Characterization of Increased Extracellular Vesicle-Mediated Tigecycline Resistance in Acinetobacter baumannii
Abstract
:1. Introduction
2. Materials and Methods
2.1. Bacterial Strains, Growth Conditions, and Chemicals
2.2. MIC Determination
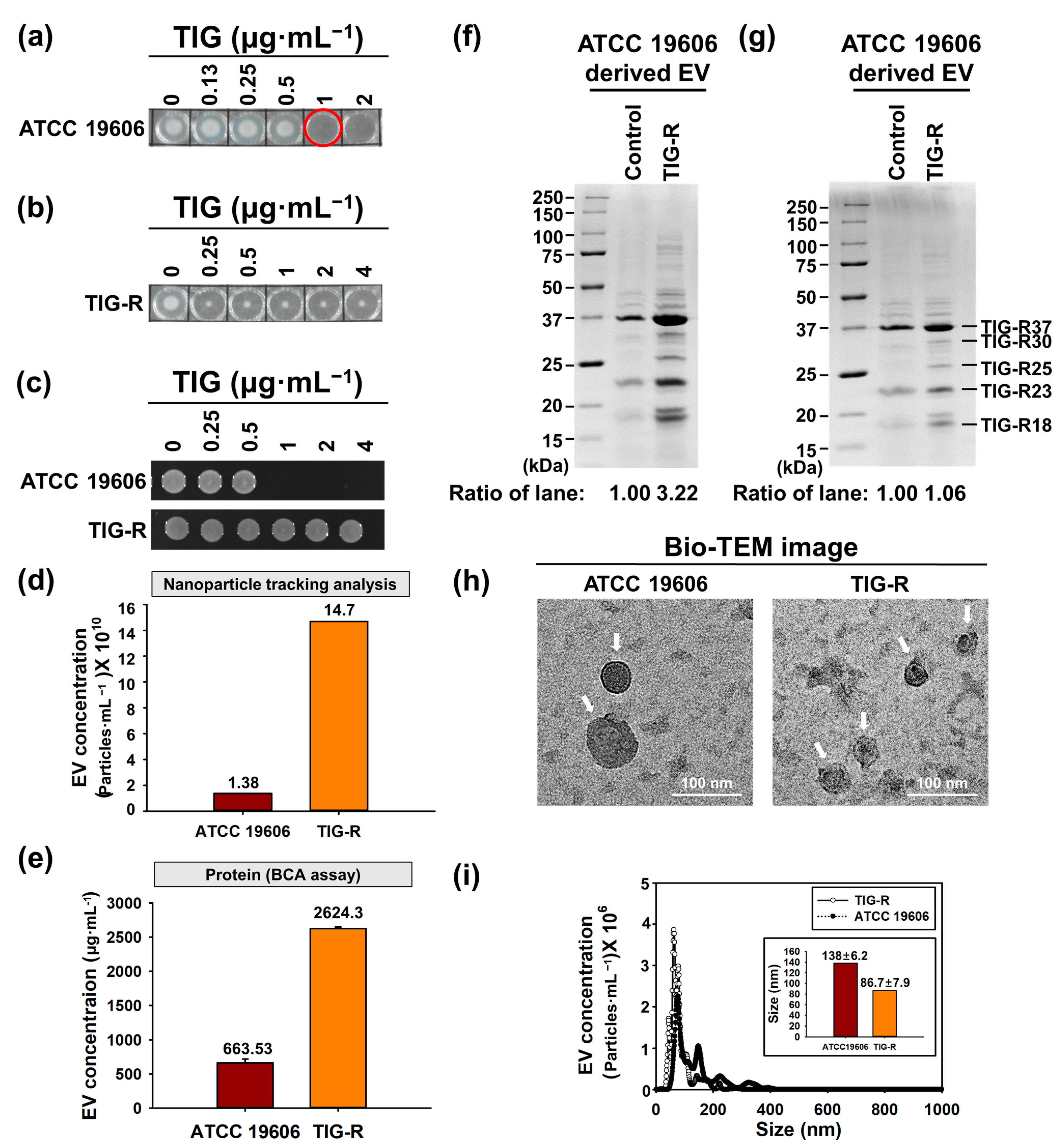
2.3. EVs Isolation
2.4. Characterization of EVs
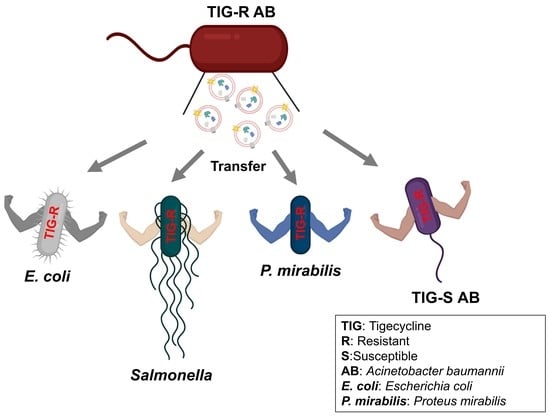
2.5. Evaluation of EV-Mediated TIG-R AB Transfer Activity to TIG-S AB and Other Bacteria
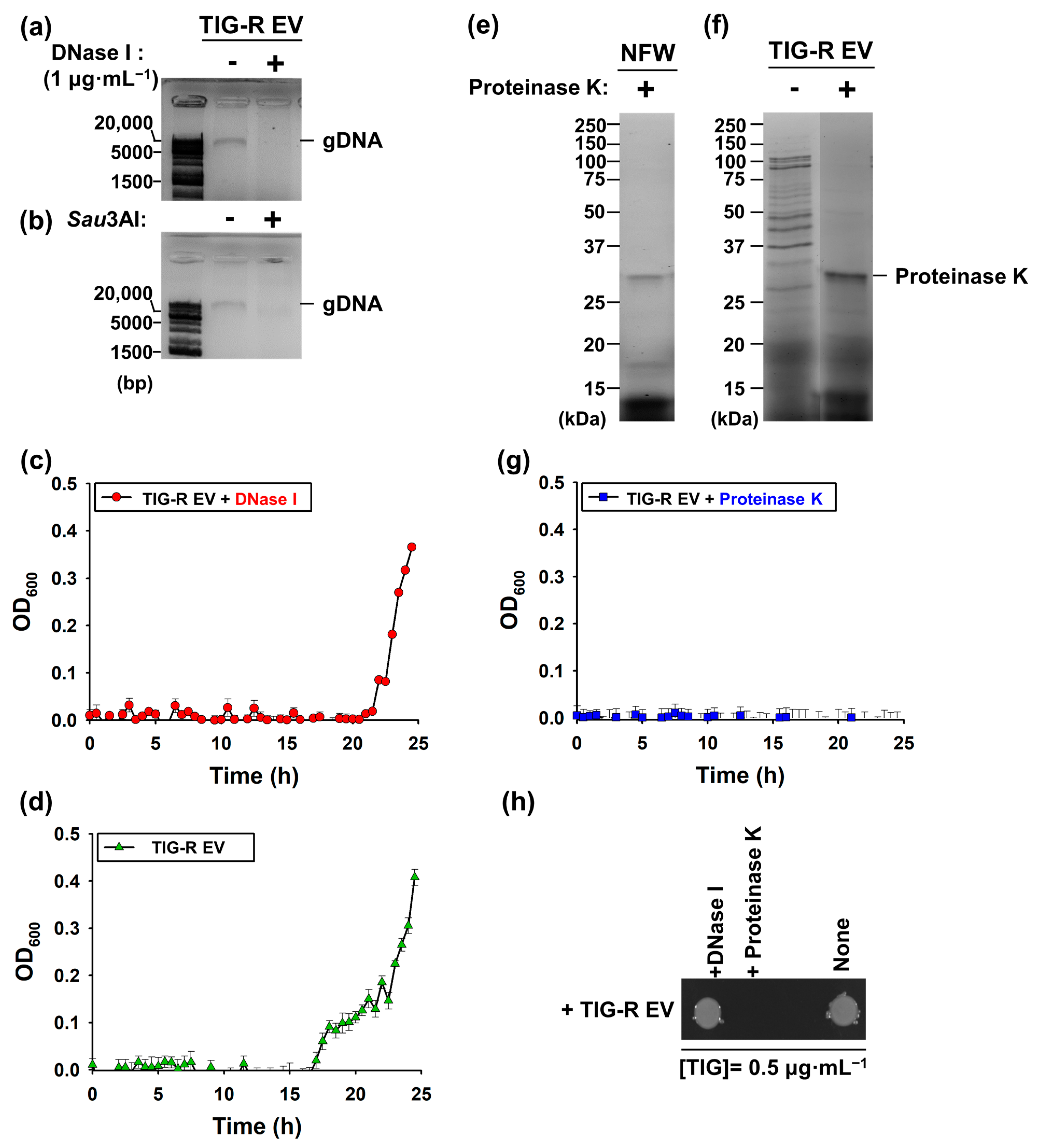
2.6. Identification of Components Involved in EV-Mediated TIG Transfer Activity
2.7. Evaluation of Recurrence Efficacy of TIG-R by TIG or TIG-R EVs
2.8. Characterization of EV Proteins
| Type of Bacteria | Strain ID | Strains | Feature | Reference |
|---|---|---|---|---|
| Gram- Negative | 17978 | Acinetobacter baumannii (AB) | Acinetobacter baumannii (AB) Bouvet and Grimont | ATCC |
| 19606 | ||||
| TIG-R | Laboratory made TIG-R ATCC 19606 | [27] | ||
| 25922 | Escherichia coli | Smooth LPS (O6 serotype reference strain) | ATCC | |
| 29906 | Proteus mirabilis | Proteus mirabilis Hauser (Type strain) | ATCC | |
| 14028S | Salmonella enterica serovar typhimurium | Wild-type Salmonella enterica serovar typhimurium; a spontaneous mutant resistant to nalidixic acid (NA) | [28] | |
| 16285 | Klebsiella pneumoniae | Klebsiella pneumoniae mcr-1 clinical isolates | NCCP | |
| Gram- Positive | 25923 | Staphylococcus aureus | Staphylococcus aureus subsp. aureus Rosenbach | ATCC |
2.9. Screening of Resistant Antibiotics by TIG-R EVs
2.10. Statistical Analysis
3. Results and Discussion
3.1. Physical Characterization of EVs Produced by Tigecycline Resistant A. baumannii
3.2. TIG-R EV as a Mediator for TIG Resistance
3.3. Proteins Are Major Determinants for EV-Mediated TIG-R Transfer
3.4. Comparison of EV-Mediated Transfer of Resistance to Antibiotics
3.5. Role of TIG-R EVs on Current Antibiotics
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- COVID-19: U.S. Impact on Antimicrobial Resistance, Special Report 2022; U.S. Department of Health and Human Services, CDC: Atlanta, GA, USA, 2022.
- World Health Organization; Regional Office for Europe. Central Asian and Eastern European Surveillance of Antimicrobial Resistance: Annual Report 2017; World Health Organization: Geneva, Switzerland; Regional Office for Europe: Copenhagen, Denmark, 2017. [Google Scholar]
- Wong, D.; Nielsen, T.B.; Bonomo, R.A.; Pantapalangkoor, P.; Luna, B.; Spellberg, B. Clinical and Pathophysiological Overview of Acinetobacter Infections: A Century of Challenges. Clin. Microbiol. Rev. 2017, 30, 409–447. [Google Scholar] [CrossRef] [PubMed]
- Curcio, D. Tigecycline in the Treatment of Community-Acquired Pneumonia. Clin. Med. Ther. 2009, 1, CMT.S2351. [Google Scholar] [CrossRef]
- Kyriakidis, I.; Vasileiou, E.; Pana, Z.D.; Tragiannidis, A. Acinetobacter baumannii Antibiotic Resistance Mechanisms. Pathogens 2021, 10, 373. [Google Scholar] [CrossRef] [PubMed]
- Capone, A.; Giannella, M.; Fortini, D.; Giordano, A.; Meledandri, M.; Ballardini, M.; Venditti, M.; Bordi, E.; Capozzi, D.; Balice, M.P.; et al. High rate of colistin resistance among patients with carbapenem-resistant Klebsiella pneumoniae infection accounts for an excess of mortality. Clin. Microbiol. Infect. 2013, 19, E23–E30. [Google Scholar] [CrossRef]
- Nordmann, P.; Cuzon, G.; Naas, T. The real threat of Klebsiella pneumoniae carbapenemase-producing bacteria. Lancet Infect. Dis. 2009, 9, 228–236. [Google Scholar] [CrossRef]
- Chen, Q.; Li, X.; Zhou, H.; Jiang, Y.; Chen, Y.; Hua, X.; Yu, Y. Decreased susceptibility to tigecycline in Acinetobacter baumannii mediated by a mutation in trm encoding SAM-dependent methyltransferase. J. Antimicrob. Chemother. 2013, 69, 72–76. [Google Scholar] [CrossRef]
- Osei Sekyere, J.; Govinden, U.; Bester, L.A.; Essack, S.Y. Colistin and tigecycline resistance in carbapenemase-producing Gram-negative bacteria: Emerging resistance mechanisms and detection methods. J. Appl. Microbiol. 2016, 121, 601–617. [Google Scholar] [CrossRef]
- Hua, X.; He, J.; Wang, J.; Zhang, L.; Zhang, L.; Xu, Q.; Shi, K.; Leptihn, S.; Shi, Y.; Fu, X.; et al. Novel tigecycline resistance mechanisms in Acinetobacter baumannii mediated by mutations in adeS, rpoB and rrf. Emerg. Microbes Infect. 2021, 10, 1404–1417. [Google Scholar] [CrossRef]
- Zhang, J.; Xie, J.; Li, H.; Wang, Z.; Yin, Y.; Wang, S.; Chen, H.; Wang, Q.; Wang, H. Genomic and Phenotypic Evolution of Tigecycline-Resistant Acinetobacter baumannii in Critically Ill Patients. Microbiol. Spectr. 2022, 10, e0159321. [Google Scholar] [CrossRef]
- Kuehn, M.J.; Kesty, N.C. Bacterial outer membrane vesicles and the host-pathogen interaction. Genes Dev. 2005, 19, 2645–2655. [Google Scholar] [CrossRef]
- Chen, J.; Zhang, H.; Wang, S.; Du, Y.; Wei, B.; Wu, Q.; Wang, H. Inhibitors of Bacterial Extracellular Vesicles. Front. Microbiol. 2022, 13, 835058. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Xiao, J.; Wang, S.; Zhou, J.; Qin, J.; Jia, Z.; Wang, Y.; Wang, Z.; Zhang, Y.; Hao, H. Research Progress on Bacterial Membrane Vesicles and Antibiotic Resistance. Int. J. Mol. Sci. 2022, 23, 11553. [Google Scholar] [CrossRef] [PubMed]
- Rumbo, C.; Fernández-Moreira, E.; Merino, M.; Poza, M.; Mendez, J.A.; Soares, N.C.; Mosquera, A.; Chaves, F.; Bou, G. Horizontal transfer of the OXA-24 carbapenemase gene via outer membrane vesicles: A new mechanism of dissemination of carbapenem resistance genes in Acinetobacter baumannii. Antimicrob. Agents Chemother. 2011, 55, 3084–3090. [Google Scholar] [CrossRef]
- Ñahui Palomino, R.A.; Vanpouille, C.; Costantini, P.E.; Margolis, L. Microbiota–host communications: Bacterial extracellular vesicles as a common language. PLoS Pathog. 2021, 17, e1009508. [Google Scholar] [CrossRef] [PubMed]
- Schwechheimer, C.; Kuehn, M.J. Outer-membrane vesicles from Gram-negative bacteria: Biogenesis and functions. Nat. Rev. Microbiol. 2015, 13, 605–619. [Google Scholar] [CrossRef]
- Park, J.; Kim, M.; Shin, B.; Kang, M.; Yang, J.; Lee, T.K.; Park, W. A novel decoy strategy for polymyxin resistance in Acinetobacter baumannii. eLife 2021, 10, e66988. [Google Scholar] [CrossRef]
- Kim, T.; Bak, G.; Lee, J.; Kim, K.S. Systematic analysis of the role of bacterial Hfq-interacting sRNAs in the response to antibiotics. J. Antimicrob. Chemother. 2015, 70, 1659–1668. [Google Scholar] [CrossRef]
- Lee, W.-H.; Choi, H.-I.; Hong, S.-W.; Kim, K.-S.; Gho, Y.S.; Jeon, S.G. Vaccination with Klebsiella pneumoniae-derived extracellular vesicles protects against bacteria-induced lethality via both humoral and cellular immunity. Exp. Mol. Med. 2015, 47, e183. [Google Scholar] [CrossRef]
- Gardiner, C.; Ferreira, Y.J.; Dragovic, R.A.; Redman, C.W.; Sargent, I.L. Extracellular vesicle sizing and enumeration by nanoparticle tracking analysis. J. Extracell. Vesicles 2013, 2, 19671. [Google Scholar] [CrossRef]
- Kim, S.W.; Seo, J.S.; Park, S.B.; Lee, A.R.; Lee, J.S.; Jung, J.W.; Chun, J.H.; Lazarte, J.M.S.; Kim, J.; Kim, J.H.; et al. Significant increase in the secretion of extracellular vesicles and antibiotics resistance from methicillin-resistant Staphylococcus aureus induced by ampicillin stress. Sci. Rep. 2020, 10, 21066. [Google Scholar] [CrossRef]
- Choi, H.; Kim, M.; Jeon, J.; Han, J.K.; Kim, K.-S. Overexpression of MicA induces production of OmpC-enriched outer membrane vesicles that protect against Salmonella challenge. Biochem. Biophys. Res. Commun. 2017, 490, 991–996. [Google Scholar] [CrossRef] [PubMed]
- Grande, R.; Celia, C.; Mincione, G.; Stringaro, A.; Di Marzio, L.; Colone, M.; Di Marcantonio, M.C.; Savino, L.; Puca, V.; Santoliquido, R.; et al. Detection and Physicochemical Characterization of Membrane Vesicles (MVs) of Lactobacillus reuteri DSM 17938. Front. Microbiol. 2017, 8, 1040. [Google Scholar] [CrossRef] [PubMed]
- Chebotar, I.V.; Konchakova, E.D.; Maianskii, A.N. Vesicle formation as a result of interaction between polymorphonuclear neutrophils and Staphylococcus aureus biofilm. J. Med. Microbiol. 2013, 62, 1153–1159. [Google Scholar] [CrossRef] [PubMed]
- The UniProt Consortium. UniProt: The Universal Protein Knowledgebase in 2023. Nucleic Acids Res. 2023, 51, D523–D531. [Google Scholar] [CrossRef]
- Cho, H.; Kim, K.-s. Repurposing of Ciclopirox to Overcome the Limitations of Zidovudine (Azidothymidine) against Multidrug-Resistant Gram-Negative Bacteria. Pharmaceutics 2022, 14, 552. [Google Scholar] [CrossRef]
- Jarvik, T.; Smillie, C.; Groisman, E.A.; Ochman, H. Short-term signatures of evolutionary change in the Salmonella enterica serovar Typhimurium 14028 genome. J. Bacteriol. 2010, 192, 560. [Google Scholar] [CrossRef]
- Fulsundar, S.; Harms, K.; Flaten, G.E.; Johnsen, P.J.; Chopade, B.A.; Nielsen, K.M. Gene transfer potential of outer membrane vesicles of Acinetobacter baylyi and effects of stress on vesiculation. Appl. Environ. Microbiol. 2014, 80, 3469–3483. [Google Scholar] [CrossRef]
- Ho, M.-H.; Chen, C.-H.; Goodwin, J.S.; Wang, B.-Y.; Xie, H. Functional Advantages of Porphyromonas gingivalis Vesicles. PLoS ONE 2015, 10, e0123448. [Google Scholar] [CrossRef]
- Bielaszewska, M.; Daniel, O.; Karch, H.; Mellmann, A. Dissemination of the blaCTX-M-15 gene among Enterobacteriaceae via outer membrane vesicles. J. Antimicrob. Chemother. 2020, 75, 2442–2451. [Google Scholar] [CrossRef]
- Marchant, P.; Carreño, A.; Vivanco, E.; Silva, A.; Nevermann, J.; Otero, C.; Araya, E.; Gil, F.; Calderón, I.L.; Fuentes, J.A. “One for All”: Functional Transfer of OMV-Mediated Polymyxin B Resistance from Salmonella enterica sv. Typhi ΔtolR and ΔdegS to Susceptible Bacteria. Front. Microbiol. 2021, 12, 672467. [Google Scholar] [CrossRef]
- Lee, A.R.; Park, S.B.; Kim, S.W.; Jung, J.W.; Chun, J.H.; Kim, J.; Kim, Y.R.; Lazarte, J.M.S.; Jang, H.B.; Thompson, K.D.; et al. Membrane vesicles from antibiotic-resistant Staphylococcus aureus transfer antibiotic-resistance to antibiotic-susceptible Escherichia coli. J. Appl. Microbiol. 2022, 132, 2746–2759. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.W.; Park, S.B.; Im, S.P.; Lee, J.S.; Jung, J.W.; Gong, T.W.; Lazarte, J.M.S.; Kim, J.; Seo, J.S.; Kim, J.H.; et al. Outer membrane vesicles from β-lactam-resistant Escherichia coli enable the survival of β-lactam-susceptible E. coli in the presence of β-lactam antibiotics. Sci. Rep. 2018, 8, 5402. [Google Scholar] [CrossRef] [PubMed]
- Cloet, T.; Momenbeitollahi, N.; Li, H. Recent advances on protein-based quantification of extracellular vesicles. Anal. Biochem. 2021, 622, 114168. [Google Scholar] [CrossRef]
- Doyle, L.M.; Wang, M.Z. Overview of Extracellular Vesicles, Their Origin, Composition, Purpose, and Methods for Exosome Isolation and Analysis. Cells 2019, 8, 727. [Google Scholar] [CrossRef] [PubMed]
- Zaborowski, M.P.; Balaj, L.; Breakefield, X.O.; Lai, C.P. Extracellular Vesicles: Composition, Biological Relevance, and Methods of Study. Bioscience 2015, 65, 783–797. [Google Scholar] [CrossRef]
- Godeux, A.-S.; Svedholm, E.; Barreto, S.; Potron, A.; Venner, S.; Charpentier, X.; Laaberki, M.-H. Interbacterial Transfer of Carbapenem Resistance and Large Antibiotic Resistance Islands by Natural Transformation in Pathogenic Acinetobacter. mBio 2022, 13, e0263121. [Google Scholar] [CrossRef] [PubMed]
- Jeon, J.H.; Jang, K.M.; Lee, J.H.; Kang, L.W.; Lee, S.H. Transmission of antibiotic resistance genes through mobile genetic elements in Acinetobacter baumannii and gene-transfer prevention. Sci. Total Environ. 2023, 857, 159497. [Google Scholar] [CrossRef]
- Munita, J.M.; Arias, C.A. Mechanisms of Antibiotic Resistance. Microbiol. Spectr. 2016, 4. [Google Scholar] [CrossRef]
- Oliver, A.; Weigel, L.M.; Rasheed, J.K.; McGowan, J.E., Jr.; Raney, P.; Tenover, F.C. Mechanisms of decreased susceptibility to cefpodoxime in Escherichia coli. Antimicrob. Agents Chemother. 2002, 46, 3829–3836. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cho, H.; Sondak, T.; Kim, K.-s. Characterization of Increased Extracellular Vesicle-Mediated Tigecycline Resistance in Acinetobacter baumannii. Pharmaceutics 2023, 15, 1251. https://doi.org/10.3390/pharmaceutics15041251
Cho H, Sondak T, Kim K-s. Characterization of Increased Extracellular Vesicle-Mediated Tigecycline Resistance in Acinetobacter baumannii. Pharmaceutics. 2023; 15(4):1251. https://doi.org/10.3390/pharmaceutics15041251
Chicago/Turabian StyleCho, Hyejin, Tesalonika Sondak, and Kwang-sun Kim. 2023. "Characterization of Increased Extracellular Vesicle-Mediated Tigecycline Resistance in Acinetobacter baumannii" Pharmaceutics 15, no. 4: 1251. https://doi.org/10.3390/pharmaceutics15041251
APA StyleCho, H., Sondak, T., & Kim, K. -s. (2023). Characterization of Increased Extracellular Vesicle-Mediated Tigecycline Resistance in Acinetobacter baumannii. Pharmaceutics, 15(4), 1251. https://doi.org/10.3390/pharmaceutics15041251