeutics for Targeting Intracellular Pathogens. Sci. Rep. 2016, 6, 20832. [Google Scholar] [CrossRef]
- Moulton, H.M.; Hase, M.C.; Smith, K.M.; Iversen, P.L. HIV Tat Peptide Enhances Cellular Delivery of Antisense Morpholino Oligomers. Antisense Nucleic Acid Drug Dev. 2003, 13, 31–43. [Google Scholar] [CrossRef]
- Li, Y.; Li, Y.; Wang, X.; Lee, R.J.; Teng, L. Fatty Acid Modified Octa-Arginine for Delivery of SiRNA. Int. J. Pharm. 2015, 495, 527–535. [Google Scholar] [CrossRef]
- Kim, S.W.; Kim, N.Y.; Choi, Y.B.; Park, S.H.; Yang, J.M.; Shin, S. RNA Interference in Vitro and in Vivo Using an Arginine Peptide/SiRNA Complex System. J. Control. Release 2010, 143, 335–343. [Google Scholar] [CrossRef]
- Meng, J.; Da, F.; Ma, X.; Wang, N.; Wang, Y.; Zhang, H.; Li, M.; Zhou, Y.; Xue, X.; Hou, Z. Antisense Growth Inhibition of Methicillin-Resistant Staphylococcus Aureus by Locked Nucleic Acid Conjugated with Cell-Penetrating Peptide as a Novel FtsZ Inhibitor. Antimicrob. Agents Chemother. 2015, 59, 914–922. [Google Scholar] [CrossRef]
- Burrer, R.; Neuman, B.W.; Ting, J.P.C.; Stein, D.A.; Moulton, H.M.; Iversen, P.L.; Kuhn, P.; Buchmeier, M.J. Antiviral Effects of Antisense Morpholino Oligomers in Murine Coronavirus Infection Models. J. Virol. 2007, 81, 5637–5648. [Google Scholar] [CrossRef]
- Geller, B.L.; Marshall-Batty, K.; Schnell, F.J.; McKnight, M.M.; Iversen, P.L.; Greenberg, D.E. Gene-Silencing Antisense Oligomers Inhibit Acinetobacter Growth in Vitro and in Vivo. J. Infect. Dis. 2013, 208, 1553–1560. [Google Scholar] [CrossRef]
- Liang, S.; He, Y.; Xia, Y.; Wang, H.; Wang, L.; Gao, R.; Zhang, M. Inhibiting the Growth of Methicillin-Resistant Staphylococcus Aureus in Vitro with Antisense Peptide Nucleic Acid Conjugates Targeting the FtsZ Gene. Int. J. Infect. Dis. 2015, 30, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Ma, W.; Lin, Y.; Xuan, W.; Iversen, P.L.; Smith, L.J.; Benchimol, S. Inhibition of P53 Expression by Peptide-Conjugated Phosphorodiamidate Morpholino Oligomers Sensitizes Human Cancer Cells to Chemotherapeutic Drugs. Oncogene 2012, 31, 1024–1033. [Google Scholar] [CrossRef] [PubMed]
- Yin, H.; Moulton, H.M.; Seow, Y.; Boyd, C.; Boutilier, J.; Iverson, P.; Wood, M.J.A. Cell-Penetrating Peptide-Conjugated Antisense Oligonucleotides Restore Systemic Muscle and Cardiac Dystrophin Expression and Function. Hum. Mol. Genet. 2008, 17, 3909–3918. [Google Scholar] [CrossRef] [PubMed]
- Wu, B.; Moulton, H.M.; Iversen, P.L.; Jiang, J.; Li, J.; Li, J.; Spurney, C.F.; Sali, A.; Guerron, A.D.; Nagaraju, K.; et al. Effective Rescue of Dystrophin Improves Cardiac Function in Dystrophin-Deficient Mice by a Modified Morpholino Oligomer. Proc. Natl. Acad. Sci. USA 2008, 105, 14814–14819. [Google Scholar] [CrossRef] [PubMed]
- Leger, A.J.; Mosquea, L.M.; Clayton, N.P.; Wu, I.-H.; Weeden, T.; Nelson, C.A.; Phillips, L.; Roberts, E.; Piepenhagen, P.A.; Cheng, S.H.; et al. Systemic Delivery of a Peptide-Linked Morpholino Oligonucleotide Neutralizes Mutant RNA Toxicity in a Mouse Model of Myotonic Dystrophy. Nucleic Acid Ther. 2013, 23, 109–117. [Google Scholar] [CrossRef] [PubMed]
- Gmeiner, W.H.; Boyacioglu, O.; Stuart, C.H.; Jennings-Gee, J.; Balaji, K.C. The Cytotoxic and Pro-Apoptotic Activities of the Novel Fluoropyrimidine F10 Towards Prostate Cancer Cells Are Enhanced by Zn2+-Chelation and Inhibiting the Serine Protease Omi/HtrA2. Prostate 2015, 75, 360–369. [Google Scholar] [CrossRef]
- Hashida, M.; Kawakami, S.; Yamashita, F. Lipid Carrier Systems for Targeted Drug and Gene Delivery. Chem. Pharm. Bull. 2005, 53, 871–880. [Google Scholar] [CrossRef]
- Echigoya, Y.; Nakamura, A.; Nagata, T.; Urasawa, N.; Lim, K.R.Q.; Trieu, N.; Panesar, D.; Kuraoka, M.; Moulton, H.M.; Saito, T. Effects of Systemic Multiexon Skipping with Peptide-Conjugated Morpholinos in the Heart of a Dog Model of Duchenne Muscular Dystrophy. Proc. Natl. Acad. Sci. USA 2017, 114, 4213–4218. [Google Scholar] [CrossRef]
- Yin, H.; Saleh, A.F.; Betts, C.; Camelliti, P.; Seow, Y.; Ashraf, S.; Arzumanov, A.; Hammond, S.; Merritt, T.; Gait, M.J. Pip5 Transduction Peptides Direct High Efficiency Oligonucleotide-Mediated Dystrophin Exon Skipping in Heart and Phenotypic Correction in Mdx Mice. Mol. Ther. 2011, 19, 1295–1303. [Google Scholar] [CrossRef]
- Betts, C.A.; Saleh, A.F.; Carr, C.A.; Hammond, S.M.; Coenen-Stass, A.M.L.; Godfrey, C.; McClorey, G.; Varela, M.A.; Roberts, T.C.; Clarke, K.; et al. Prevention of Exercised Induced Cardiomyopathy Following Pip-PMO Treatment in Dystrophic Mdx Mice. Sci. Rep. 2015, 5, 8986. [Google Scholar] [CrossRef]
- Godfrey, C.; Muses, S.; McClorey, G.; Wells, K.E.; Coursindel, T.; Terry, R.L.; Betts, C.; Hammond, S.; O’Donovan, L.; Hildyard, J. How Much Dystrophin Is Enough: The Physiological Consequences of Different Levels of Dystrophin in the Mdx Mouse. Hum. Mol. Genet. 2015, 24, 4225–4237. [Google Scholar] [CrossRef] [PubMed]
- Hammond, S.M.; Hazell, G.; Shabanpoor, F.; Saleh, A.F.; Bowerman, M.; Sleigh, J.N.; Meijboom, K.E.; Zhou, H.; Muntoni, F.; Talbot, K. Systemic Peptide-Mediated Oligonucleotide Therapy Improves Long-Term Survival in Spinal Muscular Atrophy. Proc. Natl. Acad. Sci. USA 2016, 113, 10962–10967. [Google Scholar] [CrossRef] [PubMed]
- Betts, C.; Saleh, A.F.; Arzumanov, A.A.; Hammond, S.M.; Godfrey, C.; Coursindel, T.; Gait, M.J.; Wood, M.J.A. Pip6-PMO, a New Generation of Peptide-Oligonucleotide Conjugates with Improved Cardiac Exon Skipping Activity for DMD Treatment. Mol. Ther. Acids 2012, 1, e38. [Google Scholar] [CrossRef] [PubMed]
- Blain, A.M.; Greally, E.; McClorey, G.; Manzano, R.; Betts, C.A.; Godfrey, C.; O’Donovan, L.; Coursindel, T.; Gait, M.J.; Wood, M.J. Peptide-Conjugated Phosphodiamidate Oligomer-Mediated Exon Skipping Has Benefits for Cardiac Function in Mdx and Cmah-/-Mdx Mouse Models of Duchenne Muscular Dystrophy. PLoS ONE 2018, 13, e0198897. [Google Scholar] [CrossRef]
- Tsoumpra, M.K.; Fukumoto, S.; Matsumoto, T.; Takeda, S.; Wood, M.J.A.; Aoki, Y. Peptide-Conjugate Antisense Based Splice-Correction for Duchenne Muscular Dystrophy and Other Neuromuscular Diseases. EBioMedicine 2019, 45, 630–645. [Google Scholar] [CrossRef]
- Crombez, L.; Morris, M.C.; Dufort, S.; Aldrian-Herrada, G.; Nguyen, Q.; Mc Master, G.; Coll, J.-L.; Heitz, F.; Divita, G. Targeting Cyclin B1 through Peptide-Based Delivery of SiRNA Prevents Tumour Growth. Nucleic Acids Res. 2009, 37, 4559–4569. [Google Scholar] [CrossRef]
- Morris, M.C.; Vidal, P.; Chaloin, L.; Heitz, F.; Divita, G. A New Peptide Vector for Efficient Delivery of Oligonucleotides into Mammalian Cells. Nucleic Acids Res. 1997, 25, 2730–2736. [Google Scholar] [CrossRef]
- Bartz, R.; Fan, H.; Zhang, J.; Innocent, N.; Cherrin, C.; Beck, S.C.; Pei, Y.; Momose, A.; Jadhav, V.; Tellers, D.M. Effective SiRNA Delivery and Target MRNA Degradation Using an Amphipathic Peptide to Facilitate PH-Dependent Endosomal Escape. Biochem. J. 2011, 435, 475–487. [Google Scholar] [CrossRef]
- Crombez, L.; Aldrian-Herrada, G.; Konate, K.; Nguyen, Q.N.; McMaster, G.K.; Brasseur, R.; Heitz, F.; Divita, G. A New Potent Secondary Amphipathic Cell–Penetrating Peptide for SiRNA Delivery into Mammalian Cells. Mol. Ther. 2009, 17, 95–103. [Google Scholar] [CrossRef]
- Chen, Z.; Nie, D.; Hu, Y.; Li, M.; Hou, Z.; Mao, X.; Luo, X.; Xue, X. Efficient Delivery of Antisense Oligonucleotides by an Amphipathic Cell-Penetrating Peptide in Acinetobacter Baumannii. Curr. Drug Deliv. 2019, 16, 728–736. [Google Scholar] [CrossRef]
- Vaissière, A.; Aldrian, G.; Konate, K.; Lindberg, M.F.; Jourdan, C.; Telmar, A.; Seisel, Q.; Fernandez, F.; Viguier, V.; Genevois, C. A Retro-Inverso Cell-Penetrating Peptide for SiRNA Delivery. J. Nanobiotechnol. 2017, 15, 34. [Google Scholar] [CrossRef] [PubMed]
- Cantini, L.; Attaway, C.C.; Butler, B.; Andino, L.M.; Sokolosky, M.L.; Jakymiw, A. Fusogenic-Oligoarginine Peptide-Mediated Delivery of SiRNAs Targeting the CIP2A Oncogene into Oral Cancer Cells. PLoS ONE 2013, 8, e73348. [Google Scholar] [CrossRef] [PubMed]
- Alexander-Bryant, A.A.; Zhang, H.; Attaway, C.C.; Pugh, W.; Eggart, L.; Sansevere, R.M.; Andino, L.M.; Dinh, L.; Cantini, L.P.; Jakymiw, A. Dual Peptide-Mediated Targeted Delivery of Bioactive SiRNAs to Oral Cancer Cells in Vivo. Oral Oncol. 2017, 72, 123–131. [Google Scholar] [CrossRef] [PubMed]
- El Andaloussi, S.; Lehto, T.; Mäger, I.; Rosenthal-Aizman, K.; Oprea, I.I.; Simonson, O.E.; Sork, H.; Ezzat, K.; Copolovici, D.M.; Kurrikoff, K. Design of a Peptide-Based Vector, PepFect6, for Efficient Delivery of SiRNA in Cell Culture and Systemically in Vivo. Nucleic Acids Res. 2011, 39, 3972–3987. [Google Scholar] [CrossRef]
- Aviñó, A.; Ocampo, S.M.; Caminal, C.; Perales, J.C.; Eritja, R. Stepwise Synthesis of RNA Conjugates Carrying Peptide Sequences for RNA Interference Studies. Mol. Divers. 2009, 13, 287–293. [Google Scholar] [CrossRef]
- Chiu, Y.-L.; Ali, A.; Chu, C.; Cao, H.; Rana, T.M. Visualizing a Correlation between SiRNA Localization, Cellular Uptake, and RNAi in Living Cells. Chem. Biol. 2004, 11, 1165–1175. [Google Scholar] [CrossRef]
- Moschos, S.A.; Williams, A.E.; Lindsay, M.A. Cell-Penetrating-Peptide-Mediated SiRNA Lung Delivery. Biochem. Soc. Trans. 2007, 35, 807–810. [Google Scholar] [CrossRef]
- Turner, J.J.; Jones, S.; Fabani, M.M.; Ivanova, G.; Arzumanov, A.A.; Gait, M.J. RNA Targeting with Peptide Conjugates of Oligonucleotides, SiRNA and PNA. Blood Cells Mol. Dis. 2007, 38, 1–7. [Google Scholar] [CrossRef]
- Muratovska, A.; Eccles, M.R. Conjugate for Efficient Delivery of Short Interfering RNA (SiRNA) into Mammalian Cells. FEBS Lett. 2004, 558, 63–68. [Google Scholar] [CrossRef]
- Stetsenko, D.A.; Gait, M.J. Efficient Conjugation of Peptides to Oligonucleotides by “Native Ligation. ” J. Org. Chem. 2000, 65, 4900–4908. [Google Scholar] [CrossRef]
- Singh, Y.; Defrancq, E.; Dumy, P. New Method to Prepare Peptide− Oligonucleotide Conjugates through Glyoxylic Oxime Formation. J. Org. Chem. 2004, 69, 8544–8546. [Google Scholar] [CrossRef]
- Forget, D.; Boturyn, D.; Defrancq, E.; Lhomme, J.; Dumy, P. Highly Efficient Synthesis of Peptide–Oligonucleotide Conjugates: Chemoselective Oxime and Thiazolidine Formation. Chem. Eur. J. 2001, 7, 3976–3984. [Google Scholar] [CrossRef] [PubMed]
- Ollivier, N.; Olivier, C.; Gouyette, C.; Huynh-Dinh, T.; Gras-Masse, H.; Melnyk, O. Synthesis of Oligonucleotide–Peptide Conjugates Using Hydrazone Chemical Ligation. Tetrahedron Lett. 2002, 43, 997–999. [Google Scholar] [CrossRef]
- Marchan, V.; Ortega, S.; Pulido, D.; Pedroso, E.; Grandas, A. Diels-Alder Cycloadditions in Water for the Straightforward Preparation of Peptide–Oligonucleotide Conjugates. Nucleic Acids Res. 2006, 34, e24. [Google Scholar] [CrossRef] [PubMed]
- Steven, V.; Graham, D. Oligonucleotide Conjugation to a Cell-Penetrating (TAT) Peptide by Diels–Alder Cycloaddition. Org. Biomol. Chem. 2008, 6, 3781–3787. [Google Scholar] [CrossRef]
- Gandioso, A.; Massaguer, A.; Villegas, N.; Salvans, C.; Sánchez, D.; Brun-Heath, I.; Marchán, V.; Orozco, M.; Terrazas, M. Efficient SiRNA–Peptide Conjugation for Specific Targeted Delivery into Tumor Cells. Chem. Commun. 2017, 53, 2870–2873. [Google Scholar] [CrossRef]
- Honcharenko, M.; Honcharenko, D.; Stromberg, R. Efficient Conjugation to Phosphorothioate Oligonucleotides by Cu-Catalyzed Huisgen 1, 3-Dipolar Cycloaddition. Bioconjug. Chem. 2019, 30, 1622–1628. [Google Scholar] [CrossRef] [PubMed]
- Wenska, M.; Alvira, M.; Steunenberg, P.; Stenberg, Å.; Murtola, M.; Strömberg, R. An Activated Triple Bond Linker Enables ‘Click’Attachment of Peptides to Oligonucleotides on Solid Support. Nucleic Acids Res. 2011, 39, 9047–9059. [Google Scholar] [CrossRef]
- Lonnberg, H. Solid-Phase Synthesis of Oligonucleotide Conjugates Useful for Delivery and Targeting of Potential Nucleic Acid Therapeutics. Bioconjug. Chem. 2009, 20, 1065–1094. [Google Scholar] [CrossRef]
- Lu, K.; Duan, Q.-P.; Ma, L.; Zhao, D.-X. Chemical Strategies for the Synthesis of Peptide− Oligonucleotide Conjugates. Bioconjug. Chem. 2010, 21, 187–202. [Google Scholar] [CrossRef]
- Marlin, F.; Simon, P.; Saison-Behmoaras, T.; Giovannangeli, C. Delivery of Oligonucleotides and Analogues: The Oligonucleotide Conjugate-Based Approach. Chembiochem 2010, 11, 1493–1500. [Google Scholar] [CrossRef] [PubMed]
- Terashima, T.; Ogawa, N.; Nakae, Y.; Sato, T.; Katagi, M.; Okano, J.; Maegawa, H.; Kojima, H. Gene Therapy for Neuropathic Pain through SiRNA-IRF5 Gene Delivery with Homing Peptides to Microglia. Mol. Ther. Acids 2018, 11, 203–215. [Google Scholar] [CrossRef] [PubMed]
- Terashima, T.; Ogawa, N.; Sato, T.; Katagi, M.; Nakae, Y.; Okano, J.; Maegawa, H.; Kojima, H. Advanced Technology for Gene Delivery with Homing Peptides to Spinal Cord through Systemic Circulation in Mice. Mol. Ther. Clin. Dev. 2019, 13, 474–483. [Google Scholar] [CrossRef] [PubMed]
- Klein, D.; Goldberg, S.; Theile, C.S.; Dambra, R.; Haskell, K.; Kuhar, E.; Lin, T.; Parmar, R.; Manoharan, M.; Richter, M. Centyrin Ligands for Extrahepatic Delivery of SiRNA. Mol. Ther. 2021, 29, 2053–2066. [Google Scholar] [CrossRef]
- Lim, K.R.Q.; Woo, S.; Melo, D.; Huang, Y.; Dzierlega, K.; Shah, M.N.A.; Aslesh, T.; Roshmi, R.R.; Echigoya, Y.; Maruyama, R. Development of DG9 Peptide-Conjugated Single-and Multi-Exon Skipping Therapies for the Treatment of Duchenne Muscular Dystrophy. Proc. Natl. Acad. Sci. USA 2022, 119, e2112546119. [Google Scholar] [CrossRef]
- Gait, M.J.; Arzumanov, A.A.; McClorey, G.; Godfrey, C.; Betts, C.; Hammond, S.; Wood, M.J.A. Cell-Penetrating Peptide Conjugates of Steric Blocking Oligonucleotides as Therapeutics for Neuromuscular Diseases from a Historical Perspective to Current Prospects of Treatment. Nucleic Acid Ther. 2019, 29, 1–12. [Google Scholar] [CrossRef]
- Sheikh, O.; Yokota, T. Pharmacology and Toxicology of Eteplirsen and SRP-5051 for DMD Exon 51 Skipping: An Update. Arch. Toxicol. 2022, 96, 1–9. [Google Scholar] [CrossRef]
- Wojciechowska, M.; Równicki, M.; Mieczkowski, A.; Miszkiewicz, J.; Trylska, J. Antibacterial Peptide Nucleic Acids—Facts and Perspectives. Molecules 2020, 25, 559. [Google Scholar] [CrossRef]
- Good, L.; Awasthi, S.K.; Dryselius, R.; Larsson, O.; Nielsen, P.E. Bactericidal Antisense Effects of Peptide–PNA Conjugates. Nat. Biotechnol. 2001, 19, 360–364. [Google Scholar] [CrossRef]
- Honcharenko, D.; Druceikaite, K.; Honcharenko, M.; Bollmark, M.; Tedebark, U.; Strö;mberg, R. New Alkyne and Amine Linkers for Versatile Multiple Conjugation of Oligonucleotides. ACS Omega 2020, 6, 579–593. [Google Scholar] [CrossRef]
- Martín-Nieves, V.; Fabrega, C.; Guasch, M.; Fernandez, S.; Sanghvi, Y.S.; Ferrero, M.; Eritja, R. Oligonucleotides Containing 1-Aminomethyl or 1-Mercaptomethyl-2-Deoxy-d-Ribofuranoses: Synthesis, Purification, Characterization, and Conjugation with Fluorophores and Lipids. Bioconjug. Chem. 2021, 32, 350–366. [Google Scholar] [CrossRef] [PubMed]
- Grijalvo, S.; Alagia, A.; Gargallo, R.; Eritja, R. Cellular Uptake Studies of Antisense Oligonucleotides Using G-Quadruplex-Nanostructures. The Effect of Cationic Residue on the Biophysical and Biological Properties. RSC Adv. 2016, 6, 76099–76109. [Google Scholar] [CrossRef]
- Grijalvo, S.; Clua, A.; Eres, M.; Gargallo, R.; Eritja, R. Tuning G-Quadruplex Nanostructures with Lipids. Towards Designing Hybrid Scaffolds for Oligonucleotide Delivery. Int. J. Mol. Sci. 2020, 22, 121. [Google Scholar] [CrossRef] [PubMed]
- Clua, A.; Fàbrega, C.; García-Chica, J.; Grijalvo, S.; Eritja, R. Parallel G-Quadruplex Structures Increase Cellular Uptake and Cytotoxicity of 5-Fluoro-2′-Deoxyuridine Oligomers in 5-Fluorouracil Resistant Cells. Molecules 2021, 26, 1741. [Google Scholar] [CrossRef]
- Lee, H.; Lytton-Jean, A.K.R.; Chen, Y.; Love, K.T.; Park, A.I.; Karagiannis, E.D.; Sehgal, A.; Querbes, W.; Zurenko, C.S.; Jayaraman, M. Molecularly Self-Assembled Nucleic Acid Nanoparticles for Targeted in Vivo SiRNA Delivery. Nat. Nanotechnol. 2012, 7, 389–393. [Google Scholar] [CrossRef]
- Jorge, A.F.; Aviñó, A.; Pais, A.A.C.C.; Eritja, R.; Fàbrega, C. DNA-Based Nanoscaffolds as Vehicles for 5-Fluoro-2′-Deoxyuridine Oligomers in Colorectal Cancer Therapy. Nanoscale 2018, 10, 7238–7249. [Google Scholar] [CrossRef]
- Lacroix, A.; Edwardson, T.G.W.; Hancock, M.A.; Dore, M.D.; Sleiman, H.F. Development of DNA Nanostructures for High-Affinity Binding to Human Serum Albumin. J. Am. Chem. Soc. 2017, 139, 7355–7362. [Google Scholar] [CrossRef]
- Nakagawa, O.; Ming, X.; Carver, K.; Juliano, R. Conjugation with Receptor-Targeted Histidine-Rich Peptides Enhances the Pharmacological Effectiveness of Antisense Oligonucleotides. Bioconjug. Chem. 2014, 25, 165–170. [Google Scholar] [CrossRef]
- Taskova, M.; Madsen, C.S.; Jensen, K.J.; Hansen, L.H.; Vester, B.; Astakhova, K. Antisense Oligonucleotides Internally Labeled with Peptides Show Improved Target Recognition and Stability to Enzymatic Degradation. Bioconjug. Chem. 2017, 28, 768–774. [Google Scholar] [CrossRef]
- Datta, D.; Mori, S.; Madaoui, M.; Wassarman, K.; Zlatev, I.; Manoharan, M. Aminooxy Click Chemistry as a Tool for Bis-Homo and Bis-Hetero Ligand Conjugation to Nucleic Acids. Org. Lett. 2022, 24, 4496–4501. [Google Scholar] [CrossRef]
- Lou, C.; Christensen, N.J.; Martos-Maldonado, M.C.; Midtgaard, S.R.; Ejlersen, M.; Thulstrup, P.W.; Sørensen, K.K.; Jensen, K.J.; Wengel, J. Folding Topology of a Short Coiled-coil Peptide Structure Templated by an Oligonucleotide Triplex. Chem. Eur. J. 2017, 23, 9297–9305. [Google Scholar] [CrossRef] [PubMed]
- Riccardi, C.; Fàbrega, C.; Grijalvo, S.; Vitiello, G.; D’Errico, G.; Eritja, R.; Montesarchio, D. AS1411-Decorated Niosomes as Effective Nanocarriers for Ru( <scp>iii</Scp> )-Based Drugs in Anticancer Strategies. J. Mater. Chem. B 2018, 6, 5368–5384. [Google Scholar] [PubMed]
- Carvalho, J.; Lopes-Nunes, J.; Vialet, B.; Rosado, T.; Gallardo, E.; Vale, J.; Eloy, C.; Ferreira, S.; Palmeira-de-Oliveira, R.; Campello, M.P.C. Nanoaggregate-Forming Lipid-Conjugated AS1411 Aptamer as a Promising Tumor-Targeted Delivery System of Anticancer Agents in Vitro. Nanomed. Nanotechnol. Biol. Med. 2021, 36, 102429. [Google Scholar] [CrossRef] [PubMed]
- Ghosh, P.S.; Hamilton, A.D. Noncovalent Template-Assisted Mimicry of Multiloop Protein Surfaces: Assembling Discontinuous and Functional Domains. J. Am. Chem. Soc. 2012, 134, 13208–13211. [Google Scholar] [CrossRef] [PubMed]
- Bujold, K.E.; Lacroix, A.; Sleiman, H.F. DNA Nanostructures at the Interface with Biology. Chem 2018, 4, 495–521. [Google Scholar] [CrossRef]
- Lacroix, A.; Sleiman, H.F. DNA Nanostructures: Current Challenges and Opportunities for Cellular Delivery. ACS Nano 2021, 15, 3631–3645. [Google Scholar] [CrossRef] [PubMed]
- Jorge, A.F.; Eritja, R. Overview of DNA Self-Assembling: Progresses in Biomedical Applications. Pharmaceutics 2018, 10, 268. [Google Scholar] [CrossRef]
- Goodman, R.P.; Berry, R.M.; Turberfield, A.J. The Single-Step Synthesis of a DNA Tetrahedron. Chem. Commun. 2004, 2004, 1372–1373. [Google Scholar] [CrossRef]
- Copp, W.; Pontarelli, A.; Wilds, C.J. Recent Advances of DNA Tetrahedra for Therapeutic Delivery and Biosensing. ChemBioChem 2021, 22, 2237–2246. [Google Scholar] [CrossRef]
- Yan, J.; Chen, J.; Zhang, N.; Yang, Y.; Zhu, W.; Li, L.; He, B. Mitochondria-Targeted Tetrahedral DNA Nanostructures for Doxorubicin Delivery and Enhancement of Apoptosis. J. Mater. Chem. B 2020, 8, 492–503. [Google Scholar] [CrossRef]
- Schaffert, D.H.; Okholm, A.H.; Sørensen, R.S.; Nielsen, J.S.; Tørring, T.; Rosen, C.B.; Kodal, A.L.B.; Mortensen, M.R.; Gothelf, K.V.; Kjems, J. Intracellular Delivery of a Planar DNA Origami Structure by the Transferrin-receptor Internalization Pathway. Small 2016, 12, 2634–2640. [Google Scholar] [CrossRef]
- Pascual-Gilabert, M.; López-Castel, A.; Artero, R. Myotonic Dystrophy Type 1 Drug Development: A Pipeline toward the Market. Drug Discov. Today 2021, 26, 1765–1772. [Google Scholar] [CrossRef] [PubMed]
- Herbert, B.-S.; Gellert, G.C.; Hochreiter, A.; Pongracz, K.; Wright, W.E.; Zielinska, D.; Chin, A.C.; Harley, C.B.; Shay, J.W.; Gryaznov, S.M. Lipid Modification of GRN163, an N3′→ P5′ Thio-Phosphoramidate Oligonucleotide, Enhances the Potency of Telomerase Inhibition. Oncogene 2005, 24, 5262–5268. [Google Scholar] [CrossRef] [PubMed]
- Tran, P.; Weldemichael, T.; Liu, Z.; Li, H. Delivery of Oligonucleotides: Efficiency with Lipid Conjugation and Clinical Outcome. Pharmaceutics 2022, 14, 342. [Google Scholar] [CrossRef] [PubMed]
- Yuen, M.F.; Schiefke, I.; Yoon, J.H.; Ahn, S.H.; Heo, J.; Kim, J.H. RNA Interference Therapy with ARC-520 Results in Prolonged HBsAg Response in Patients with Chronic Hepatitis B Infection. Hepatology 2020, 72, 19–31. [Google Scholar] [CrossRef] [PubMed]
| Name | Sequence | Type | Application | Ref. |
|---|---|---|---|---|
| Hydrophobic | ||||
| P4 | LGAQSNF | ASO (PS) | SSO(2OMe) for DMD | [140] |
| Polycationic | ||||
| TAT | RKKRRQRRR | siRNA | Neurodegenerative pathologies Lung and Malignant glioma | [141,142,143] |
| ASO (PNA/PMO) | bacteria & nematodecell lines | [144,145] | ||
| pAnt | RQIKIWFQNRRMKWKKGGC | ASO (PNA/PS) | bacteria & nematodecell lines | [129,144] |
| PolyArg | Rn (n = 6–18) optimal 8 to 10 | siRNA | Neurons and different cancers | [146,147] |
| (KFF)3K | KFFKFFKFFK | ASO (PNA/LNA) | Anti-bacterial | [144,148] |
| (RXR)4 | RXRRXRRXRRXRXR | ASO (PMO) | Anti-viral anti-bacterial | [144,149,150,151,152] |
| B peptide | (RXRRBR)2XB | ASO (PMO) | SSO of DMD DM1 | [153,154,155,156,157,158] |
| Pip5e | RXRRBRRXR-ILFQY-RXRBRXRB | ASO (PMO) | SSO of DMD, SMA | [159] |
| Pip6a | RXRRBRRXR-YQFLI-RXRBRXRB | ASO (PMO) | SSO of DMD, SMA | [160,161,162,163] |
| Pip6b | RXRRBRRXR-IQFLI-RXRBRXRB | ASO (PMO) | SSO of DMD, SMA | [164,165] |
| Amphipathic | ||||
| MPG-8 | GALFLGFLGAAGSTMGAWSQPKKKRK | siRNA | Xenograft tumor model | [166] |
| ASO | Mammalian cells | [167] | ||
| Pep-1 | KETWWETWWTEWSQPKKRK | siRNA | Cells | [168] |
| CADY | GLWRALWRLLRSLWRLLWRA | siRNA | Several cancer cells | [169] |
| ASO | Anti-bacterial | [170] | ||
| RICK | KWLLRWLSRLLRWLARWLG | siRNA | H glioblastoma cells | [171] |
| 599 | GLFEAIEGFIENGWEGMIDGWY(G)4(R)9K | siRNA | Oral cancer | [172,173] |
| Pepfect6 | AGYLLGK(ε-Mtt)INLKALAALAKKIL | siRNA | Cell lines (various) | [174] |
| Therapeutic Target/Disease | Name | Conjugate | Sponsor | Status |
|---|---|---|---|---|
| DMD, Exon51 | SRP-5051 | Peptide-PMO | Sarepta therapeutics | Phase II |
| DMD, Exon 51 | PGN-EDO51 | Peptide EDO-PMO | PepGen | Phase I |
| DMD, Exon 53 | PGN-EDO53 | Peptide EDO-PMO | PepGen | preclinical |
| DM1, DMPK | PGN-EDODM1 | Peptide EDO-PMO | PepGen | preclinical |
| DMD, Exon 44 | ENTR-601-44 | Peptide-PMO | Entrada therapeutics | precilinal |
| DMD, Exon 44 | ENTR-601-45 | Peptide-PMO | Entrada therapeutics | precilinal |
| DM1 | ENTR-701-CUG | Peptide EEV-PMO | Entrada therapeutics | precilinal |
| DM1, MBNL1 | Pip6a-PMI-CAG7 | CPP peptide-PMO | Oxford University | preclinical |
| Telomerase | GRN163-L, imetelstat | Palmitate-ASO | Geron Corporation | Phase III |
| Chronic Hepatitis B | ARC-520-HBV | Two cholesterol-siRNAs | Arrowhead pharmaceuticals | Phase II |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fàbrega, C.; Aviñó, A.; Navarro, N.; Jorge, A.F.; Grijalvo, S.; Eritja, R. Lipid and Peptide-Oligonucleotide Conjugates for Therapeutic Purposes: From Simple Hybrids to Complex Multifunctional Assemblies. Pharmaceutics 2023, 15, 320. https://doi.org/10.3390/pharmaceutics15020320
Fàbrega C, Aviñó A, Navarro N, Jorge AF, Grijalvo S, Eritja R. Lipid and Peptide-Oligonucleotide Conjugates for Therapeutic Purposes: From Simple Hybrids to Complex Multifunctional Assemblies. Pharmaceutics. 2023; 15(2):320. https://doi.org/10.3390/pharmaceutics15020320
Chicago/Turabian StyleFàbrega, Carme, Anna Aviñó, Natalia Navarro, Andreia F. Jorge, Santiago Grijalvo, and Ramon Eritja. 2023. "Lipid and Peptide-Oligonucleotide Conjugates for Therapeutic Purposes: From Simple Hybrids to Complex Multifunctional Assemblies" Pharmaceutics 15, no. 2: 320. https://doi.org/10.3390/pharmaceutics15020320
APA StyleFàbrega, C., Aviñó, A., Navarro, N., Jorge, A. F., Grijalvo, S., & Eritja, R. (2023). Lipid and Peptide-Oligonucleotide Conjugates for Therapeutic Purposes: From Simple Hybrids to Complex Multifunctional Assemblies. Pharmaceutics, 15(2), 320. https://doi.org/10.3390/pharmaceutics15020320