Non-Covalent Linkage of Helper Functions to Dumbbell-Shaped DNA Vectors for Targeted Delivery
Abstract
:1. Introduction
2. Material and Methods
2.1. Plasmid Construction
2.2. Oligonucleotides
2.3. Dumbbell Vector Production
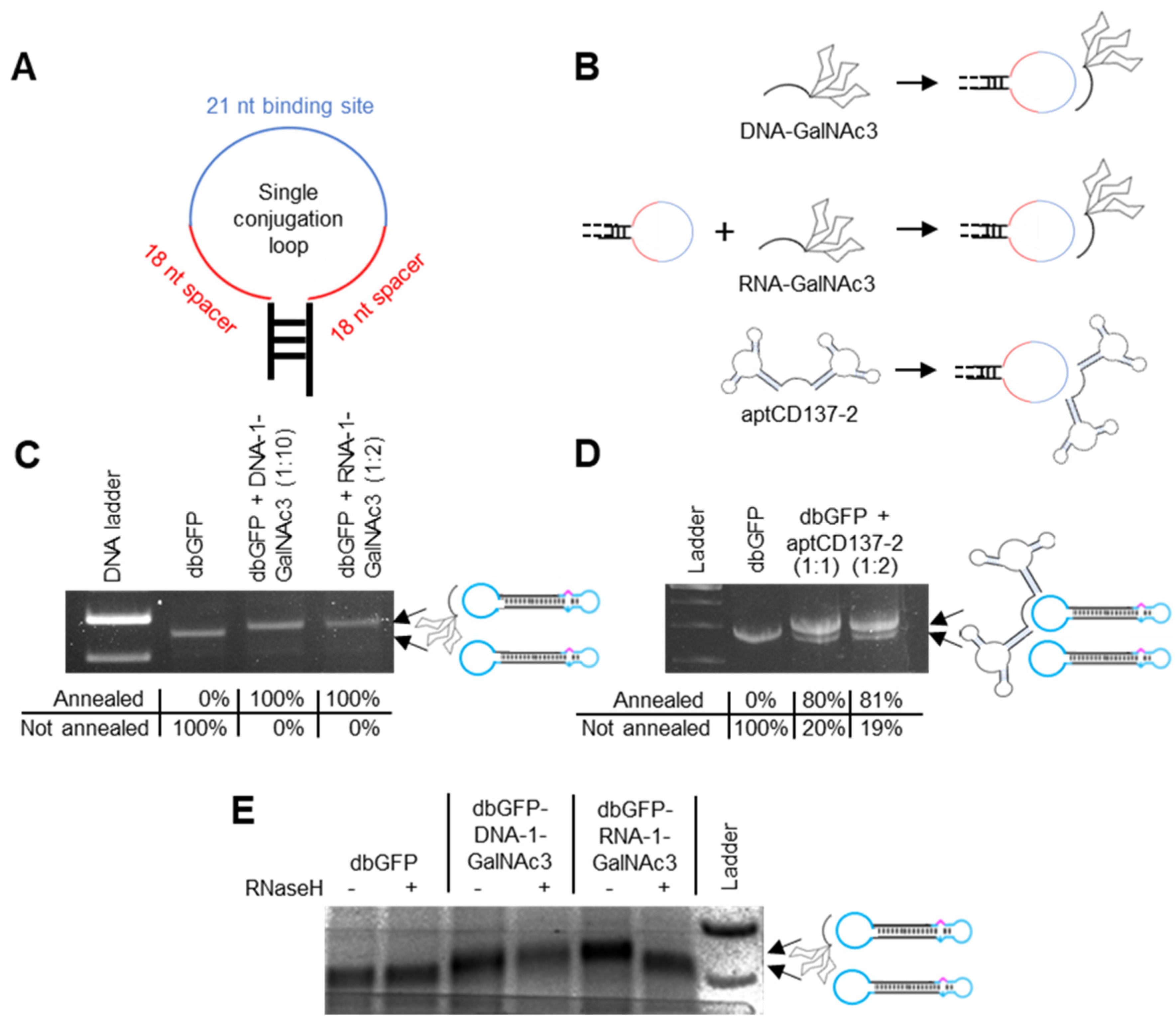
3. Formation of Dumbbell Conjugates
3.1. Dumbbell-GalNAc3-Conjugates
3.2. Dumbbell-aptCD137-2-Conjugates
4. Tissue Cell Culture
5. Uptake of Dumbbell-GalNAc3-Conjugates from the Tissue Cell Culture Medium
5.1. Uptake of MaxGFP Dumbbell-GalNAc3-Conjugates
5.2. Uptake of HSVtk Dumbbell-GalNAc3 or -2GalNAc3-Conjugates
6. qPCR Quantification of Uptaken MaxGFP Dumbbell-GalNAc3 DNA
7. Lipofection of HepG2 Cells
8. Flow Cytometry
9. Preparation of Ganciclovir Working Stock
10. AlamarBlue® Cell Viability Assay
Statistical Analysis
11. Results
Increasing the Size of One Dumbbell Vector Loop Does Not Impair Gene Expression
12. Non-Covalent Conjugation of Dumbbell Vector DNA with GalNAc3 and aptCD137-2 Residues via Complementary Base Pairing
13. GalNAc3-RNA but Not -DNA Linkers Are Cleavable by RNaseH
14. Dumbbell-GalNAc3 Conjugates Are Taken up by Hepatoblastoma-Derived Human Tissue Culture Cells Triggering MaxGFP Expression
15. HSVtk Expressing Dumbbells Featured with Two GalNAc3 Residues at One Conjugation Loop Triggered Death of Hepatoblastoma-Derived Human Tissue Culture Cells upon Ganciclovir Treatment
16. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Kay, M.A. State-of-the-art gene-based therapies: The road ahead. Nat. Rev. Genet. 2011, 12, 316–328. [Google Scholar] [CrossRef] [PubMed]
- Mok, P.L.; Cheong, S.K.; Leong, C.F.; Chua, K.H.; Ainoon, O. Extended and stable gene expression via nucleofection of MIDGE construct into adult human marrow mesenchymal stromal cells. Cytotechnology 2012, 64, 203–216. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kaur, T.; Slavcev, R.A.; Wettig, S.D. Addressing the Challenge: Current and Future Directions in Ovarian Cancer Therapy. Curr. Gene Ther. 2009, 9, 434–458. [Google Scholar] [CrossRef] [PubMed]
- López-Fuertes, L.; Pérez-Jiménez, E.; Vila-Coro, A.J.; Sack, F.; Moreno, S.; Konig, S.; Junghans, C.; Wittig, B.; Timón, M.; Esteban, M. DNA vaccination with linear minimalistic (MIDGE) vectors confers protection against Leishmania major infection in mice. Vaccine 2002, 21, 247–257. [Google Scholar] [CrossRef] [PubMed]
- Schakowski, F.; Gorschlüter, M.; Junghans, C.; Schroff, M.; Buttgereit, P.; Ziske, C.; Schöttker, B.; König-Merediz, S.A.; Sauerbruch, T.; Wittig, B.; et al. A Novel Minimal-Size Vector (MIDGE) Improves Transgene Expression in Colon Carcinoma Cells and Avoids Transfection of Undesired DNA. Mol. Ther. 2001, 3, 793–800. [Google Scholar] [CrossRef] [PubMed]
- Zanta, M.A.; Belguise-Valladier, P.; Behr, J.-P. Gene delivery: A single nuclear localization signal peptide is sufficient to carry DNA to the cell nucleus. Proc. Natl. Acad. Sci. USA 1999, 96, 91–96. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jiang, X.; Yu, H.; Teo, C.R.; Tan, G.; Goh, S.C.; Patel, P.; Chua, Y.K.; Hameed, N.B.S.; Bertoletti, B.; Patzel, V. Advanced design of dumbbell-shaped genetic minimal vectors improves non-coding and coding RNA expression. Mol. Ther. 2016, 24, 1581–1591. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zogg, H.; Singh, R.; Ro, S. Current Advances in RNA Therapeutics for Human Diseases. Int. J. Mol. Sci. 2022, 23, 2736. [Google Scholar] [CrossRef] [PubMed]
- Dassie, J.P.; Liu, X.; Thomas, G.S.; Whitaker, R.M.; Thiel, K.W.; Stockdale, K.R.; Meyerholz, D.K.; McCaffrey, A.P.; McNamara, J.O.; Giangrande, P.H. Systemic administration of optimized aptamer-siRNA chimeras promotes regression of PSMA-expressing tumors. Nat. Biotechnol. 2009, 27, 839–849. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, J.; Li, H.; Zhang, J.; Piotr, S.; Rossi, J. Development of Cell-type specific anti-HIV gp120 aptamers for siRNA delivery. J. Vis. Exp. 2011, 52, 2954. [Google Scholar] [CrossRef] [PubMed]
- Kawaguchi, K.; Kuhlenschmidt, M.; Roseman, S.; Lee, Y.C. Synthesis of some cluster galactosides and their effect on the hepatic galactose binding system. Arch. Biochem. Biophys. 1980, 205, 388–395. [Google Scholar] [CrossRef] [PubMed]
- Pasto, F.; Kolonias, D.; McNamara, J.O.; Gilboa, E. Targeting 4-1BB costimulation to disseminated tumor lesions with bi-specific oligonucleotide aptamers. Mol. Therapy. 2011, 19, 1878–1886. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Poddar, S.; Loh, P.S.; Ooi, Z.H.; Osman, F.; Eul, J.; Patzel, V. RNA structure design improves activity and specificity of trans-splicing triggered cell death in a suicide gene therapy approach. Mol. Ther. Nucleic Acids 2018, 11, 41–56. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yu, H.; Jiang, X.; Tan, K.T.; Hang, L.; Patzel, V. Efficient Production of Superior Dumbbell-Shaped DNA Minimal Vectors for Small Hairpin RNA Expression. Nucleic Acids Res. 2015, 43, e120. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cost, G.J. Enzymatic ligation assisted by nucleases: Simultaneous ligation and digestion promote the ordered assembly of DNA. Nat. Protoc. 2007, 2, 2198–2202. [Google Scholar] [CrossRef] [PubMed]
- Taki, M.; Kato, Y.; Miyagishi, M.; Takagi, Y.; Taira, K. Small-Interfering-RNA Expression in Cells Based on an Efficiently Constructed Dumbbell-Shaped DNA. Angew. Chem. Int. Ed. 2004, 43, 3160–3163. [Google Scholar] [CrossRef] [PubMed]
- Taki, M.; Kato, Y.; Miyagishi, M.; Takagi, Y.; Sano, M.; Taira, K.A. Direct and efficient synthesis method for dumbell-shaped linear DNA using PCR in vitro. Nucleic Acids Symp. Ser. 2003, 3, 191. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Loh, P.S.; Healthy Longevity Translational Research Programme and Department of Microbiology and Immunology, Yong Loo Lin School of Medicine, National University of Singapore, Singapore; Patzel, V.; Healthy Longevity Translational Research Programme and Department of Microbiology and Immunology, Yong Loo Lin School of Medicine, National University of Singapore, Singapore. Fluorescence microscopy images. Unpublished work, 2022. [Google Scholar]
- Touraine, R.L.; Ishii-Morita, H.; Ramsey, W.J.; Blaese, R.M. The bystander effect in the HSVtk/ganciclovir system and its relationship to gap junctional communication. Gene Ther. 1998, 5, 1705–1711. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zuker, M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003, 31, 3406–3415. [Google Scholar] [CrossRef] [PubMed]




Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Loh, P.S.; Patzel, V. Non-Covalent Linkage of Helper Functions to Dumbbell-Shaped DNA Vectors for Targeted Delivery. Pharmaceutics 2023, 15, 370. https://doi.org/10.3390/pharmaceutics15020370
Loh PS, Patzel V. Non-Covalent Linkage of Helper Functions to Dumbbell-Shaped DNA Vectors for Targeted Delivery. Pharmaceutics. 2023; 15(2):370. https://doi.org/10.3390/pharmaceutics15020370
Chicago/Turabian StyleLoh, Pei She, and Volker Patzel. 2023. "Non-Covalent Linkage of Helper Functions to Dumbbell-Shaped DNA Vectors for Targeted Delivery" Pharmaceutics 15, no. 2: 370. https://doi.org/10.3390/pharmaceutics15020370





