Chitosan Surface-Modified PLGA Nanoparticles Loaded with Cranberry Powder Extract as a Potential Oral Delivery Platform for Targeting Colon Cancer Cells
Abstract
:1. Introduction
2. Materials and Methods
2.1. Statistical Design and Optimization of CBPE-PLGA NPs
2.2. Preparation of CBPE-PLGA NPs
2.3. In Vitro Characterization of the Prepared CBPE-PLGA NPs
2.3.1. Calculation of EE%
2.3.2. Estimation of PS, PDI, and ZP
2.4. Optimization of the Prepared CBPE-PLGA NPs
2.5. Surface Modification of the Optimum CBPE-PLGA NPs Formula Using Chitosan
2.6. In Vitro Characterization of the Optimized CBPE-PLGA and CS-CBPE-PLGA NPs
2.6.1. Transmission Electron Microscopy (TEM)
2.6.2. Attenuated Total Reflection–Fourier Transform Infrared (ATR-FTIR)
2.6.3. In Vitro Release Study
2.6.4. Ex Vivo Permeation Study
2.6.5. Stability Study
2.7. Cell-Based Antitumor Activity
2.7.1. MTT Cytotoxicity Assay
2.7.2. Caspase 3 Protein Level Assay
2.7.3. Vascular Endothelial Growth Factor (VEGF) Protein Level Assay
2.7.4. Signal Transducer and Activator of Transcription-3 (STAT-3) Assay
3. Results and Discussion
3.1. Factorial Design Outcomes
3.1.1. Influence of Formulation Variables on EE% (Y1)
3.1.2. Influence of Formulation Variables on PS (Y2)
3.1.3. Influence of Formulation Variables on PDI (Y3)
3.1.4. Influence of Formulation Variables on ZP (Y4)
3.2. Optimization of CBPE-PLGA NPs
3.3. Surface-Modification of the Optimum CBPE-PLGA NPs
3.4. In Vitro Characterization of the Optimized CBPE-PLGA NPs (F4) and CS-CBPE-PLGA NPs (CS-F4)
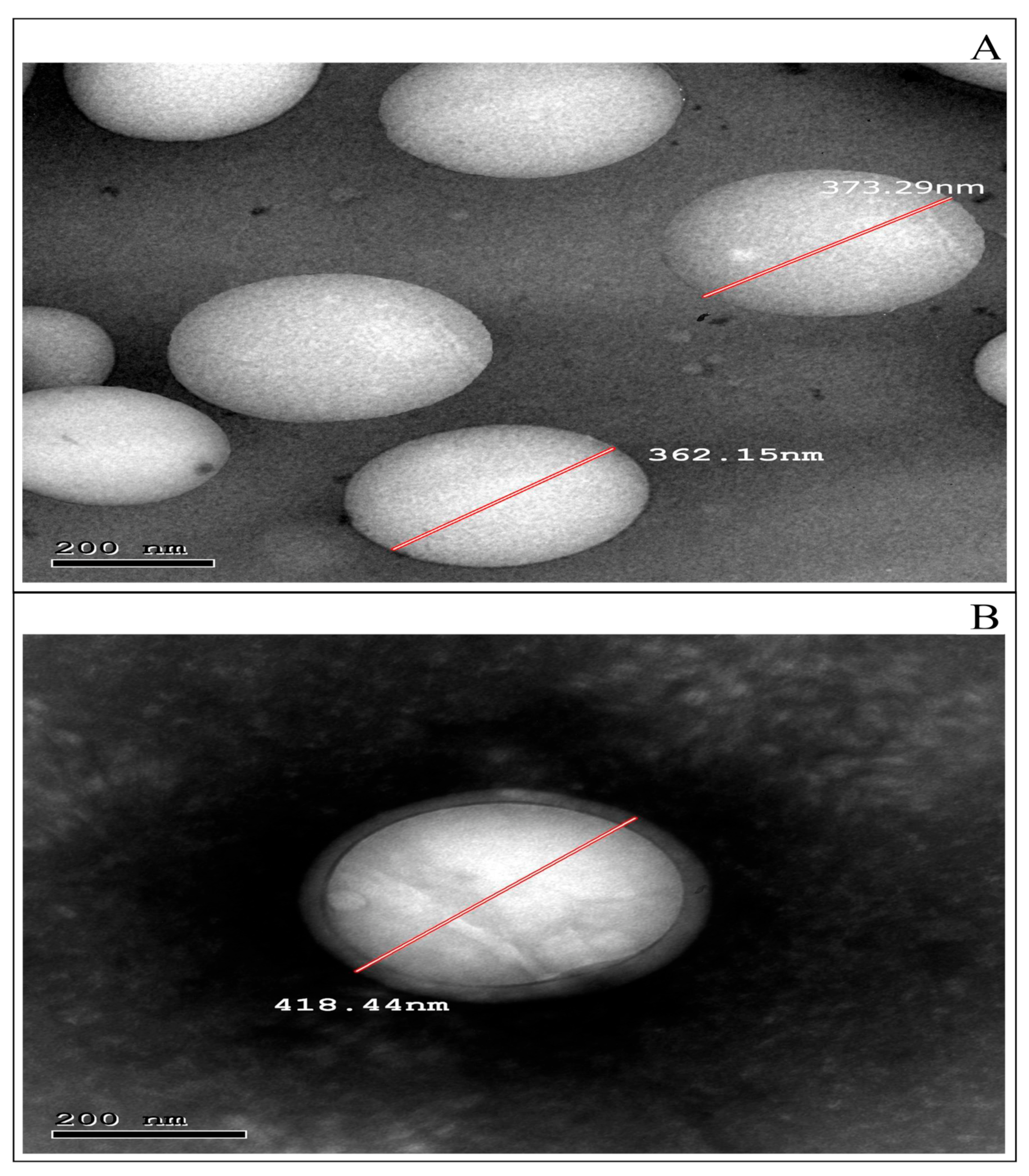
3.4.1. TEM Examination
3.4.2. ATR-FTIR Examination
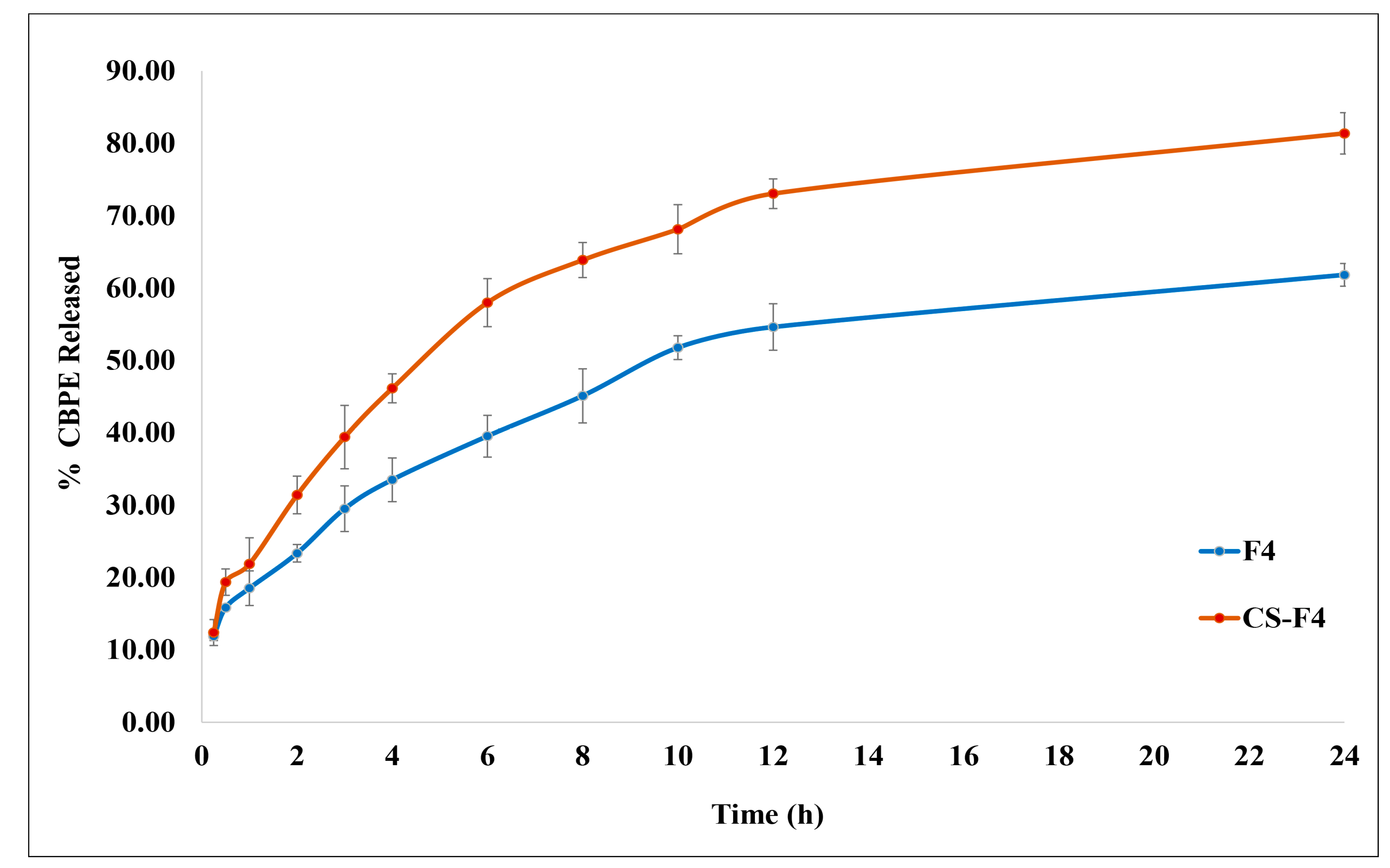
3.4.3. In Vitro CBPE Release Study
3.4.4. Ex Vivo Permeation Study
3.4.5. Stability Study
3.4.6. In Vitro Antitumor Activity
Cytotoxic Activity Determination Assay (IC50)
Effect on Caspase-3 Protein Level
Effect on VEGF Protein Level
Effect on STAT-3 Protein Level
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Darroudi, M.; Gholami, M.; Rezayi, M.; Khazaei, M. An overview and bibliometric analysis on the colorectal cancer therapy by magnetic functionalized nanoparticles for the responsive and targeted drug delivery. J. Nanobiotechnol. 2021, 19, 399. [Google Scholar] [CrossRef] [PubMed]
- Cruz-Nova, P.; Ancira-Cortez, A.; Ferro-Flores, G.; Ocampo-García, B.; Gibbens-Bandala, B. Controlled-release nanosystems with a dual function of targeted therapy and radiotherapy in colorectal cancer. Pharmaceutics 2022, 14, 1095. [Google Scholar] [CrossRef] [PubMed]
- Yi, L.; Wang, Q.; Luo, H.; Lei, D.; Tang, Z.; Lei, S.; Xiao, H. Bioactive components from Gracilaria rubra with growth inhibition on HCT116 colon cancer cells and anti-inflammatory capacity in RAW 264.7 macrophages. Front. Nutr. 2022, 9, 856282. [Google Scholar] [CrossRef] [PubMed]
- Krasteva, N.; Georgieva, M. Promising therapeutic strategies for colorectal cancer treatment based on nanomaterials. Pharmaceutics 2022, 14, 1213. [Google Scholar] [CrossRef]
- Blumberg, J.B.; Camesano, T.A.; Cassidy, A.; Kris-Etherton, P.; Howell, A.; Manach, C.; Ostertag, L.M.; Sies, H.; Skulas-Ray, A.; Vita, J.A. Cranberries and their bioactive constituents in human health. Adv. Nutr. 2013, 4, 618–632. [Google Scholar] [CrossRef]
- Caldas, A.P.S.; Coelho, O.G.L.; Bressan, J. Cranberry antioxidant power on oxidative stress, inflammation and mitochondrial damage. Int. J. Food Prop. 2018, 21, 582–592. [Google Scholar] [CrossRef]
- Caillet, S.; Côté, J.; Doyon, G.; Sylvain, J.F.; Lacroix, M. Antioxidant and antiradical properties of cranberry juice and extracts. Food Res. Int. 2011, 44, 1408–1413. [Google Scholar] [CrossRef]
- Wu, X.; Song, M.; Cai, X.; Neto, C.; Tata, A.; Han, Y.; Wang, Q.; Tang, Z.; Xiao, H. Chemopreventive effects of whole cranberry (Vaccinium macrocarpon) on colitis-associated colon tumorigenesis. Mol. Nutr. Food Res. 2018, 62, 1800942. [Google Scholar] [CrossRef]
- Mathison, B.D.; Kimble, L.L.; Kaspar, K.L.; Khoo, C.; Chew, B.P. Consumption of cranberry beverage improved endogenous antioxidant status and protected against bacteria adhesion in healthy humans: A randomized controlled trial. Nutr. Res. 2014, 34, 420–427. [Google Scholar] [CrossRef]
- Soliman, S.M.; Mosallam, S.; Mamdouh, M.A.; Hussein, M.A.; Abd El-Halim, S.M. Design and optimization of cranberry extract loaded bile salt augmented liposomes for targeting of MCP-1/STAT3/VEGF signaling pathway in DMN-intoxicated liver in rats. Drug Deliv. 2022, 29, 427–439. [Google Scholar] [CrossRef]
- Niculescu, A.G.; Grumezescu, A.M. Novel tumor-targeting nanoparticles for cancer treatment—A review. Int. J. Mol. Sci. 2022, 23, 5253. [Google Scholar] [CrossRef]
- Hamad, K.; Kaseem, M.; Yang, H.W.; Deri, F.; Ko, Y.G. Properties and medical applications of polylactic acid: A review. Express Polym. Lett. 2015, 9, 435–455. [Google Scholar] [CrossRef]
- Badran, M.M.; Mady, M.M.; Ghannam, M.M.; Shakeel, F. Preparation and characterization of polymeric nanoparticles surface modified with chitosan for target treatment of colorectal cancer. Int. J. Biol. Macromol. 2017, 95, 643–649. [Google Scholar] [CrossRef]
- Joshi, G.; Kumar, A.; Sawant, K. Enhanced bioavailability and intestinal uptake of Gemcitabine HCl loaded PLGA nanoparticles after oral delivery. Eur. J. Pharm. Sci. 2014, 60, 80–89. [Google Scholar] [CrossRef]
- Begines, B.; Ortiz, T.; Pérez-Aranda, M.; Martínez, G.; Merinero, M.; Argüelles-Arias, F.; Alcudia, A. Polymeric nanoparticles for drug delivery: Recent developments and future prospects. Nanomaterials 2020, 10, 1403. [Google Scholar] [CrossRef]
- Hu, C.M.J.; Aryal, S.; Zhang, L. Nanoparticle-assisted combination therapies for effective cancer treatment. Ther. Deliv. 2010, 1, 323–334. [Google Scholar] [CrossRef]
- Ernest, U.; Chen, H.Y.; Xu, M.J.; Taghipour, Y.D.; Bin Asad, M.H.H.; Rahimi, R.; Murtaza, G. Anti-cancerous potential of polyphenol-loaded polymeric nanotherapeutics. Molecules 2018, 23, 2787. [Google Scholar] [CrossRef]
- Khuroo, T.; Verma, D.; Talegaonkar, S.; Padhi, S.; Panda, A.K.; Iqbal, Z. Topotecan-tamoxifen duple PLGA polymeric nanoparticles: Investigation of in vitro, in vivo and cellular uptake potential. Int. J. Pharm. 2014, 473, 384–394. [Google Scholar] [CrossRef]
- Wang, M.; Zhang, Y.; Feng, J.; Gu, T.; Dong, Q.; Yang, X.; Sun, Y.; Wu, Y.; Chen, Y.; Kong, W. Preparation, characterization, and in vitro and in vivo investigation of chitosan-coated poly (d,l-lactide-co-glycolide) nanoparticles for intestinal delivery of exendin-4. Int. J. Nanomed. 2013, 8, 1141–1154. [Google Scholar] [CrossRef]
- Danhier, F.; Ansorena, E.; Silva, J.M.; Coco, R.; Le Breton, A.; Préat, V. PLGA-based nanoparticles: An overview of biomedical applications. J. Control. Release 2012, 161, 505–522. [Google Scholar] [CrossRef]
- Öztürk, A.A.; Yenilmez, E.; Özarda, M.G. Clarithromycin-loaded poly (Lactic- co-glycolic Acid) (PLGA) nanoparticles for oral administration: Effect of polymer molecular weight and surface modification with chitosan on formulation, nanoparticle characterization and antibacterial effects. Polymers 2019, 11, 1632. [Google Scholar] [CrossRef] [PubMed]
- Pereira, M.C.; Oliveira, D.A.; Hill, L.E.; Zambiazi, R.C.; Borges, C.D.; Vizzotto, M.; Mertens-Talcott, S.; Talcott, S.; Gomes, C.L. Effect of nanoencapsulation using PLGA on antioxidant and antimicrobial activities of guabiroba fruit phenolic extract. Food Chem. 2018, 240, 396–404. [Google Scholar] [CrossRef] [PubMed]
- Lu, B.; Lv, X.; Le, Y. Chitosan-modified PLGA nanoparticles for control-released drug delivery. Polymers 2019, 11, 304. [Google Scholar] [CrossRef] [PubMed]
- Abd El-Halim, S.M.; Abdelbary, G.A.; Amin, M.M.; Zakaria, M.Y.; Shamsel-Din, H.A.; Ibrahim, A.B. Stabilized oral nanostructured lipid carriers of Adefovir Dipivoxil as a potential liver targeting: Estimation of liver function panel and uptake following intravenous injection of radioiodinated indicator. DARU J. Pharm. Sci. 2020, 28, 517–532. [Google Scholar] [CrossRef]
- Xiao, B.; Han, M.K.; Viennois, E.; Wang, L.; Zhang, M.; Si, X.; Merlin, D. Hyaluronic acid-functionalized polymeric nanoparticles for colon cancer-targeted combination chemotherapy. Nanoscale 2015, 7, 17745–17755. [Google Scholar] [CrossRef]
- Al-Nemrawi, N.K.; Okour, A.R.; Dave, R.H. Surface modification of PLGA nanoparticles using chitosan: Effect of molecular weight, concentration, and degree of deacetylation. Adv. Polym. Technol. 2018, 37, 3066–3075. [Google Scholar] [CrossRef]
- Zakaria, M.Y.; Georghiou, P.E.; Banoub, J.H.; Beshay, B.Y. Inclusion of a phytomedicinal flavonoid in biocompatible surface-modified chylomicron mimic nanovesicles with improved oral bioavailability and virucidal activity: Molecular modeling and pharmacodynamic studies. Pharmaceutics 2022, 14, 905. [Google Scholar] [CrossRef]
- Soliman, S.M.; Sheta, N.M.; Ibrahim, B.M.M.; El-Shawwa, M.M.; El-Halim, S.M.A. Novel intranasal drug delivery: Geraniol charged polymeric mixed micelles for targeting cerebral insult as a result of ischaemia/reperfusion. Pharmaceutics 2020, 12, 76. [Google Scholar] [CrossRef]
- Weng, J.; Tong, H.H.Y.; Chow, S.F. In vitro release study of the polymeric drug nanoparticles: Development and validation of a novel method. Pharmaceutics 2020, 12, 732. [Google Scholar] [CrossRef]
- Amin, M.M.; El Gazayerly, O.N.; Abd El-Gawad, N.A.; Abd El-Halim, S.M.; El-Awdan, S.A. Effect of formulation variables on design, in vitro evaluation of valsartan SNEDDS and estimation of its antioxidant effect in adrenaline-induced acute myocardial infarction in rats. Pharm. Dev. Technol. 2016, 21, 909–920. [Google Scholar] [CrossRef]
- De Lima, I.A.; Khalil, N.M.; Tominaga, T.T.; Lechanteur, A.; Sarmento, B.; Mainardes, R.M. Mucoadhesive chitosan-coated PLGA nanoparticles for oral delivery of ferulic acid. Artif. Cells Nanomed. Biotechnol. 2018, 46, 993–1002. [Google Scholar] [CrossRef]
- Alam, M.A.; Al-Jenoobi, F.I.; Al-Mohizea, A.M. Everted gut sac model as a tool in pharmaceutical research: Limitations and applications. J. Pharm. Pharmacol. 2012, 64, 326–336. [Google Scholar] [CrossRef]
- Abd El-Halim, S.M.; Mamdouh, M.A.; Eid, S.M.; Ibrahim, B.M.M.; Labib, D.A.A.; Soliman, S.M. The potential synergistic activity of zolmitriptan combined in new self-nanoemulsifying drug delivery systems: Atr-ftir real-time fast dissolution monitoring and pharmacodynamic assessment. Int. J. Nanomed. 2021, 16, 6395–6412. [Google Scholar] [CrossRef]
- Vidal-Casanella, O.; Arias-Alpizar, K.; Núñez, O.; Saurina, J. Extraction and characterization of flavanol-rich nutraceuticals based on high-performance liquid chromatography. Separations 2022, 9, 87. [Google Scholar] [CrossRef]
- Mady, O.Y.; Donia, A.A.; Al-Shoubki, A.A.; Qasim, W. Paracellular pathway enhancement of metformin hydrochloride via molecular dispersion in span 60 microparticles. Front. Pharmacol. 2019, 10, 713. [Google Scholar] [CrossRef]
- Feng, C.; Wang, Z.; Jiang, C.; Kong, M.; Zhou, X.; Li, Y.; Cheng, X.; Chen, X. Chitosan/o-carboxymethyl chitosan nanoparticles for efficient and safe oral anticancer drug delivery: In vitro and in vivo evaluation. Int. J. Pharm. 2013, 457, 158–167. [Google Scholar] [CrossRef]
- Deng, P.; Athary Abdulhaleem, M.F.; Masoud, R.E.; Alamoudi, W.M.; Zakaria, M.Y. Employment of PEGylated ultra-deformable transferosomes for transdermal delivery of tapentadol with boosted bioavailability and analgesic activity in post-surgical pain. Int. J. Pharm. 2022, 628, 122274. [Google Scholar] [CrossRef]
- MTT Assay Protocol for Cell Viability and Proliferation. Available online: https://www.sigmaaldrich.com/EG/en/technical-documents/protocol/cell-culture-and-cell-culture-analysis/cell-counting-and-health-analysis/cell-proliferation-kit-i-mtt (accessed on 2 August 2022).
- Sandwich ELISA, Abcam Co. Available online: https://www.abcam.com/ps/pdf/protocols/Sandwich_ELISA.pdf (accessed on 2 August 2022).
- Recombinant anti-Caspase-3 Antibody [EPR17664-153]-BSA and Azide Free (Capture) (ab281235), Abcam Co. Available online: https://www.abcam.com/caspase-3-antibody-epr17664-153-bsa-and-azide-free-capture-ab281235.html (accessed on 3 August 2022).
- Ab209882 Mouse VEGF Simplestep ELISA® Kit, Version 3a, Abcam Co. Available online: https://www.abcam.com/ps/products/209/ab209882/documents/Mouse-VEGF-ELISA-kit-protocol-book-v3a-ab209882(website).pdf (accessed on 3 August 2022).
- Ab126459 − STAT3 (pY705) + total STAT3 ELISA Kit, Abcam Co. Available online: https://www.abcam.com/ps/products/126/ab126459/documents/ab126459-STAT3-pY705-total-STAT3-ELISA-Kit-v3a(website).pdf (accessed on 3 August 2022).
- Fatima, S.; Iqbal, Z.; Panda, A.K.; Samim, M.; Talegaonkar, S.; Ahmad, F.J. Polymeric nanoparticles as a platform for permeability enhancement of class III drug amikacin. Colloids Surf. B Biointerfaces 2018, 169, 206–213. [Google Scholar] [CrossRef]
- Ge, S.; Wang, E.; Li, J.; Tang, B.Z. Aggregation-induced emission boosting the study of polymer science. Macromol. Rapid Commun. 2022, 43, 2200080. [Google Scholar] [CrossRef]
- Park, P.I.P.; Jonnalagadda, S. Predictors of glass transition in the biodegradable polylactide and poly-lactide-co-glycolide polymers. J. Appl. Polym. Sci. 2006, 100, 1983–1987. [Google Scholar] [CrossRef]
- Lee, J.S.; Chae, G.S.; Khang, G.; Kim, M.S.; Cho, S.H.; Lee, H.B. The effect of Gamma irradiation on PLGA and release behavior of BCNU from PLGA wafer. Macromol. Res. 2003, 11, 352–356. [Google Scholar] [CrossRef]
- Psimadas, D.; Georgoulias, P.; Valotassiou, V.; Loudos, G. Molecular nanomedicine towards cancer: 111In-labeled nanoparticles. J. Pharm. Sci. 2012, 101, 2271–2280. [Google Scholar] [CrossRef] [PubMed]
- Park, K.; Otte, A.; Sharifi, F.; Garner, J.; Skidmore, S.; Park, H.; Jhon, Y.K.; Qin, B.; Wang, Y. Potential roles of the glass transition temperature of plga microparticles in drug release kinetics. Mol. Pharm. 2021, 18, 18–32. [Google Scholar] [CrossRef] [PubMed]
- El Hady, W.E.A.; Mohamed, E.A.; El-Aazeem Soliman, O.A.; El-Sabbagh, H.M. In vitro-in vivo evaluation of chitosan-PLGA nanoparticles for potentiated gastric retention and anti-ulcer activity of diosmin. Int. J. Nanomed. 2019, 14, 7191–7213. [Google Scholar] [CrossRef] [PubMed]
- Manchanda, R.; Fernandez-Fernandez, A.; Nagesetti, A.; McGoron, A.J. Preparation and characterization of a polymeric (PLGA) nanoparticulate drug delivery system with simultaneous incorporation of chemotherapeutic and thermo-optical agents. Colloids Surf. B Biointerfaces 2010, 75, 260–267. [Google Scholar] [CrossRef]
- Zakeri-Milani, P.; Loveymi, B.D.; Jelvehgari, M.; Valizadeh, H. The characteristics and improved intestinal permeability of vancomycin PLGA-nanoparticles as colloidal drug delivery system. Colloids Surf. B Biointerfaces 2013, 103, 174–181. [Google Scholar] [CrossRef]
- Sharma, N.; Madan, P.; Lin, S. Effect of process and formulation variables on the preparation of parenteral paclitaxel-loaded biodegradable polymeric nanoparticles: A co-surfactant study. Asian J. Pharm. Sci. 2016, 11, 404–416. [Google Scholar] [CrossRef]
- Haggag, Y.A.; Osman, M.A.; El-Gizawy, S.A.; Goda, A.E.; Shamloula, M.M.; Faheem, A.M.; McCarron, P.A. Polymeric nano-encapsulation of 5-fluorouracil enhances anti-cancer activity and ameliorates side effects in solid Ehrlich Carcinoma-bearing mice. Biomed. Pharmacother. 2018, 105, 215–224. [Google Scholar] [CrossRef]
- Budhian, A.; Siegel, S.J.; Winey, K.I. Haloperidol-loaded PLGA nanoparticles: Systematic study of particle size and drug content. Int. J. Pharm. 2007, 336, 367–375. [Google Scholar] [CrossRef]
- Rafiei, P.; Haddadi, A. A robust systematic design: Optimization and preparation of polymeric nanoparticles of PLGA for docetaxel intravenous delivery. Mater. Sci. Eng. C 2019, 104, 109950. [Google Scholar] [CrossRef]
- Todaro, B.; Moscardini, A.; Luin, S. Pioglitazone-loaded PLGA nanoparticles: Towards the most reliable synthesis method. Int. J. Mol. Sci. 2022, 23, 2522. [Google Scholar] [CrossRef]
- Tefas, L.R.; Tomuţă, I.; Achim, M.; Vlase, L. Development and optimization of quercetin-loaded plga nanoparticles by experimental design. Clujul Med. 2015, 88, 214–223. [Google Scholar] [CrossRef]
- Jamil, A.; Aamir Mirza, M.; Anwer, M.K.; Thakur, P.S.; Alshahrani, S.M.; Alshetaili, A.S.; Telegaonkar, S.; Panda, A.K.; Iqbal, Z. Co-delivery of gemcitabine and simvastatin through PLGA polymeric nanoparticles for the treatment of pancreatic cancer: In-vitro characterization, cellular uptake, and pharmacokinetic studies. Drug Dev. Ind. Pharm. 2019, 45, 745–753. [Google Scholar] [CrossRef]
- Radwan, R.; Abdelkader, A.; Fathi, H.A.; Elsabahy, M.; Fetih, G.; El-Badry, M. Development and evaluation of Letrozole-loaded hyaluronic acid/chitosan-coated poly(d,l-lactide-co-glycolide) nanoparticles. J. Pharm. Innov. 2022, 17, 572–583. [Google Scholar] [CrossRef]
- Mehrotra, A.; Kumar Pandit, J. Preparation and characterization and biodistribution studies of Lomustine loaded PLGA nanoparticles by interfacial deposition method. J. Nanomedine. Biotherapeutic Discov. 2015, 5, 1000138. [Google Scholar] [CrossRef]
- Büyükköroğlu, G.; Şenel, B.; Başaran, E.; Yenilmez, E.; Yazan, Y. Preparation and in vitro evaluation of vaginal formulations including siRNA and paclitaxel-loaded SLNs for cervical cancer. Eur. J. Pharm. Biopharm. 2016, 109, 174–183. [Google Scholar] [CrossRef]
- Anwer, K.E.; Abd El-Sattar, N.E.A.; Shamaa, M.M.; Zakaria, M.Y.; Beshay, B.Y. Design, green synthesis and tailoring of vitamin E TPGS augmented niosomal nano-carrier of Pyrazolopyrimidines as potential anti-liver and breast cancer agents with accentuated oral bioavailability. Pharmaceuticals 2022, 15, 330. [Google Scholar] [CrossRef]
- Haggag, Y.A.; Abosalha, A.K.; Tambuwala, M.M.; Osman, E.Y.; El-Gizawy, S.A.; Essa, E.A.; Donia, A.A. Polymeric nanoencapsulation of zaleplon into PLGA nanoparticles for enhanced pharmacokinetics and pharmacological activity. Biopharm. Drug Dispos. 2021, 42, 12–23. [Google Scholar] [CrossRef]
- Alkholief, M.; Kalam, M.A.; Anwer, M.K.; Alshamsan, A. Effect of solvents, stabilizers and the concentration of stabilizers on the physical properties of poly(D,L-lactide-co-glycolide) nanoparticles: Encapsulation, in vitro release of indomethacin and cytotoxicity against HepG2-cell. Pharmaceutics 2022, 14, 870. [Google Scholar] [CrossRef]
- Wang, Y.; Li, P.; Kong, L. Chitosan-modified PLGA nanoparticles with versatile surface for improved drug delivery. AAPS PharmSciTech 2013, 14, 585–592. [Google Scholar] [CrossRef] [Green Version]
- Shi, Y.; Xue, J.; Jia, L.; Du, Q.; Niu, J.; Zhang, D. Surface-modified PLGA nanoparticles with chitosan for oral delivery of tolbutamide. Colloids Surf. B Biointerfaces 2018, 161, 67–72. [Google Scholar] [CrossRef] [PubMed]
- Dandamudi, M.; McLoughlin, P.; Behl, G.; Rani, S.; Coffey, L.; Chauhan, A.; Kent, D.; Fitzhenry, L. Chitosan-coated PLGA nanoparticles encapsulating triamcinolone acetonide as a potential candidate for sustained ocular drug delivery. Pharmaceutics 2021, 13, 1590. [Google Scholar] [CrossRef] [PubMed]
- Ahmad, N.; Alam, M.A.; Ahmad, R.; Naqvi, A.A.; Ahmad, F.J. Preparation and characterization of surface-modified PLGA-polymeric nanoparticles used to target treatment of intestinal cancer. Artif. Cells Nanomed. Biotechnol. 2018, 46, 432–446. [Google Scholar] [CrossRef] [PubMed]
- Aldawsari, H.M.; Alhakamy, N.A.; Padder, R.; Husain, M.; Md, S. Preparation and characterization of chitosan coated plga nanoparticles of resveratrol: Improved stability, antioxidant and apoptotic activities in H1299 lung cancer cells. Coatings 2020, 10, 439. [Google Scholar] [CrossRef]
- Ashour, A.A.; Raafat, D.; El-Gowelli, H.M.; El-Kamel, A.H. Green synthesis of silver nanoparticles using cranberry powder aqueous extract: Characterization and antimicrobial properties. Int. J. Nanomed. 2015, 10, 7207–7221. [Google Scholar] [CrossRef]
- Namiesnik, J.; Vearasilp, K.; Leontowicz, H.; Leontowicz, M.; Ham, K.S.; Kang, S.G.; Park, Y.K.; Arancibia-Avila, P.; Toledo, F.; Gorinstein, S. Comparative assessment of two extraction procedures for determination of bioactive compounds in some berries used for daily food consumption. Int. J. Food Sci. Technol. 2014, 49, 337–346. [Google Scholar] [CrossRef]
- Andronie, L.; Pop, I.; Rotaru, A.; Sobolu, R.; Igori, B.; Coroian, A.; Longodor, A.L.; Marchis, Z. Physico-Chemical Analysis of Cranberry Using Ft-Ir. Sci. Pap. Ser. B Hortic. 2020, LXIV, 623–627. [Google Scholar]
- Blasi, P.; D’Souza, S.S.; Selmin, F.; DeLuca, P.P. Plasticizing effect of water on poly(lactide-co-glycolide). J. Control. Release 2005, 108, 1–9. [Google Scholar] [CrossRef]
- Park, K.; Skidmore, S.; Hadar, J.; Garner, J.; Park, H.; Otte, A.; Soh, B.K.; Yoon, G.; Yu, D.; Yun, Y.; et al. Injectable, long-acting PLGA formulations: Analyzing PLGA and understanding microparticle formation. J. Control. Release 2019, 304, 125–134. [Google Scholar] [CrossRef]
- Liu, G.; McEnnis, K. Glass transition temperature of plga particles and the influence on drug delivery applications. Polymers 2022, 14, 993. [Google Scholar] [CrossRef]
- Lappe, S.; Mulac, D.; Langer, K. Polymeric nanoparticles—Influence of the glass transition temperature on drug release. Int. J. Pharm. 2017, 517, 338–347. [Google Scholar] [CrossRef]
- Chatzitaki, A.T.; Jesus, S.; Karavasili, C.; Andreadis, D.; Fatouros, D.G.; Borges, O. Chitosan-coated PLGA nanoparticles for the nasal delivery of ropinirole hydrochloride: In vitro and ex vivo evaluation of efficacy and safety. Int. J. Pharm. 2020, 589, 119776. [Google Scholar] [CrossRef]
- Cortés, H.; Hernández-Parra, H.; Bernal-Chávez, S.A.; Del Prado-Audelo, M.L.; Caballero-Florán, I.H.; Borbolla-Jiménez, F.V.; González-Torres, M.; Magaña, J.J.; Leyva-Gómez, G. Non-ionic surfactants for stabilization of polymeric nanoparticles for biomedical uses. Materials 2021, 14, 3197. [Google Scholar] [CrossRef]
- Maiti, S. Nanometric biopolymer devices for oral delivery of macromolecules with clinical significance. In Multifunctional Systems for Combined Delivery, Biosensing and Diagnostics; Grumezescu, A.M., Ed.; Elsevier Inc.: Amsterdam, The Netherlands, 2017; pp. 109–138. ISBN 9780323527255. [Google Scholar]




| Factors (Independent Variables) | Levels | |
|---|---|---|
| X1: L/G ratio | 50:50 | 85:15 |
| X2: PLGA Weight (mg) | 20 | 50 |
| X3: PVA Conc in IAP (%) | 0 | 1 |
| Responses (Dependent Variables) | Desirability Constraints | |
| Y1: EE% | Maximize | |
| Y2: PS (nm) | Minimize | |
| Y3: PDI | In the range | |
| Y4: ZP | In the range | |
| Formulations | X1 L/G Ratio | X2 PLGA Weight (mg) | X3 PVA Conc in IAP (%) | Y1 EE% | Y2 PS (nm) | Y3 PDI | Y4 ZP (mV) |
|---|---|---|---|---|---|---|---|
| F1 | PLGA 50:50 | 20 | 0 | 48.30 ± 2.21 | 259.87 ± 4.68 | 0.496 ± 0.01 | −8.81 ± 0.41 |
| F2 | PLGA 50:50 | 50 | 0 | 56.82 ± 2.36 | 345.10 ± 9.17 | 0.431 ± 0.040 | −8.57 ± 0.48 |
| F3 | PLGA 50:50 | 20 | 1 | 58.38 ± 2.57 | 284.87 ± 8.11 | 0.453 ± 0.048 | −4.98 ± 0.25 |
| F4 | PLGA 50:50 | 50 | 1 | 72.30 ± 2.86 | 370.10 ± 10.31 | 0.398 ± 0.001 | −5.40 ± 0.21 |
| F4-CS | PLGA 50:50 | 50 | 1 | 74.74 ± 1.48 | 417.67 ± 6.77 | 0.323 ± 0.019 | +21.63 ± 2.46 |
| F5 | PLGA 85:15 | 20 | 0 | 41.95 ± 2.84 | 265.87 ± 4.15 | 0.481 ± 0.001 | −8.32 ± 0.56 |
| F6 | PLGA 85:15 | 50 | 0 | 51.61 ± 2.16 | 365.90 ± 6.79 | 0.406 ± 0.025 | −7.13 ± 0.94 |
| F7 | PLGA 85:15 | 20 | 1 | 53.12 ± 2.72 | 417.67 ± 6.77 | 0.379 ± 0.007 | −4.97 ± 0.33 |
| F8 | PLGA 85:15 | 50 | 1 | 57.83 ± 1.99 | 423.10 ± 8.36 | 0.331 ± 0.016 | −5.78 ± 0.74 |
| (A) Responses | R2 | Adjusted R2 | Predicted R2 | Adequate Precision | Significant Factors |
|---|---|---|---|---|---|
| EE (%) | 0.921 | 0.893 | 0.842 | 17.99 | X1, X2, X3 |
| PS (nm) | 0.952 | 0.935 | 0.905 | 21.83 | X1, X2, X3 |
| PDI | 0.861 | 0.812 | 0.724 | 12.86 | X1, X2, X3 |
| ZP (mV) | 0.914 | 0.884 | 0.828 | 13.71 | X3 |
| (B) Response | Y1 EE% | Y2 PS | Y3 PDI | Y4 ZP | |
| Observed values | 72.30 | 370.10 | 0.398 | −5.40 | |
| Predicted values | 71.01 | 358.28 | 0.4 | −5.56 | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mostafa, M.M.; Amin, M.M.; Zakaria, M.Y.; Hussein, M.A.; Shamaa, M.M.; Abd El-Halim, S.M. Chitosan Surface-Modified PLGA Nanoparticles Loaded with Cranberry Powder Extract as a Potential Oral Delivery Platform for Targeting Colon Cancer Cells. Pharmaceutics 2023, 15, 606. https://doi.org/10.3390/pharmaceutics15020606
Mostafa MM, Amin MM, Zakaria MY, Hussein MA, Shamaa MM, Abd El-Halim SM. Chitosan Surface-Modified PLGA Nanoparticles Loaded with Cranberry Powder Extract as a Potential Oral Delivery Platform for Targeting Colon Cancer Cells. Pharmaceutics. 2023; 15(2):606. https://doi.org/10.3390/pharmaceutics15020606
Chicago/Turabian StyleMostafa, Mona M., Maha M. Amin, Mohamed Y. Zakaria, Mohammed Abdalla Hussein, Marium M. Shamaa, and Shady M. Abd El-Halim. 2023. "Chitosan Surface-Modified PLGA Nanoparticles Loaded with Cranberry Powder Extract as a Potential Oral Delivery Platform for Targeting Colon Cancer Cells" Pharmaceutics 15, no. 2: 606. https://doi.org/10.3390/pharmaceutics15020606







