A New Strategy for Nucleic Acid Delivery and Protein Expression Using Biocompatible Nanohydrogels of Predefined Sizes
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Synthesis of NHGs
2.3. Labelling of NHGs
2.4. Cell Culture
2.5. Plasmid Preparation
2.6. Cell Proliferation Assays
2.7. In Vitro Transfection
2.8. In Vitro Transfection (Passing Method)
2.9. In Vivo Analysis
2.10. Statistical Analysis
3. Results and Discussion
3.1. In Vitro Experiments
3.1.1. Cell Proliferation
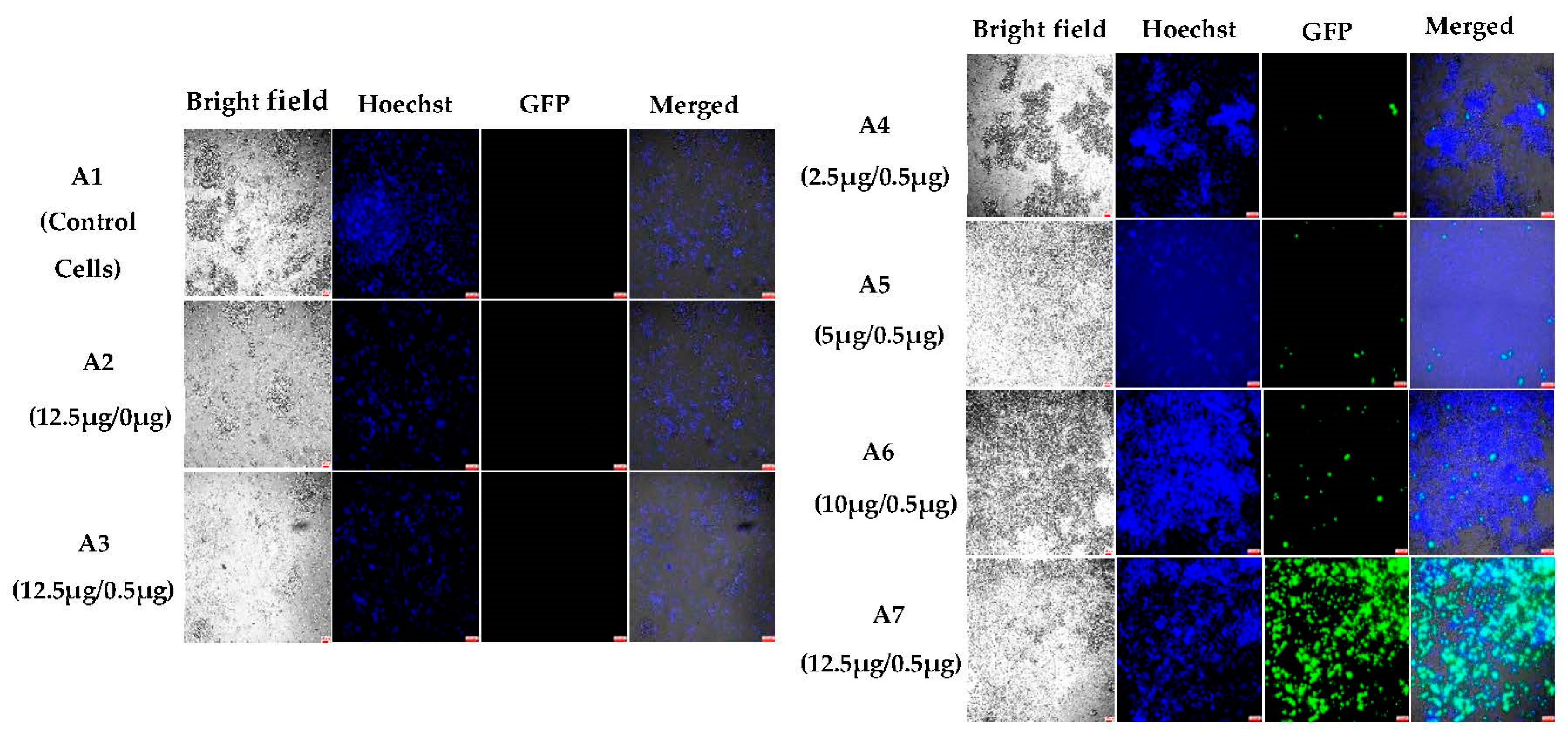
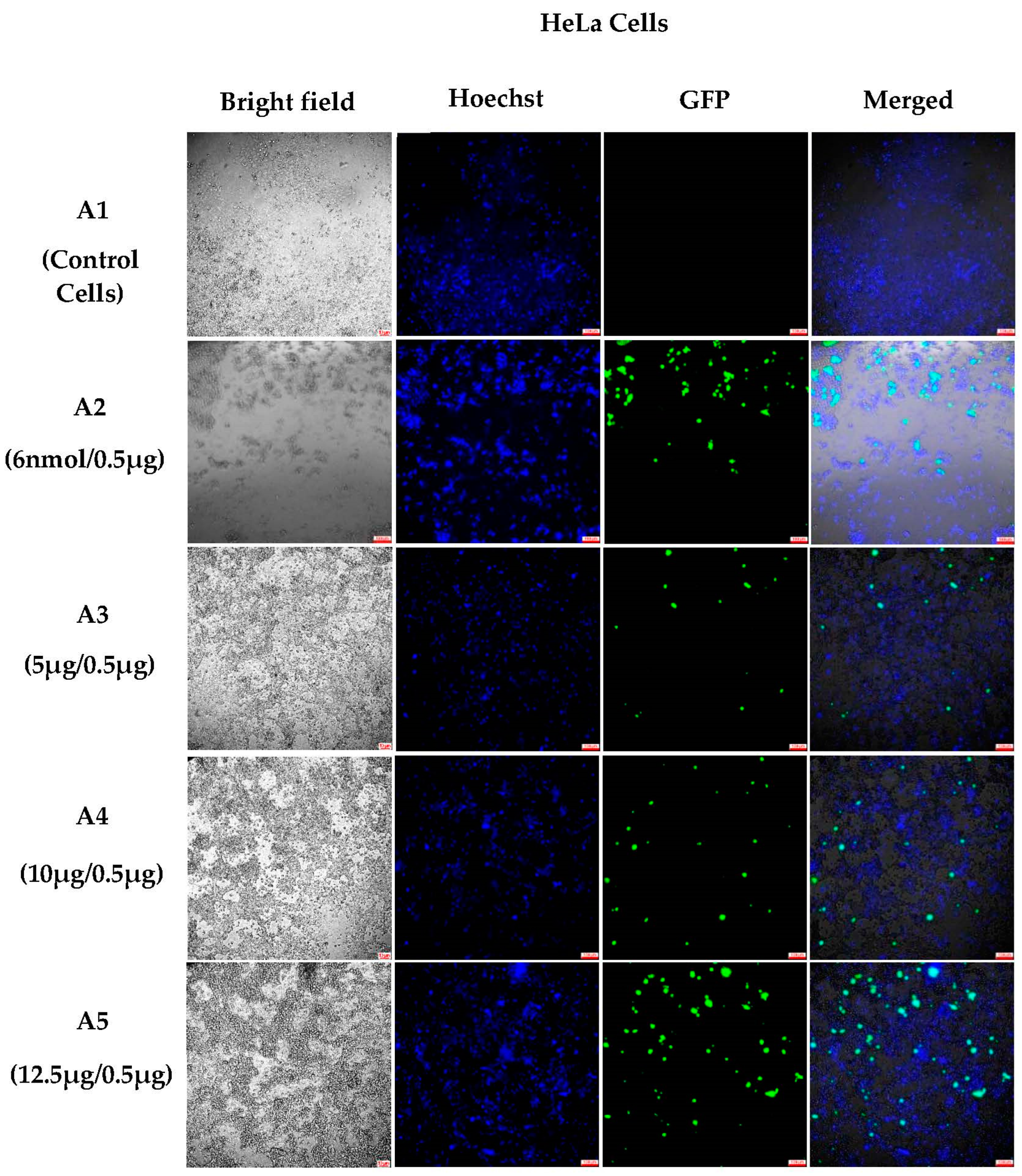
3.1.2. In Vitro Transfections
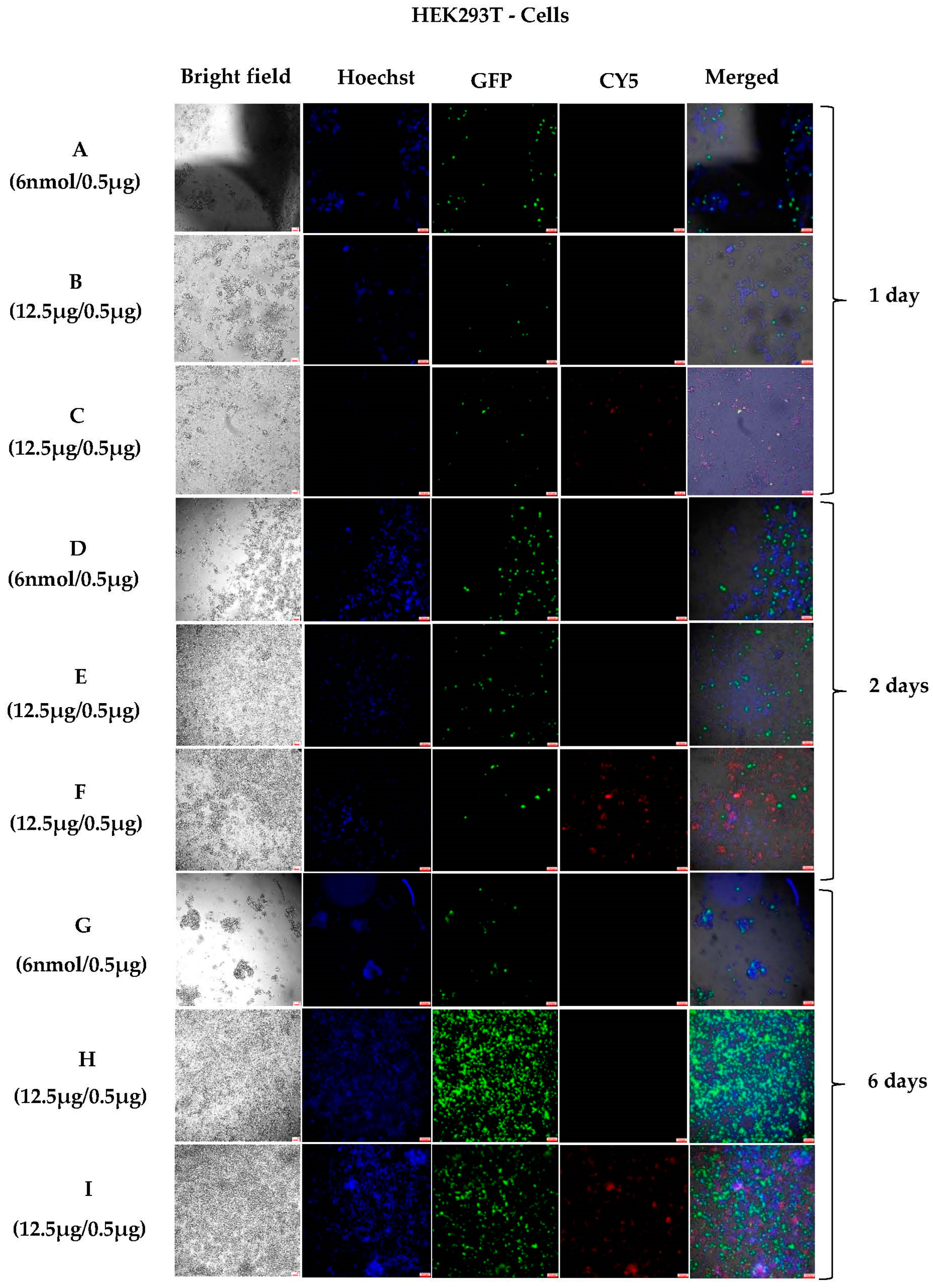
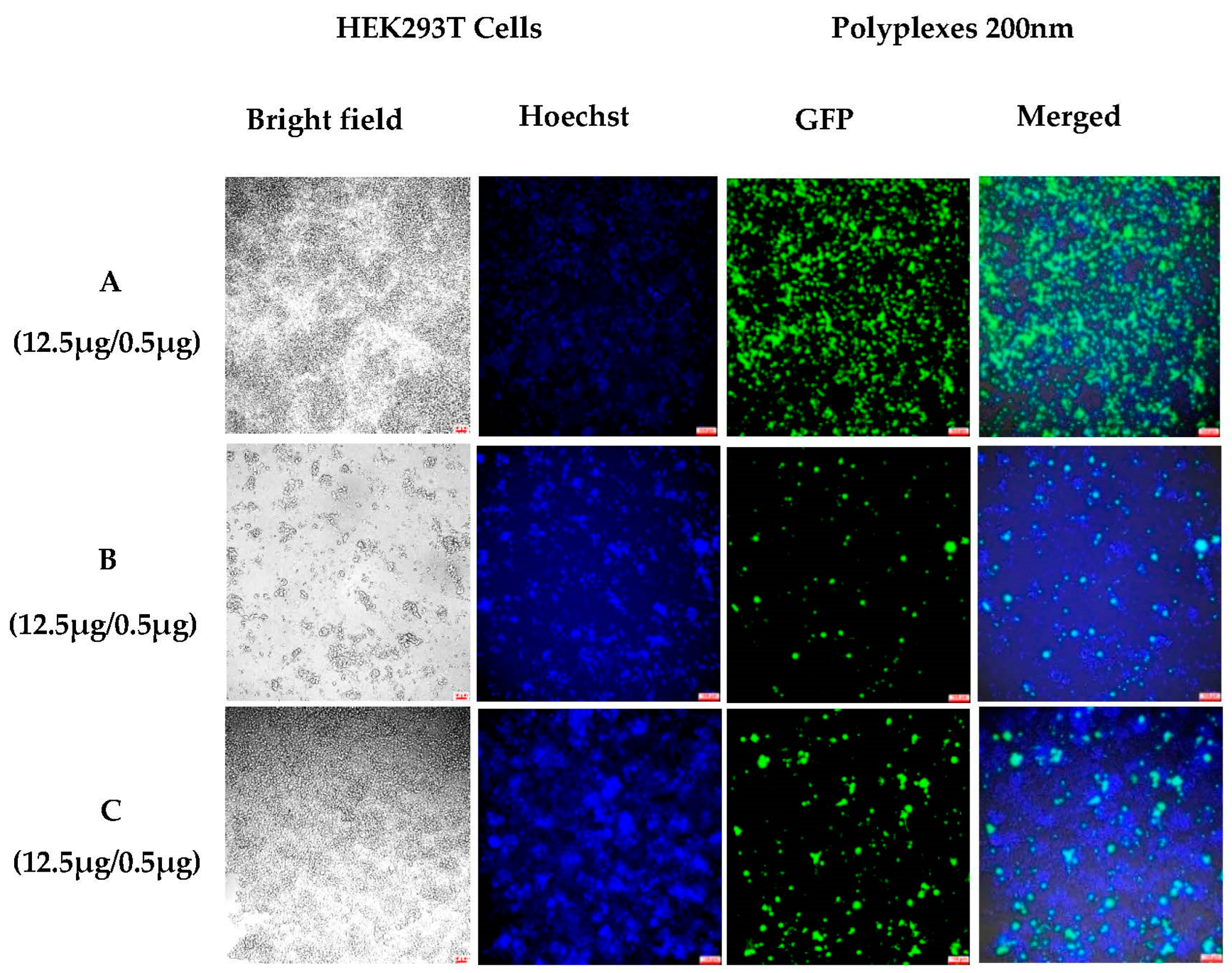
3.1.3. Sustained In Vitro Transfection after Cell Passing Using 200 nm Polyplexes
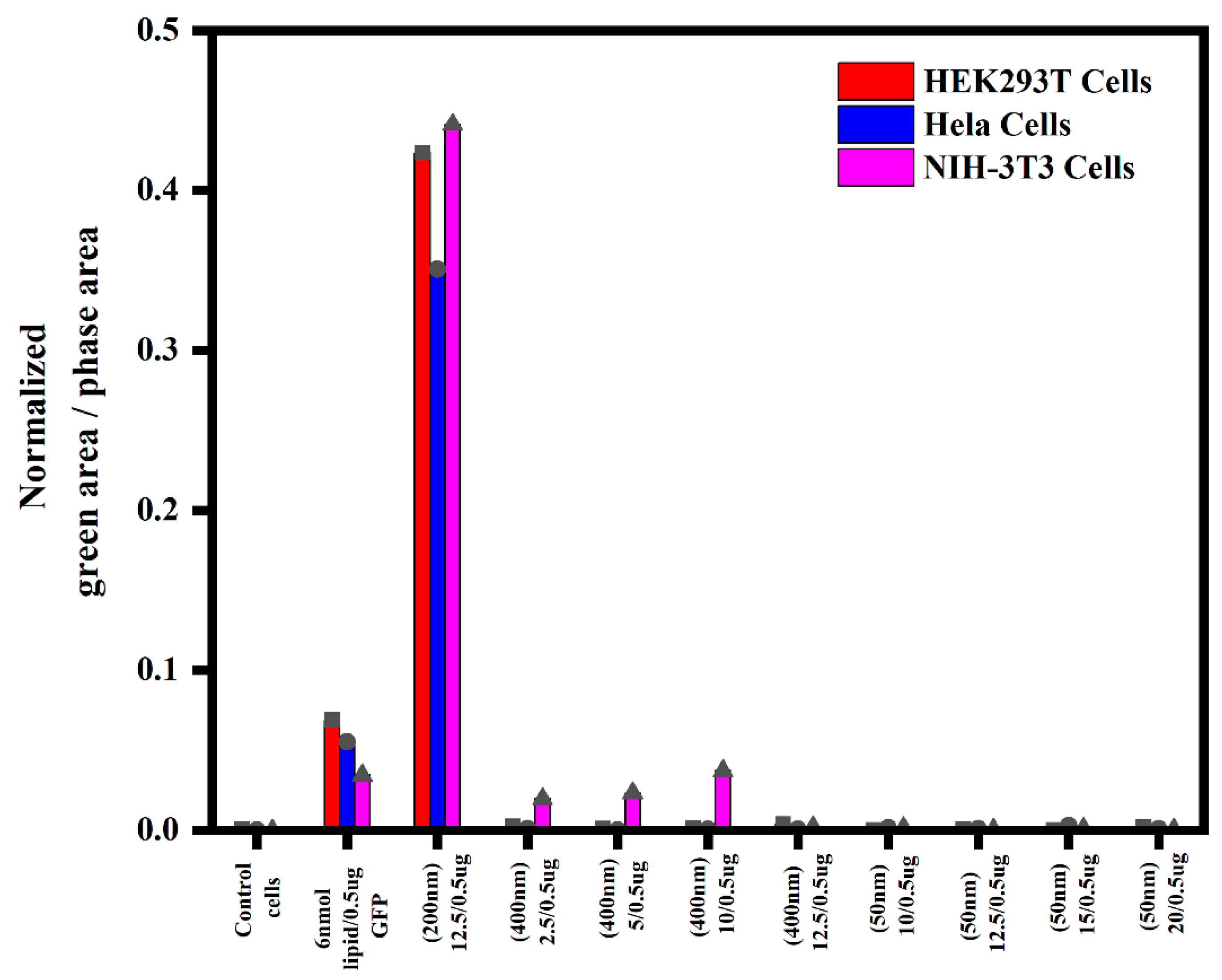
3.1.4. Semi-Quantitative Analysis of In Vitro Transfections in Various Cell Lines Using an Incucyte Microscope
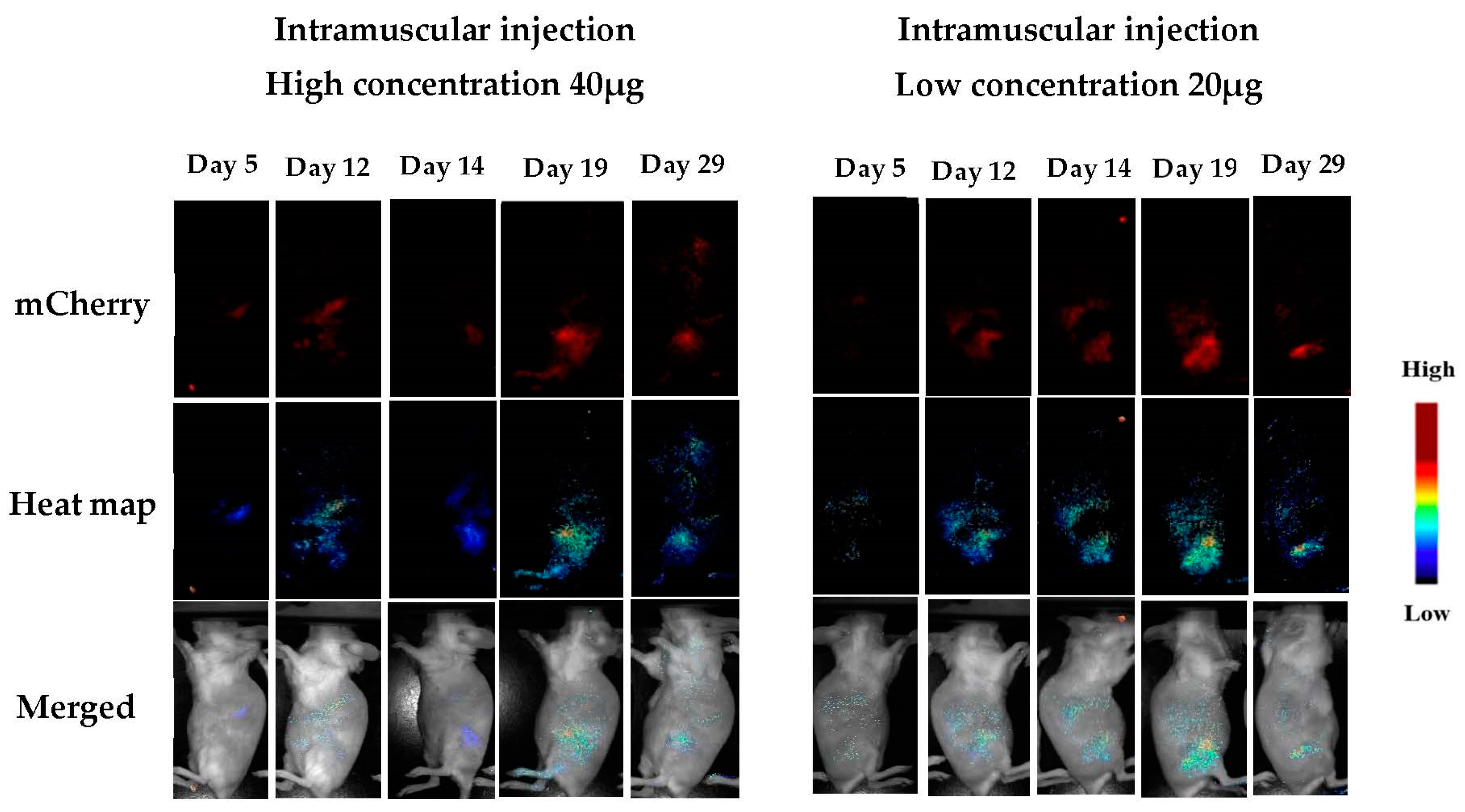
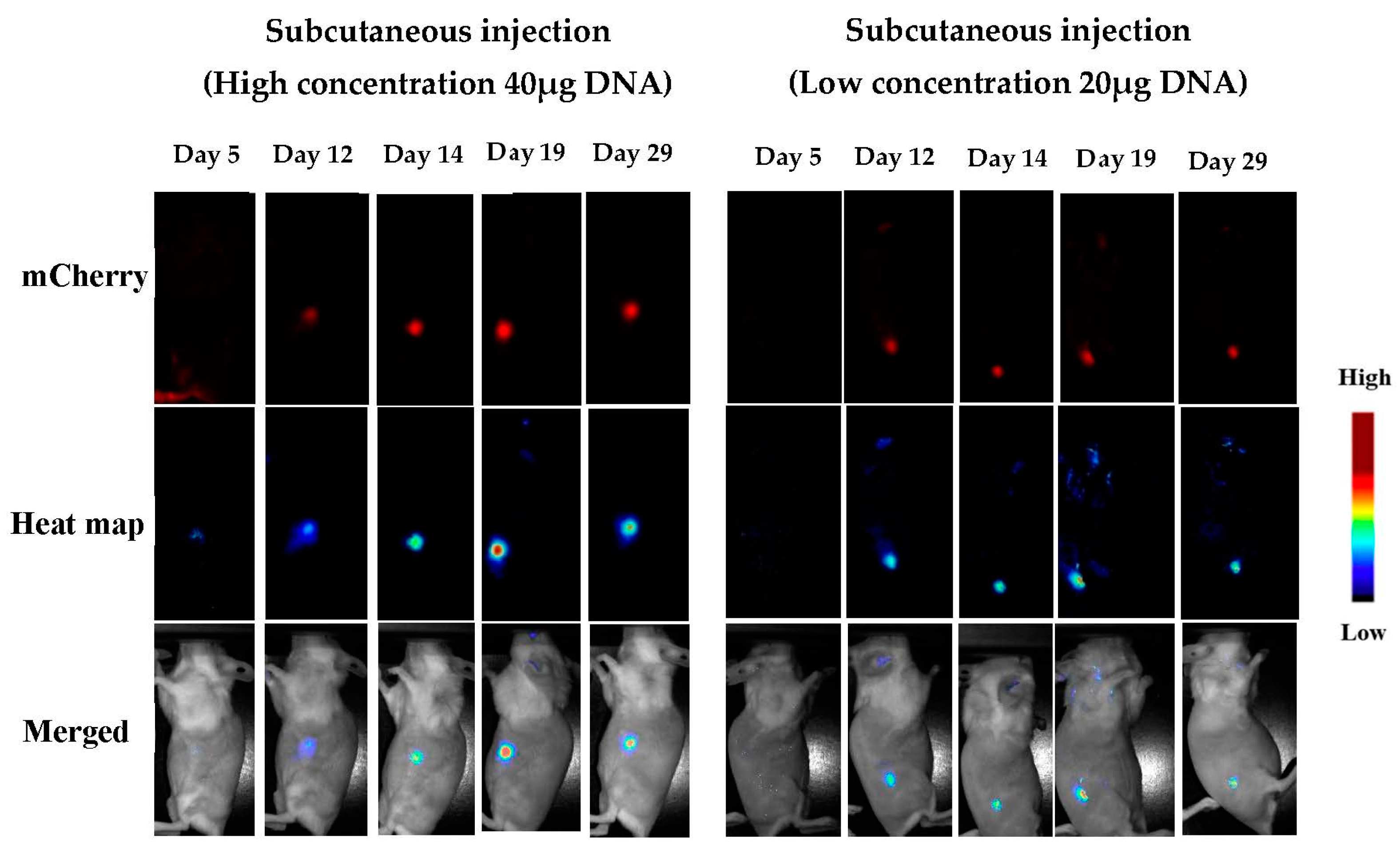
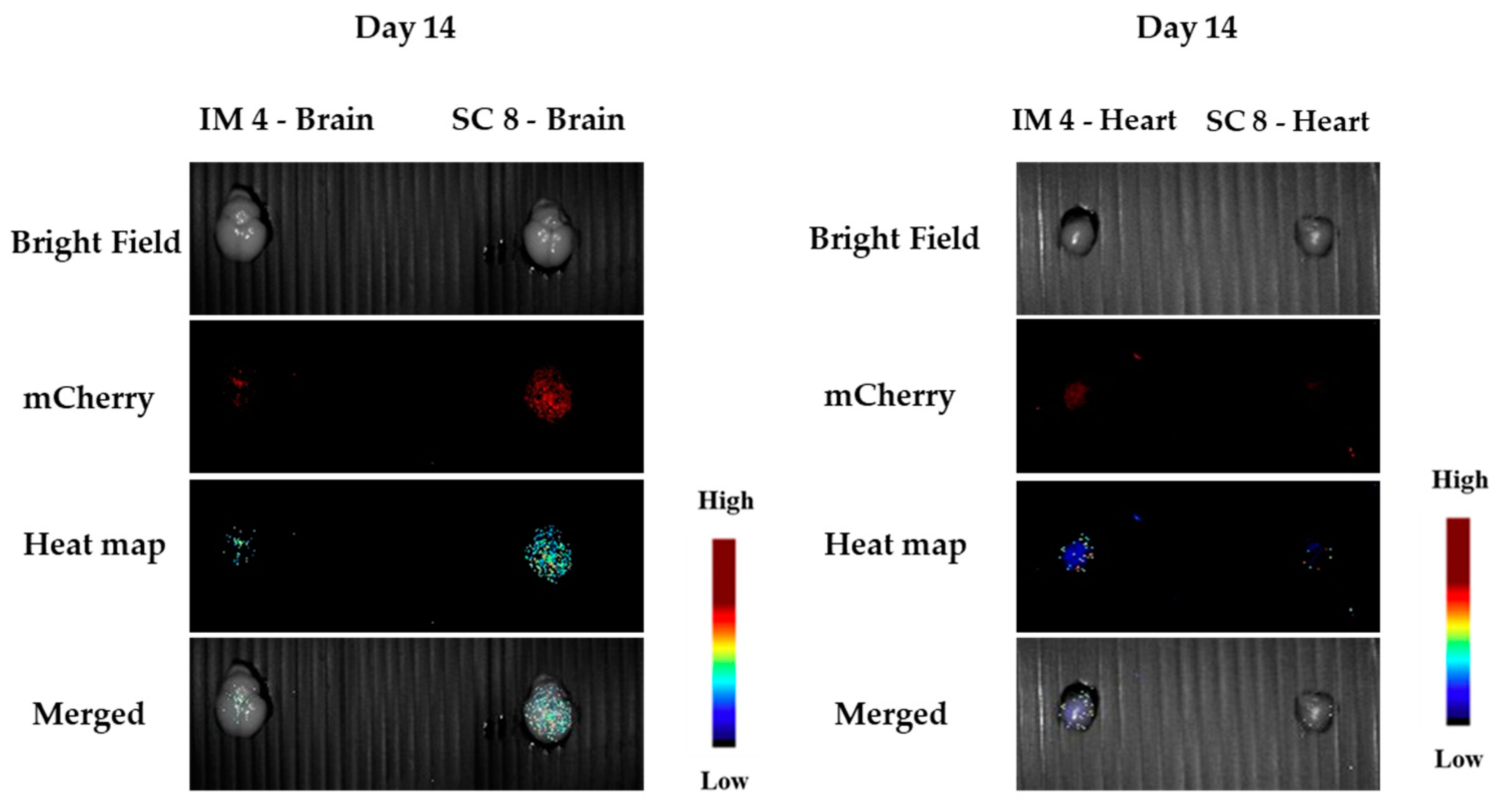
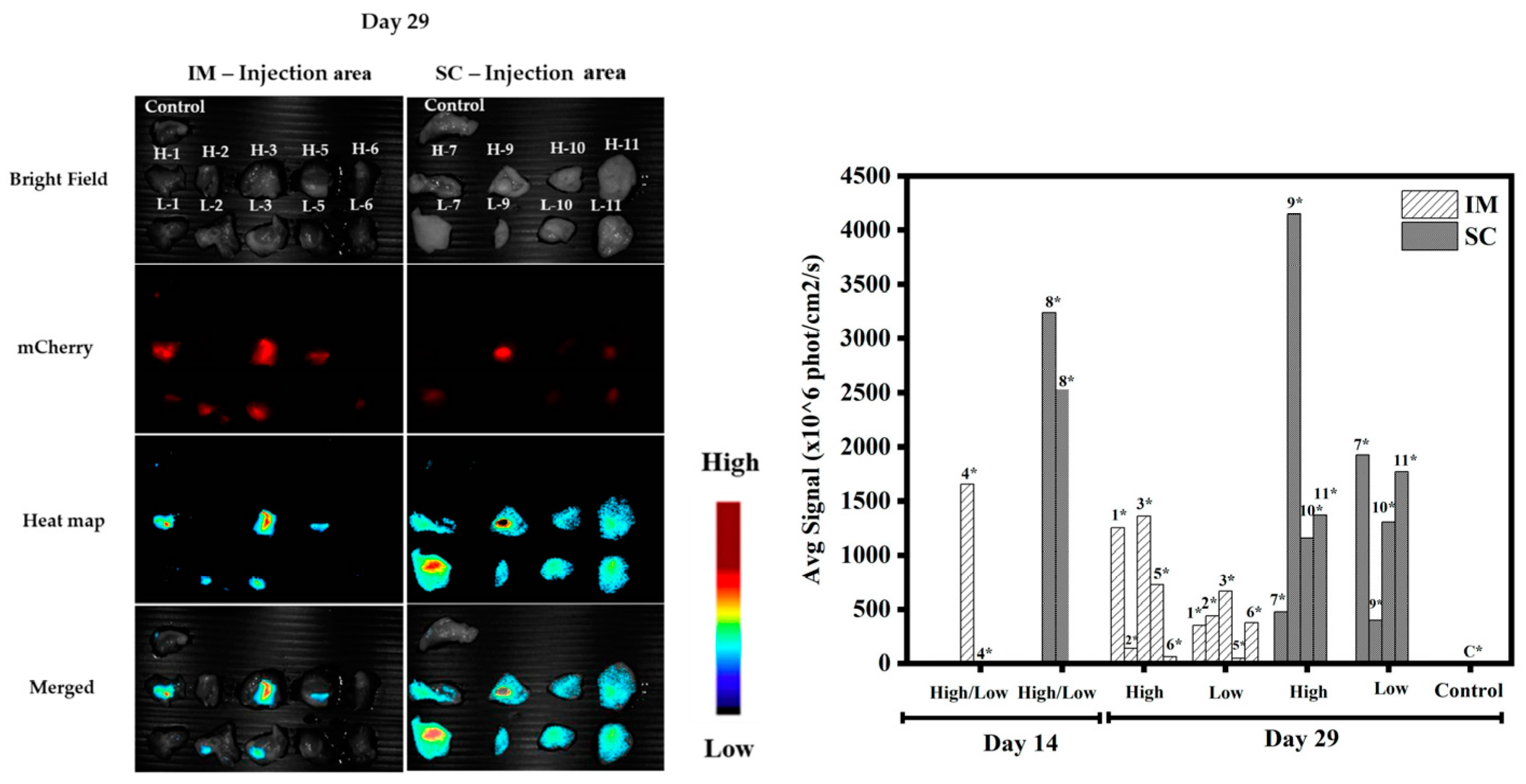
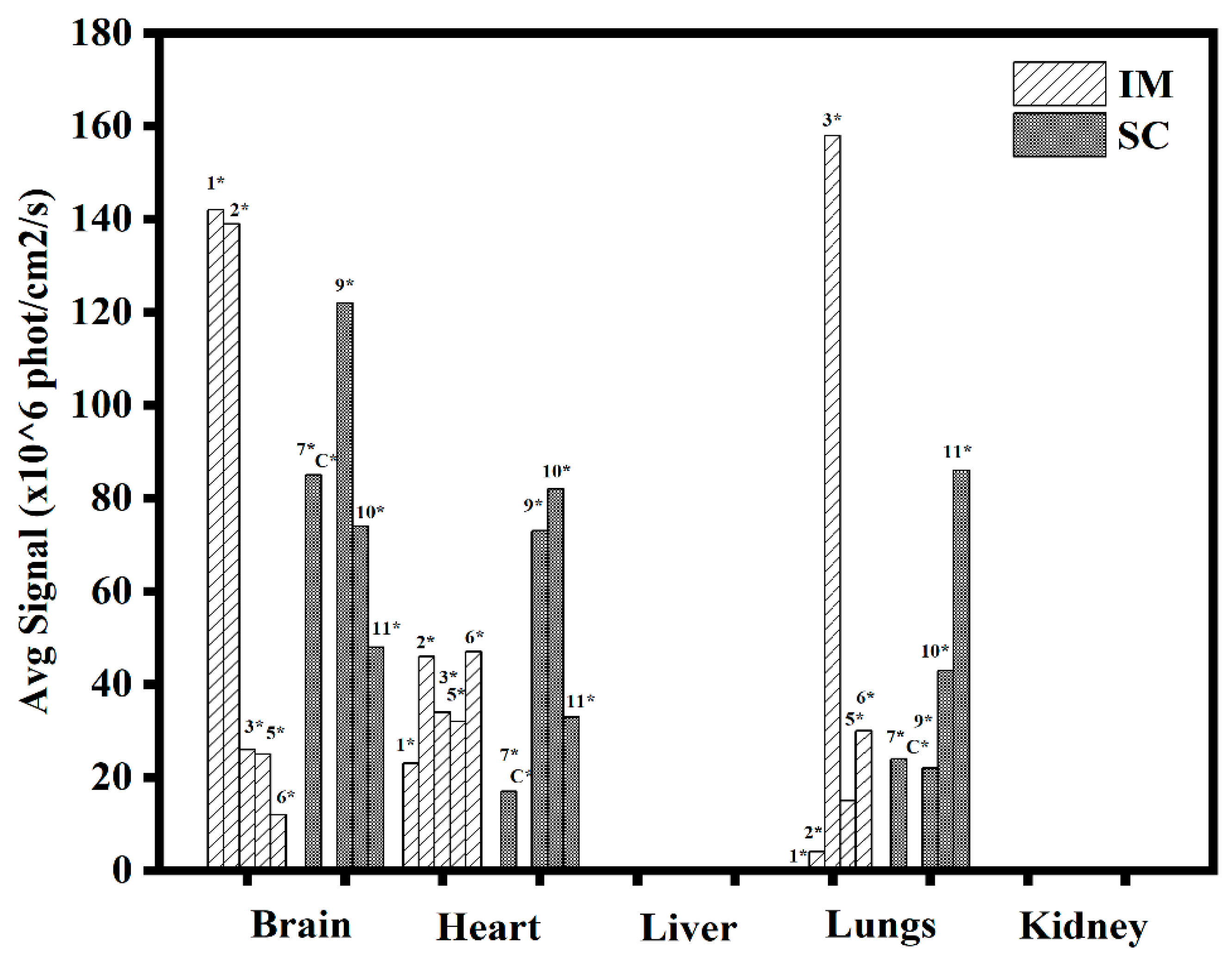
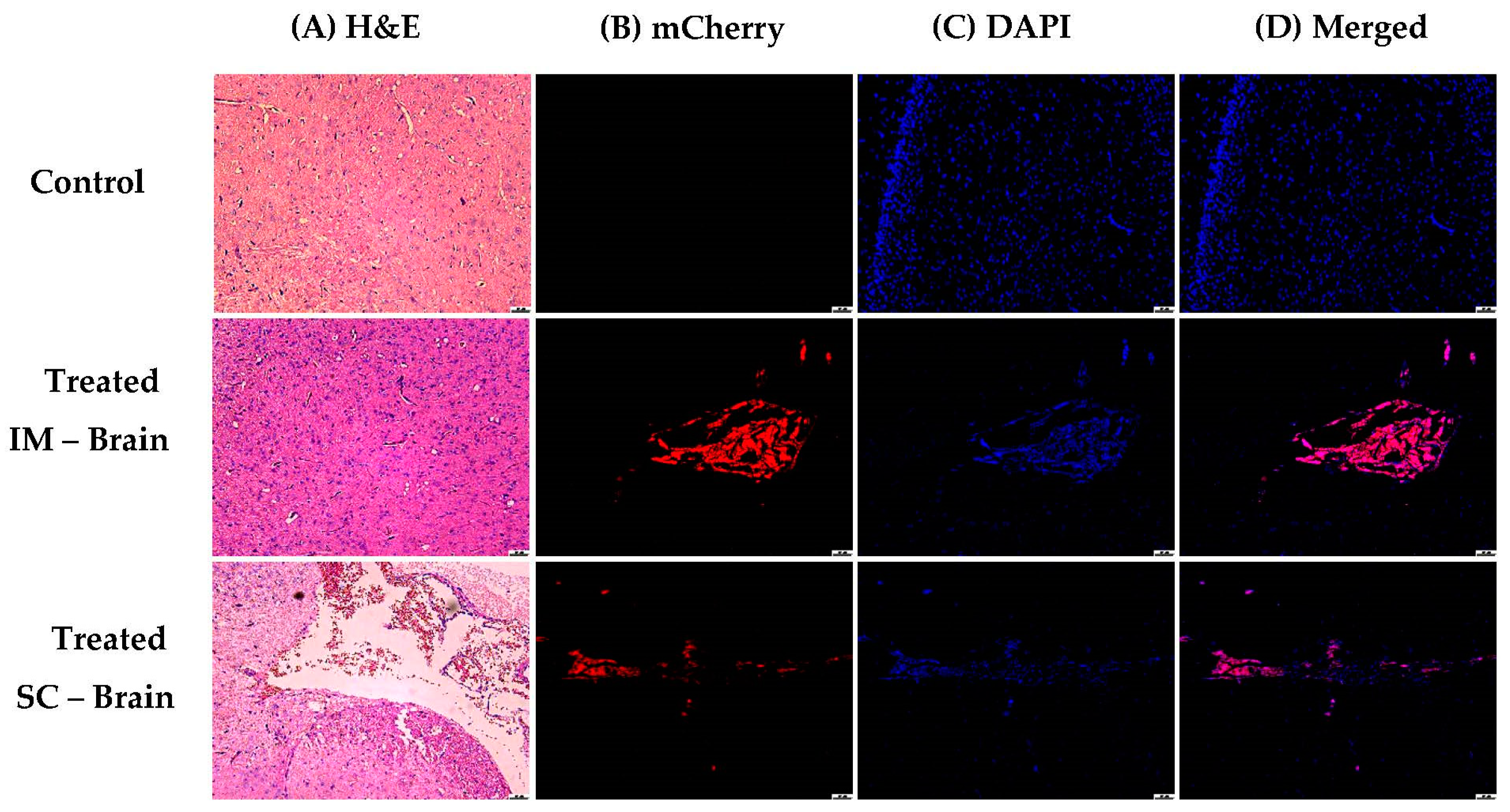
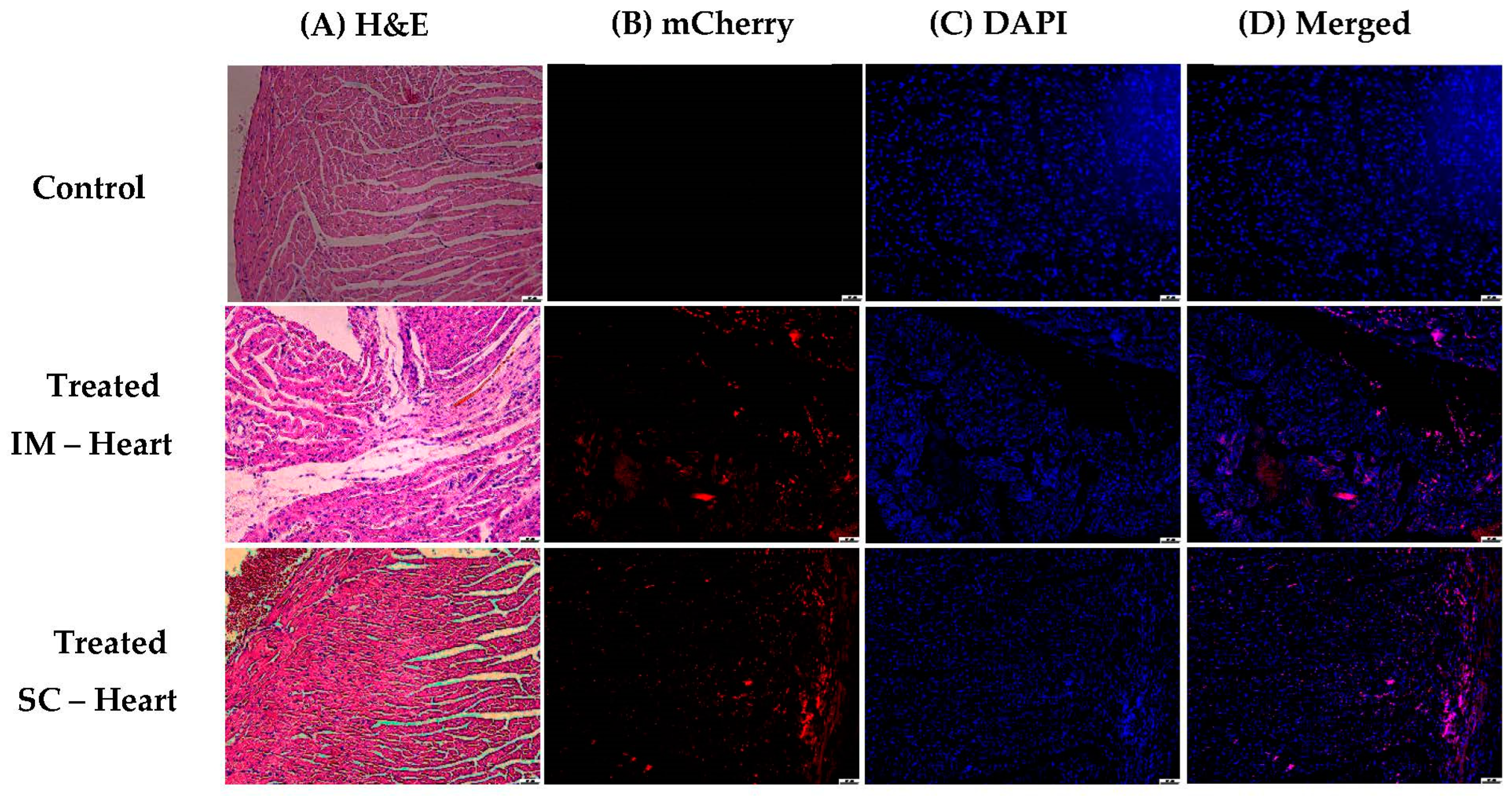
3.1.5. In Vivo Transfections
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- Song, X.; Liu, C.; Wang, N.; Huang, H.; He, S.; Gong, C.; Wei, Y. Delivery of CRISPR/Cas systems for cancer gene therapy and immunotherapy. Adv. Drug Deliv. Rev. 2021, 168, 158–180. [Google Scholar] [CrossRef]
- Elsworth, J.D. Parkinson’s disease treatment: Past, present, and future. J. Neural Transm. 2020, 127, 785–791. [Google Scholar] [CrossRef] [PubMed]
- Abreu, N.J.; Waldrop, M.A. Overview of gene therapy in spinal muscular atrophy and Duchenne muscular dystrophy. Pediatr. Pulmonol. 2021, 56, 710–720. [Google Scholar] [CrossRef] [PubMed]
- Arora, S.; Layek, B.; Singh, J. Design and Validation of Liposomal ApoE2 Gene Delivery System to Evade Blood-Brain Barrier for Effective Treatment of Alzheimer’s Disease. Mol. Pharm. 2021, 18, 714–725. [Google Scholar] [CrossRef]
- Wadhwa, A.; Aljabbari, A.; Lokras, A.; Foged, C.; Thakur, A. Opportunities and challenges in the delivery of mrna-based vaccines. Pharmaceutics 2020, 12, 102. [Google Scholar] [CrossRef] [Green Version]
- Shi, B.; Zheng, M.; Tao, W.; Chung, R.; Jin, D.; Ghaffari, D.; Farokhzad, O.C. Challenges in DNA Delivery and Recent Advances in Multifunctional Polymeric DNA Delivery Systems. Biomacromolecules 2017, 18, 2231–2246. [Google Scholar] [CrossRef]
- Gheibi-Hayat, S.M.; Jamialahmadi, K. Antisense Oligonucleotide (AS-ODN) Technology: Principle, Mechanism and Challenges. Biotechnol. Appl. Biochem. 2021, 68, 1086–1094. [Google Scholar] [CrossRef] [PubMed]
- Jiao, Y.; Xia, Z.L.; Ze, L.J.; Jing, H.; Xin, B.; Fu, S. Research Progress of nucleic acid delivery vectors for gene therapy. Biomed. Microdevices 2020, 22, 10. [Google Scholar] [CrossRef] [PubMed]
- Ghosh, S.; Brown, A.M.; Jenkins, C.; Campbell, K. Viral Vector Systems for Gene Therapy: A Comprehensive Literature Review of Progress and Biosafety Challenges. Appl. Biosaf. 2020, 25, 7–18. [Google Scholar] [CrossRef]
- Yahya, E.B.; Alqadhi, A.M. Recent trends in cancer therapy: A review on the current state of gene delivery. Life Sci. 2021, 269, 119087. [Google Scholar] [CrossRef]
- Song, H.; Wang, G.; He, B.; Li, L.; Li, C.; Lai, Y.; Xu, X.; Gu, Z. Cationic lipid-coated PEI/DNA polyplexes with improved efficiency and reduced cytotoxicity for gene delivery into mesenchymal stem cells. Int. J. Nanomed. 2012, 7, 4637–4648. [Google Scholar]
- Joubert, D.; Van Zyl, J.; Hawtrey, A.; Ariatti, M. A note on poly-L-lysine-mediated gene transfer in HeLa cells. Drug Deliv. J. Deliv. Target. Ther. Agents 2003, 10, 209–211. [Google Scholar] [CrossRef]
- Elzes, M.R.; Akeroyd, N.; Engbersen, J.F.J.; Paulusse, J.M.J. Disulfide-functional poly(amido amine)s with tunable degradability for gene delivery. J. Control. Release 2016, 244, 357–365. [Google Scholar] [CrossRef] [PubMed]
- Liu, T.; Wu, X.; Wang, Y.; Hou, X.; Jiang, G.; Wu, T.; Xie, H.; Xie, M. CD-PLLD co-delivering docetaxel and MMP-9 siRNA plasmid for nasopharyngeal carcinoma therapy in vivo. Mol. Med. Rep. 2017, 16, 1383–1388. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, J.; Lee, S.; Kwon, Y.E.; Kim, Y.J.; Choi, J.S. Gene Delivery by PAMAM Dendrimer Conjugated with the Nuclear Localization Signal Peptide Derived from Influenza B Virus Nucleoprotein. Macromol. Res. 2019, 27, 360–368. [Google Scholar] [CrossRef]
- Chen, P.; Qiu, M.; Deng, C.; Meng, F.; Zhang, J.; Cheng, R.; Zhong, Z. pH-Responsive Chimaeric Pepsomes Based on Asymmetric Poly(ethylene glycol)-b-Poly(L-leucine)-b-Poly(L-glutamic acid) Triblock Copolymer for Efficient Loading and Active Intracellular Delivery of Doxorubicin Hydrochloride. Biomacromolecules 2015, 16, 1322–1330. [Google Scholar] [CrossRef]
- Yilmaz, Z.E.; Jérôme, C. Polyphosphoesters: New Trends in Synthesis and Drug Delivery Applications. Macromol. Biosci. 2016, 16, 1745–1761. [Google Scholar] [CrossRef]
- Yang, J.; Zhang, Q.; Chang, H.; Cheng, Y. Surface-Engineered Dendrimers in Gene Delivery. Chem. Rev. 2015, 115, 5274–5300. [Google Scholar] [CrossRef]
- Olden, B.R.; Cheng, Y.; Yu, J.L.; Pun, S.H. Cationic polymers for non-viral gene delivery to human T cells. J. Control. Release 2018, 282, 140–147. [Google Scholar] [CrossRef]
- Zhi, D.; Bai, Y.; Yang, J.; Cui, S.; Zhao, Y.; Chen, H.; Zhang, S. A review on cationic lipids with different linkers for gene delivery. Adv. Colloid. Interface Sci. 2018, 253, 117–140. [Google Scholar] [CrossRef]
- Dowaidar, M.; Abdelhamid, H.N.; Hällbrink, M.; Freimann, K.; Kurrikoff, K.; Zou, X.; Langel, Ü. Magnetic Nanoparticle Assisted Self-assembly of Cell Penetrating Peptides-Oligonucleotides Complexes for Gene Delivery. Sci. Rep. 2017, 7, 11. [Google Scholar] [CrossRef] [Green Version]
- Amani, A.; Kabiri, T.; Shafiee, S.; Hamidi, A. Preparation and characterization of PLA-PEG-PLA/PEI/DNA nanoparticles for improvement of transfection efficiency and controlled release of DNA in gene delivery systems. Iran. J. Pharm. Res. 2019, 18, 125–141. [Google Scholar]
- Salameh, J.W.; Zhou, L.; Ward, S.M.; Santa Chalarca, C.F.; Emrick, T.; Figueiredo, M.L. Polymer-mediated gene therapy: Recent advances and merging of delivery techniques. Wiley Interdiscip Rev. Nanomed. Nanobiotechnol. 2020, 12, e1598. [Google Scholar] [CrossRef]
- Mukalel, A.J.; Riley, R.S.; Zhang, R.; Mitchell, M.J. Nanoparticles for nucleic acid delivery: Applications in cancer immunotherapy. Cancer Lett. 2019, 458, 102–112. [Google Scholar] [CrossRef]
- Zhang, X.; Zhang, K.; Haag, R. Multi-stage, charge conversional, stimuli-responsive nanogels for therapeutic protein delivery. Biomater. Sci. 2015, 3, 1487–1496. [Google Scholar] [CrossRef]
- Niu, Z.; Conejos-Sánchez, I.; Griffin, B.T.; O’Driscoll, C.M.; Alonso, M.J. Lipid-based nanocarriers for oral peptide delivery. Adv. Drug Deliv. Rev. 2016, 106, 337–354. [Google Scholar] [CrossRef]
- Zamboni, C.G.; Kozielski, K.L.; Vaughan, H.J.; Nakata, M.M.; Kim, J.; Higgins, L.J.; Pomper, M.G. Green JJ. Polymeric nanoparticles as cancer-specific DNA delivery vectors to human hepatocellular carcinoma. J. Control. Release 2017, 263, 18–28. [Google Scholar] [CrossRef]
- Yuan, Y.; Zhang, C.J.; Liu, B. A Photoactivatable AIE Polymer for Light-Controlled Gene Delivery: Concurrent Endo/Lysosomal Escape and DNA Unpacking. Angew Chemie-Int Ed. 2015, 54, 11419–11423. [Google Scholar] [CrossRef]
- Eswaran, L.; Kazimirsky, G.; Byk, G. New Biocompatible Nanohydrogels of Predefined Sizes for Complexing Nucleic Acids. Pharmaceutics 2023, 15, 332. [Google Scholar] [CrossRef]
- Byk, G.; Dubertret, C.; Escriou, V.; Frederic, M.; Jaslin, G.; Rangara, R.; Pitard, B.; Crouzet, J.; Wils, P.; Schwartz, B.; et al. Synthesis, activity, and structure-activity relationship studies of novel cationic lipids for DNA transfer. J. Med. Chem. 1998, 41, 224–235. [Google Scholar] [CrossRef]
- Jin, J.; Zhu, L.; Chen, M.; Xu, H.; Wang, H.; Feng, X.; Zhu, X.; Zhou, Q. The optimal choice of medication administration route regarding intravenous, intramuscular, and subcutaneous injection. Patient Prefer. Adherence 2015, 9, 923–942. [Google Scholar] [PubMed] [Green Version]
- Pepe, S.; Gregory, A.T.; Robert Denniss, A. Myocarditis, Pericarditis and Cardiomyopathy After COVID-19 Vaccination. Heart Lung Circ. 2021, 30, 1425–1429. [Google Scholar] [CrossRef] [PubMed]
- Heymans, S.; Cooper, L.T. Myocarditis after COVID-19 mRNA vaccination: Clinical observations and potential mechanisms. Nat. Rev. Cardiol. 2022, 19, 75–77. [Google Scholar] [CrossRef] [PubMed]







Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Eswaran, L.; Kazimirsky, G.; Yehuda, R.; Byk, G. A New Strategy for Nucleic Acid Delivery and Protein Expression Using Biocompatible Nanohydrogels of Predefined Sizes. Pharmaceutics 2023, 15, 961. https://doi.org/10.3390/pharmaceutics15030961
Eswaran L, Kazimirsky G, Yehuda R, Byk G. A New Strategy for Nucleic Acid Delivery and Protein Expression Using Biocompatible Nanohydrogels of Predefined Sizes. Pharmaceutics. 2023; 15(3):961. https://doi.org/10.3390/pharmaceutics15030961
Chicago/Turabian StyleEswaran, Lakshmanan, Gila Kazimirsky, Ronen Yehuda, and Gerardo Byk. 2023. "A New Strategy for Nucleic Acid Delivery and Protein Expression Using Biocompatible Nanohydrogels of Predefined Sizes" Pharmaceutics 15, no. 3: 961. https://doi.org/10.3390/pharmaceutics15030961
APA StyleEswaran, L., Kazimirsky, G., Yehuda, R., & Byk, G. (2023). A New Strategy for Nucleic Acid Delivery and Protein Expression Using Biocompatible Nanohydrogels of Predefined Sizes. Pharmaceutics, 15(3), 961. https://doi.org/10.3390/pharmaceutics15030961







