Recombinant Proteins for Assembling as Nano- and Micro-Scale Materials for Drug Delivery: A Host Comparative Overview
Abstract
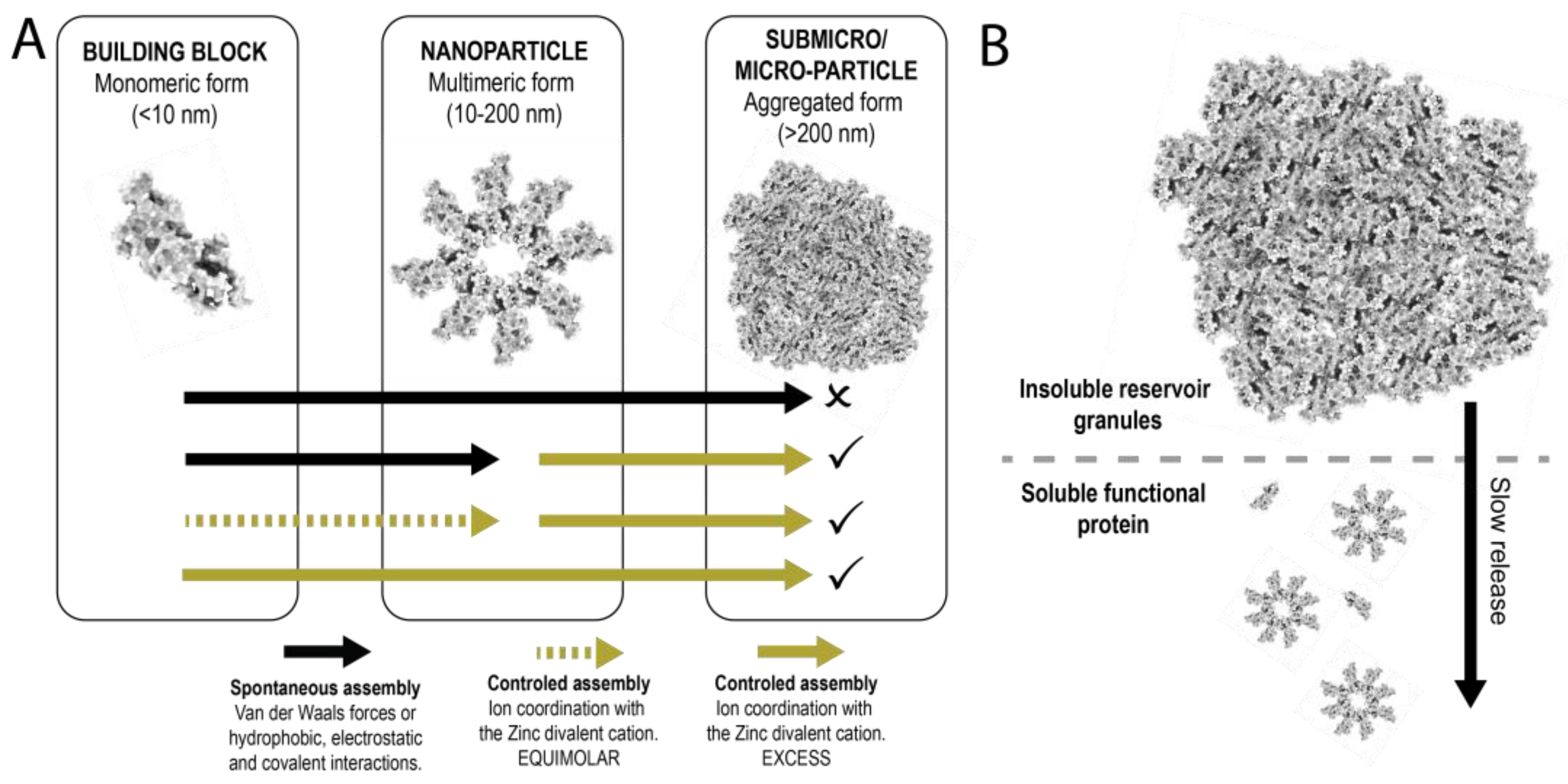
1. Introduction
2. Materials and Methods
2.1. Genetic Design
2.2. Protein Production in Bacteria
2.3. Protein Production in Sf9 Cells
2.4. Protein Production in Mammalian Cells
2.5. Electrophoresis and Western Blot
2.6. Electron Microscopy
2.7. Determination of Material Volume
2.8. Formation of Microparticles
2.9. Analysis of Protein Release
3. Results
3.1. Protein Production
3.2. Formation of Nanoparticles
3.3. Formation and Disintegration of Microparticles
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A

| Refolding Buffer | Step 1 | Step 2 | Step 3 | Step 4 | Storage Buffer | |
|---|---|---|---|---|---|---|
| Phosphate buffer pH 8 | 20 mM | 20 mM | 20 mM | 20 mM | 20 mM | 20 mM |
| EDTA | 0.18 mM | - | - | - | - | - |
| Gluthathione red. | 1.9 mM | - | - | - | - | - |
| Gluthatione ox | 0.9 mM | - | - | - | - | - |
| L-Arginine | 0.5 M | - | - | - | 75 mM | 75 mM |
| Urea | 2 M | 1 M | 0.5 M | 0.02 M | - | - |
| Sucrose | - | - | 292 mM | 292 mM | 233 mM | 233 mM |
| Polysorbate 20 | - | - | 0.25 mM | 0.25 mM | 0.25 mM | - |
References
- Hu, X.; Cebe, P.; Weiss, A.S.; Omenetto, F.; Kaplan, D.L. Protein-Based Composite Materials. Mater. Today 2012, 15, 208–215. [Google Scholar] [CrossRef]
- Qian, Z.G.; Pan, F.; Xia, X.X. Synthetic Biology for Protein-Based Materials. Curr. Opin. Biotechnol. 2020, 65, 197–204. [Google Scholar] [CrossRef] [PubMed]
- Korpi, A.; Anaya-Plaza, E.; Välimäki, S.; Kostiainen, M. Highly Ordered Protein Cage Assemblies: A Toolkit for New Materials. Wiley Interdiscip. Rev. Nanomed. Nanobiotechnol. 2020, 12, e1578. [Google Scholar] [CrossRef] [PubMed]
- Harrington, M.J.; Fratzl, P. Natural Load-Bearing Protein Materials. Prog. Mater. Sci. 2021, 120, 100767. [Google Scholar] [CrossRef]
- Lendel, C.; Solin, N. Protein Nanofibrils and Their Use as Building Blocks of Sustainable Materials. RSC Adv. 2021, 11, 39188–39215. [Google Scholar] [CrossRef]
- López-Laguna, H.; Sánchez, J.; Unzueta, U.; Mangues, R.; Vázquez, E.; Villaverde, A. Divalent Cations: A Molecular Glue for Protein Materials Trends in Biochemical Sciences An Official Publication of the International Union of Biochemistry and Molecul ar Biology. Trends Biochem. Sci. 2020, 45, 992–1003. [Google Scholar] [CrossRef]
- Corchero, J.L.; Vázquez, E.; García-Fruitós, E.; Ferrer-Miralles, N.; Villaverde, A. Recombinant Protein Materials for Bioengineering and Nanomedicine. Nanomedicine 2014, 9, 2817–2828. [Google Scholar] [CrossRef]
- Cano-Garrido, O.; Serna, N.; Unzueta, U.; Parladé, E.; Mangues, R.; Villaverde, A.; Vázquez, E. Protein Scaffolds in Human Clinics. Biotechnol. Adv. 2022, 61, 108032. [Google Scholar] [CrossRef]
- Cicciù, M.; Fiorillo, L.; Cervino, G.; Habal, M.B. Bone Morophogenetic Protein Application as Grafting Materials for Bone Regeneration in Craniofacial Surgery: Current Application and Future Directions. J. Craniofacial Surg. 2021, 32, 787–793. [Google Scholar] [CrossRef]
- Farrar, D.F. Bone Adhesives for Trauma Surgery: A Review of Challenges and Developments. Int. J. Adhes. Adhes. 2012, 33, 89–97. [Google Scholar] [CrossRef]
- Bello, A.B.; Kim, D.; Kim, D.; Park, H.; Lee, S.H. Engineering and Functionalization of Gelatin Biomaterials: From Cell Culture to Medical Applications. Tissue Eng. Part B Rev. 2020, 26, 164–180. [Google Scholar] [CrossRef] [PubMed]
- Kermanizadeh, A.; Balharry, D.; Wallin, H.; Loft, S.; Møller, P. Nanomaterial Translocation-the Biokinetics, Tissue Accumulation, Toxicity and Fate of Materials in Secondary Organs-A Review. Crit. Rev. Toxicol. 2015, 45, 837–872. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Qin, Z.; Zeng, W.; Yang, T.; Cao, Y.; Mei, C.; Kuang, Y. Toxicity Assessment of Nanoparticles in Various Systems and Organs. Nanotechnol. Rev. 2017, 6, 279–289. [Google Scholar] [CrossRef]
- Sahu, S.C.; Hayes, A.W. Toxicity of Nanomaterials Found in Human Environment. Toxicol. Res. Appl. 2017, 1, 2397847317726352. [Google Scholar] [CrossRef]
- de Pinho Favaro, M.T.; Atienza-Garriga, J.; Martínez-Torró, C.; Parladé, E.; Vázquez, E.; Corchero, J.L.; Ferrer-Miralles, N.; Villaverde, A. Recombinant Vaccines in 2022: A Perspective from the Cell Factory. Microb. Cell Fact. 2022, 21, 203. [Google Scholar] [CrossRef]
- Parladé, E.; Voltà-Durán, E.; Cano-Garrido, O.; Sánchez, J.M.; Unzueta, U.; López-Laguna, H.; Serna, N.; Cano, M.; Rodríguez-Mariscal, M.; Vazquez, E.; et al. An In Silico Methodology That Facilitates Decision Making in the Engineering of Nanoscale Protein Materials. Int. J. Mol. Sci. 2022, 23, 4958. [Google Scholar] [CrossRef]
- López-Laguna, H.; Voltà-Durán, E.; Parladé, E.; Villaverde, A.; Vázquez, E.; Unzueta, U. Insights on the Emerging Biotechnology of Histidine-Rich Peptides. Biotechnol. Adv. 2022, 54, 107817. [Google Scholar] [CrossRef]
- Voltà-Durán, E.; Sánchez, J.M.; López-Laguna, H.; Parladé, E.; Sánchez-García, L.; Sánchez-Chardi, A.; de Marco, A.; Unzueta, U.; Vázquez, E.; Villaverde, A. The Spectrum of Building Block Conformers Sustains the Biophysical Properties of Clinically-Oriented Self-Assembling Protein Nanoparticles. Sci. China Mater. 2022, 65, 1662–1670. [Google Scholar] [CrossRef]
- López-Laguna, H.; Sánchez, J.M.; Carratalá, J.V.; Rojas-Peña, M.; Sánchez-García, L.; Parladé, E.; Sánchez-Chardi, A.; Voltà-Durán, E.; Serna, N.; Cano-Garrido, O.; et al. Biofabrication of Functional Protein Nanoparticles through Simple His-Tag Engineering. ACS Sustain. Chem. Eng. 2021, 9, 12341–12354. [Google Scholar] [CrossRef] [PubMed]
- Sanchez, J.M.; López-Laguna, H.; Serna, N.; Unzueta, U.; Clop, P.D.; Villaverde, A.; Vazquez, E. Engineering the Performance of Artificial Inclusion Bodies Built of Catalytic β-Galactosidase. ACS Sustain. Chem. Eng. 2021, 9, 2552–2558. [Google Scholar] [CrossRef]
- López-Laguna, H.; Sánchez-García, L.; Serna, N.; Voltà-Durán, E.; Sánchez, J.M.; Sánchez-Chardi, A.; Unzueta, U.; Łoś, M.; Villaverde, A.; Vázquez López-Laguna, E.H.; et al. 2001885 (1 of 8) Engineering Protein Nanoparticles Out from Components of the Human Microbiome. Small 2020, 16, 2001885. [Google Scholar] [CrossRef] [PubMed]
- Voltà-Durán, E.; Serna, N.; Sánchez-García, L.; Aviñó, A.; Sánchez, J.M.; López-Laguna, H.; Cano-Garrido, O.; Casanova, I.; Mangues, R.; Eritja, R.; et al. Design and Engineering of Tumor-Targeted, Dual-Acting Cytotoxic Nanoparticles. Acta Biomater. 2021, 119, 312–322. [Google Scholar] [CrossRef] [PubMed]
- Pallarès, V.; Núñez, Y.; Sánchez-García, L.; Falgàs, A.; Serna, N.; Unzueta, U.; Gallardo, A.; Alba-Castellón, L.; Álamo, P.; Sierra, J.; et al. Antineoplastic Effect of a Diphtheria Toxin-Based Nanoparticle Targeting Acute Myeloid Leukemia Cells Overexpressing CXCR4. J. Control. Release 2021, 335, 117–129. [Google Scholar] [CrossRef] [PubMed]
- Rioja-Blanco, E.; Arroyo-Solera, I.; Álamo, P.; Casanova, I.; Gallardo, A.; Unzueta, U.; Serna, N.; Sánchez-García, L.; Quer, M.; Villaverde, A.; et al. Self-Assembling Protein Nanocarrier for Selective Delivery of Cytotoxic Polypeptides to CXCR4+ Head and Neck Squamous Cell Carcinoma Tumors. Acta Pharm. Sin. B 2022, 12, 2578–2591. [Google Scholar] [CrossRef]
- Serna, N.; Álamo, P.; Ramesh, P.; Vinokurova, D.; Sánchez-García, L.; Unzueta, U.; Gallardo, A.; Céspedes, M.V.; Vázquez, E.; Villaverde, A.; et al. Nanostructured Toxins for the Selective Destruction of Drug-Resistant Human CXCR4+ Colorectal Cancer Stem Cells. J. Control. Release 2020, 320, 96–104. [Google Scholar] [CrossRef] [PubMed]
- Falgàs, A.; Pallarès, V.; Unzueta, U.; Núñez, Y.; Sierra, J.; Gallardo, A.; Alba-Castellón, L.; Mangues, M.A.; Álamo, P.; Villaverde, A.; et al. Specific Cytotoxic Effect of an Auristatin Nanoconjugate towards Cxcr4+ Diffuse Large B-Cell Lymphoma Cells. Int. J. Nanomed. 2021, 16, 1869. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Z.; Huang, Y.; Wang, J.; Lin, H.; Cao, F.; Li, S.; Li, Y.; Li, Z.; Liu, X. A Self-Assembling CXCR4-Targeted Pyroptosis Nanotoxin for Melanoma Therapy. Biomater. Sci. 2023, 11, 2200–2210. [Google Scholar] [CrossRef]
- Chen, T.Y.; Cheng, W.J.; Horng, J.C.; Hsu, H.Y. Artificial Peptide-Controlled Protein Release of Zn2+-Triggered, Self-Assembled Histidine-Tagged Protein Microparticle. Colloids Surf B Biointerfaces 2020, 187, 110644. [Google Scholar] [CrossRef]
- López-Laguna, H.; Parladé, E.; Álamo, P.; Sánchez, J.M.; Voltà-Durán, E.; Serna, N.; Sánchez-García, L.; Cano-Garrido, O.; Sánchez-Chardi, A.; Villaverde, A.; et al. In Vitro Fabrication of Microscale Secretory Granules. Adv. Funct.Mater. 2021, 31, 2100914. [Google Scholar] [CrossRef]
- Sánchez, J.M.; López-Laguna, H.; Álamo, P.; Serna, N.; Sánchez-Chardi, A.; Nolan, V.; Cano-Garrido, O.; Casanova, I.; Unzueta, U.; Vazquez, E.; et al. Artificial Inclusion Bodies for Clinical Development. Adv. Sci. 2020, 7, 1902420. [Google Scholar] [CrossRef]
- Álamo, P.; Parladé, E.; López-Laguna, H.; Voltà-Durán, E.; Unzueta, U.; Vazquez, E.; Mangues, R.; Villaverde, A. Ion-Dependent Slow Protein Release from in vivo Disintegrating Micro-Granules. Drug Deliv. 2021, 28, 2383–2391. [Google Scholar] [CrossRef] [PubMed]
- Serna, N.; Falgàs, A.; García-León, A.; Unzueta, U.; Núñez, Y.; Sánchez-Chardi, A.; Martínez-Torró, C.; Mangues, R.; Vazquez, E.; Casanova, I.; et al. Time-Prolonged Release of Tumor-Targeted Protein–MMAE Nanoconjugates from Implantable Hybrid Materials. Pharmaceutics 2022, 14, 192. [Google Scholar] [CrossRef]
- Zhao, L.; Luo, L.; Meng, F. Metal-Mediated Nanobody Assemblies as Potent Alleviators of Human Islet Amyloid Polypeptide Aggregation. Mater. Chem. Front. 2023. [Google Scholar] [CrossRef]
- Serna, N.; Cano-Garrido, O.; Sánchez, J.M.; Sánchez-Chardi, A.; Sánchez-García, L.; López-Laguna, H.; Fernández, E.; Vázquez, E.; Villaverde, A. Release of Functional Fibroblast Growth Factor-2 from Artificial Inclusion Bodies. J. Control. Release 2020, 327, 61–69. [Google Scholar] [CrossRef] [PubMed]
- López-Laguna, H.; Unzueta, U.; Conchillo-Solé, O.; Sánchez-Chardi, A.; Pesarrodona, M.; Cano-Garrido, O.; Voltà, E.; Sánchez-García, L.; Serna, N.; Saccardo, P.; et al. Assembly of Histidine-Rich Protein Materials Controlled through Divalent Cations. Acta Biomater. 2019, 83, 257–264. [Google Scholar] [CrossRef] [PubMed]
- Jacob, R.S.; Anoop, A.; Maji, S.K. Protein Nanofibrils as Storage Forms of Peptide Drugs and Hormones. Adv. Exp. Med. Biol. 2019, 1174, 265–290. [Google Scholar] [CrossRef] [PubMed]
- Jacob, R.S.; Das, S.; Ghosh, S.; Anoop, A.; Jha, N.N.; Khan, T.; Singru, P.; Kumar, A.; Maji, S.K. Amyloid Formation of Growth Hormone in Presence of Zinc: Relevance to Its Storage in Secretory Granules. Sci. Rep. 2016, 6, 1–18. [Google Scholar] [CrossRef] [PubMed]
- Céspedes, M.V.; Cano-Garrido, O.; Álamo, P.; Sala, R.; Gallardo, A.; Serna, N.; Falgàs, A.; Voltà-Durán, E.; Casanova, I.; Sánchez-Chardi, A.; et al. Engineering Secretory Amyloids for Remote and Highly Selective Destruction of Metastatic Foci. Adv. Mater. 2020, 32, 1907348. [Google Scholar] [CrossRef] [PubMed]
- Sánchez, J.M.; Carratalá, J.V.; Serna, N.; Unzueta, U.; Nolan, V.; Sánchez-Chardi, A.; Voltà-Durán, E.; López-Laguna, H.; Ferrer-Miralles, N.; Villaverde, A.; et al. The Poly-Histidine Tag H6 Mediates Structural and Functional Properties of Disintegrating, Protein-Releasing Inclusion Bodies. Pharmaceutics 2022, 14, 602. [Google Scholar] [CrossRef] [PubMed]
- Soragni, A.; Maji, S.K.; Riek, R. Toward a Comprehension of Functional Aggregation into Amyloids in Pituitary Secretory Granules. Amyloid 2010, 17, 41. [Google Scholar]
- Maji, S.K.; Perrin, M.H.; Sawaya, M.R.; Jessberger, S.; Vadodaria, K.; Rissman, R.A.; Singru, P.S.; Nilsson, K.P.R.; Simon, R.; Schubert, D.; et al. Functional Amyloids as Natural Storage of Peptide Hormones in Pituitary Secretory Granules. Science 2009, 325, 328–332. [Google Scholar] [CrossRef] [PubMed]
- Tripathi, N.K.; Shrivastava, A. Recent Developments in Bioprocessing of Recombinant Proteins: Expression Hosts and Process Development. Front. Bioeng. Biotechnol. 2019, 7, 420. [Google Scholar] [CrossRef] [PubMed]
- Andersen, D.C.; Krummen, L. Recombinant Protein Expression for Therapeutic Applications. Curr. Opin. Biotechnol. 2002, 13, 117–123. [Google Scholar] [CrossRef] [PubMed]
- Chen, R. Bacterial Expression Systems for Recombinant Protein Production: E. Coli and Beyond. Biotechnol. Adv. 2012, 30, 1102–1107. [Google Scholar] [CrossRef]
- O’Flaherty, R.; Bergin, A.; Flampouri, E.; Mota, L.M.; Obaidi, I.; Quigley, A.; Xie, Y.; Butler, M. Mammalian Cell Culture for Production of Recombinant Proteins: A Review of the Critical Steps in Their Biomanufacturing. Biotechnol. Adv. 2020, 43, 107552. [Google Scholar] [CrossRef]
- Zahrl, R.J.; Peña, D.A.; Mattanovich, D.; Gasser, B. Systems Biotechnology for Protein Production in Pichia Pastoris. FEMS Yeast Res. 2017, 17, Issue 7, fox068. [Google Scholar] [CrossRef]
- Corchero, J.L.; Gasser, B.; Resina, D.; Smith, W.; Parrilli, E.; Vázquez, F.; Abasolo, I.; Giuliani, M.; Jäntti, J.; Ferrer, P.; et al. Unconventional Microbial Systems for the Cost-Efficient Production of High-Quality Protein Therapeutics. Biotechnol. Adv. 2013, 31, 140–153. [Google Scholar] [CrossRef] [PubMed]
- Merlin, M.; Gecchele, E.; Capaldi, S.; Pezzotti, M.; Avesani, L. Comparative Evaluation of Recombinant Protein Production in Different Biofactories: The Green Perspective. Biomed. Res. Int. 2014, 2014, 136419. [Google Scholar] [CrossRef]
- Gecchele, E.; Merlin, M.; Brozzetti, A.; Falorni, A.; Pezzotti, M.; Avesani, L. A Comparative Analysis of Recombinant Protein Expression in Different Biofactories: Bacteria, Insect Cells and Plant Systems. J. Vis. Exp. 2015, 2015, e52459. [Google Scholar] [CrossRef]
- Sevastsyanovich, Y.; Alfasi, S.; Cole, J. Recombinant Protein Production: A Comparative View on Host Physiology. N. Biotechnol. 2009, 25, 175–180. [Google Scholar] [CrossRef]
- Gasser, B.; Saloheimo, M.; Rinas, U.; Dragosits, M.; Rodríguez-Carmona, E.; Baumann, K.; Giuliani, M.; Parrilli, E.; Branduardi, P.; Lang, C.; et al. Protein Folding and Conformational Stress in Microbial Cells Producing Recombinant Proteins: A Host Comparative Overview. Microb. Cell Fact. 2008, 7, 11. [Google Scholar] [CrossRef]
- Dragosits, M.; Frascotti, G.; Bernard-Granger, L.; Vázquez, F.; Giuliani, M.; Baumann, K.; Rodríguez-Carmona, E.; Tokkanen, J.; Parrilli, E.; Wiebe, M.G.; et al. Influence of Growth Temperature on the Production of Antibody Fab Fragments in Different Microbes: A Host Comparative Analysis. Biotechnol. Prog. 2011, 27, 38–46. [Google Scholar] [CrossRef]
- Yang, J.; Wang, W.; Chen, Z.; Lu, S.; Yang, F.; Bi, Z.; Bao, L.; Mo, F.; Li, X.; Huang, Y.; et al. A Vaccine Targeting the RBD of the S Protein of SARS-CoV-2 Induces Protective Immunity. Nature 2020, 586, 572–577. [Google Scholar] [CrossRef] [PubMed]
- He, Y.; Qi, J.; Xiao, L.; Shen, L.; Yu, W.; Hu, T. Purification and Characterization of the Receptor-Binding Domain of SARS-CoV-2 Spike Protein from Escherichia Coli. Eng. Life Sci. 2021, 21, 453–460. [Google Scholar] [CrossRef] [PubMed]
- Puente-Massaguer, E.; Strobl, F.; Grabherr, R.; Striedner, G.; Lecina, M.; Gòdia, F. PEI-Mediated Transient Transfection of High Five Cells at Bioreactor Scale for HIV-1 VLP Production. Nanomaterials 2020, 10, 1580. [Google Scholar] [CrossRef] [PubMed]
- Puente-Massaguer, E.; Gòdia, F.; Lecina, M. Development of a Non-Viral Platform for Rapid Virus-like Particle Production in Sf9 Cells. J. Biotechnol. 2020, 322, 43–53. [Google Scholar] [CrossRef] [PubMed]
- Corchero, J.L.; Mendoza, R.; Lorenzo, J.; Rodríguez-Sureda, V.; Domínguez, C.; Vázquez, E.; Ferrer-Miralles, N.; Villaverde, A. Integrated Approach to Produce a Recombinant, His-Tagged Human α-Galactosidase a in Mammalian Cells. Biotechnol. Prog. 2011, 27, 1206–1217. [Google Scholar] [CrossRef]
- Chang, G.D.; Chen, C.J.; Lin, C.Y.; Chen, H.C.; Chen, H. Improvement of Glycosylation in Insect Cells with Mammalian Glycosyltransferases. J. Biotechnol. 2003, 102, 61–71. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez-Carmona, E.; Mendoza, R.; Ruiz-Cánovas, E.; Ferrer-Miralles, N.; Abasolo, I.; Schwartz, S.; Villaverde, A.; Corchero, J.L. A Novel Bio-Functional Material Based on Mammalian Cell Aggresomes. Appl. Microbiol. Biotechnol. 2015, 99, 7079–7088. [Google Scholar] [CrossRef] [PubMed]
- Rueda, F.; Céspedes, M.V.; Conchillo-Solé, O.; Sánchez-Chardi, A.; Seras-Franzoso, J.; Cubarsi, R.; Gallardo, A.; Pesarrodona, M.; Ferrer-Miralles, N.; Daura, X.; et al. Bottom-Up Instructive Quality Control in the Biofabrication of Smart Protein Materials. Adv. Mater. 2015, 27, 1–7. [Google Scholar] [CrossRef]
- Céspedes, M.V.; Unzueta, U.; Tatkiewicz, W.; Sánchez-Chardi, A.; Conchillo-Solé, O.; Álamo, P.; Xu, Z.; Casanova, I.; Corchero, J.L.; Pesarrodona, M.; et al. In Vivo Architectonic Stability of Fully de Novo Designed Protein-Only Nanoparticles. ACS Nano 2014, 8, 4166–4176. [Google Scholar] [CrossRef] [PubMed]
- Unzueta, U.; Ferrer-Miralles, N.; Cedano, J.; Zikung, X.; Pesarrodona, M.; Saccardo, P.; García-Fruitós, E.; Domingo-Espín, J.; Kumar, P.; Gupta, K.C.; et al. Non-Amyloidogenic Peptide Tags for the Regulatable Self-Assembling of Protein-Only Nanoparticles. Biomaterials 2012, 33, 8714–8722. [Google Scholar] [CrossRef] [PubMed]
- Schellenberg, M.J.; Petrovich, R.M.; Malone, C.C.; Williams, R.S. Selectable High-Yield Recombinant Protein Production in Human Cells Using a GFP/YFP Nanobody Affinity Support. Protein Sci. 2018, 27, 1083–1092. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Zou, X.; Li, T.; Wang, X.; Yuan, W.; Chen, Y.; Han, W. Enhanced Production of Secretory Glycoprotein VSTM1-v2 with Mouse IgGκ Signal Peptide in Optimized HEK293F Transient Transfection. J. Biosci. Bioeng. 2016, 121, 133–139. [Google Scholar] [CrossRef]
- Subedi, G.P.; Johnson, R.W.; Moniz, H.A.; Moremen, K.W.; Barb, A. High Yield Expression of Recombinant Human Proteins with the Transient Transfection of HEK293 Cells in Suspension. J. Vis. Exp. 2015, 2015, e53568. [Google Scholar] [CrossRef]
- Amiram, M.; Luginbuhl, K.M.; Li, X.; Feinglos, M.N.; Chilkoti, A. Injectable Protease-Operated Depots of Glucagon-like Peptide-1 Provide Extended and Tunable Glucose Control. Proc. Natl. Acad. Sci. USA 2013, 110, 2792–2797. [Google Scholar] [CrossRef]
- Nieschlag, E. Testosterone Treatment Comes of Age: New Options for Hypogonadal Men. Clin. Endocrinol. 2006, 65, 275–281. [Google Scholar] [CrossRef]
- Wang, Z.; Guo, J.; Ning, J.; Feng, X.; Liu, X.; Sun, J.; Chen, X.; Lu, F.; Gao, W. One-Month Zero-Order Sustained Release and Tumor Eradication after a Single Subcutaneous Injection of Interferon Alpha Fused with a Body-Temperature-Responsive Polypeptide. Biomater. Sci. 2019, 7, 104–112. [Google Scholar] [CrossRef]
- Detsi, A.; Kavetsou, E.; Kostopoulou, I.; Pitterou, I.; Pontillo, A.R.N.; Tzani, A.; Christodoulou, P.; Siliachli, A.; Zoumpoulakis, P. Nanosystems for the Encapsulation of Natural Products: The Case of Chitosan Biopolymer as a Matrix. Pharmaceutics 2020, 12, 669. [Google Scholar] [CrossRef]
- García-González, C.A.; Sosnik, A.; Kalmár, J.; De Marco, I.; Erkey, C.; Concheiro, A.; Alvarez-Lorenzo, C. Aerogels in Drug Delivery: From Design to Application. J. Control. Release 2021, 332, 40–63. [Google Scholar] [CrossRef]
- Cid, A.G.; Sonvico, F.; Bettini, R.; Colombo, P.; Gonzo, E.; Jimenez-Kairuz, A.F.; Bermúdez, J.M. Evaluation of the Drug Release Kinetics in Assembled Modular Systems Based on the Dome Matrix Technology. J. Pharm. Sci. 2020, 109, 2819–2826. [Google Scholar] [CrossRef] [PubMed]
- Kajdič, S.; Planinšek, O.; Gašperlin, M.; Kocbek, P. Electrospun Nanofibers for Customized Drug-Delivery Systems. J. Drug Deliv. Sci. Technol. 2019, 51, 672–681. [Google Scholar] [CrossRef]
- Seuring, C.; Verasdonck, J.; Gath, J.; Ghosh, D.; Nespovitaya, N.; Wälti, M.A.; Maji, S.K.; Cadalbert, R.; Güntert, P.; Meier, B.H.; et al. The Three-Dimensional Structure of Human β-Endorphin Amyloid Fibrils. Nat. Struct. Mol. Biol. 2020, 27, 1178–1184. [Google Scholar] [CrossRef] [PubMed]
- Arey, B.J. The Role of Glycosylation in Receptor Signaling. Glycosylation 2012, 26, 50262. [Google Scholar]
- Hilairet, S.; Foord, S.M.; Marshall, F.H.; Bouvier, M. Protein-Protein Interaction and Not Glycosylation Determines the Binding Selectivity of Heterodimers between the Calcitonin Receptor-like Receptor and the Receptor Activity-Modifying Proteins. J. Biol. Chem. 2001, 276, 29575–29581. [Google Scholar] [CrossRef] [PubMed]
- Rudd, P.M.; Dwek, R.A. Glycosylation: Heterogeneity and the 3D Structure of Proteins. Crit. Rev. Biochem. Mol. Biol. 1997, 32, 1–100. [Google Scholar] [CrossRef]
- Fonseca-Maldonado, R.; Vieira, D.S.; Alponti, J.S.; Bonneil, E.; Thibault, P.; Ward, R.J. Engineering the Pattern of Protein Glycosylation Modulates the Thermostability of a GH11 Xylanase. J. Biol. Chem. 2013, 288, 25522–25534. [Google Scholar] [CrossRef]
- Sola, R.J.; Griebenow, K. Effects of Glycosylate on the Stability of Protein Pharmaceuticals. J. Pharm. Sci. 2009, 98, 1223–1245. [Google Scholar] [CrossRef]



| Host | Productivity (mg/L) | Purity (%) | Molecular Mass (kDa) | Proteolysis (Y/N) | Spontaneous Formation of Nanoparticles (Y/N; size, nm) | Formation of Microparticles (Y/N) | Release of Nanoparticles (Y/N; Size, nm) |
|---|---|---|---|---|---|---|---|
| E. coli | 40 | >90 | 28.6 | N | Y (37.9) | Y | Y (23.3–317.4) |
| sf9 | 1.50–1.64 | >98 | 31.1 | N | N | Y | Y (291.9–868.0) |
| HEK293F | 0.15 | <60 ii | 35.5 (33.1 i) | N | N | Y | Y (ND) |
| Expi293 | 0.38 | <60 ii | 35.8 (32.6 i) | N | N | Y | Y (11.1–205.8) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Corchero, J.L.; Favaro, M.T.P.; Márquez-Martínez, M.; Lascorz, J.; Martínez-Torró, C.; Sánchez, J.M.; López-Laguna, H.; de Souza Ferreira, L.C.; Vázquez, E.; Ferrer-Miralles, N.; et al. Recombinant Proteins for Assembling as Nano- and Micro-Scale Materials for Drug Delivery: A Host Comparative Overview. Pharmaceutics 2023, 15, 1197. https://doi.org/10.3390/pharmaceutics15041197
Corchero JL, Favaro MTP, Márquez-Martínez M, Lascorz J, Martínez-Torró C, Sánchez JM, López-Laguna H, de Souza Ferreira LC, Vázquez E, Ferrer-Miralles N, et al. Recombinant Proteins for Assembling as Nano- and Micro-Scale Materials for Drug Delivery: A Host Comparative Overview. Pharmaceutics. 2023; 15(4):1197. https://doi.org/10.3390/pharmaceutics15041197
Chicago/Turabian StyleCorchero, José Luis, Marianna T. P. Favaro, Merce Márquez-Martínez, Jara Lascorz, Carlos Martínez-Torró, Julieta M. Sánchez, Hèctor López-Laguna, Luís Carlos de Souza Ferreira, Esther Vázquez, Neus Ferrer-Miralles, and et al. 2023. "Recombinant Proteins for Assembling as Nano- and Micro-Scale Materials for Drug Delivery: A Host Comparative Overview" Pharmaceutics 15, no. 4: 1197. https://doi.org/10.3390/pharmaceutics15041197
APA StyleCorchero, J. L., Favaro, M. T. P., Márquez-Martínez, M., Lascorz, J., Martínez-Torró, C., Sánchez, J. M., López-Laguna, H., de Souza Ferreira, L. C., Vázquez, E., Ferrer-Miralles, N., Villaverde, A., & Parladé, E. (2023). Recombinant Proteins for Assembling as Nano- and Micro-Scale Materials for Drug Delivery: A Host Comparative Overview. Pharmaceutics, 15(4), 1197. https://doi.org/10.3390/pharmaceutics15041197






