Extracellular Vesicles and Their Mimetics: A Comparative Study of Their Pharmacological Activities and Immunogenicity Profiles
Abstract
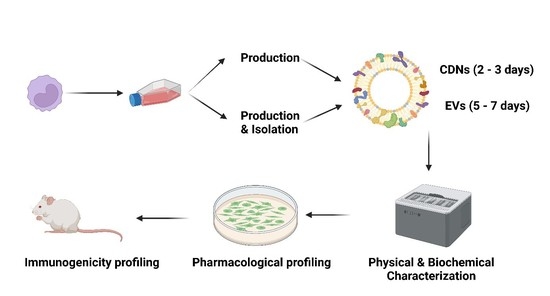
:1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Cell Culture
2.3. Animal Work
2.4. Isolation of U937 Extracellular Vesicles (EVs)
2.5. Production of U937 Cell-Derived Nanovesicles (CDNs)
2.6. Physical Characterisation of EVs and CDNs
2.7. Transmission Electron Microscopy Imaging
2.8. Proteomic Analysis
2.9. Lipidomic Analysis
2.10. miRNA Profiling
2.11. Nitric Oxide Assay
2.12. Inflammatory Cytokines Production
2.13. Antioxidant Levels
2.14. In Vivo Immunological Profiling
2.15. Statistical Analysis
3. Results
3.1. Physical Characterisation of U937 EVs and CDNs
3.2. Biochemical Composition of U937 EVs and CDNs
3.3. Pharmacological Activity of U937 EVs and U937 CDNs on Macrophages
3.4. Antioxidant Levels and Enzyme Activities
3.5. In Vivo Immune Cell Profiling
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Théry, C.; Witwer, K.W.; Aikawa, E.; Alcaraz, M.J.; Anderson, J.D.; Andriantsitohaina, R.; Antoniou, A.; Arab, T.; Archer, F.; Atkin-Smith, G.K.; et al. Minimal information for studies of extracellular vesicles 2018 (MISEV2018): A position statement of the International Society for Extracellular Vesicles and update of the MISEV2014 guidelines. J. Extracell. Vesicles 2018, 7, 1535750. [Google Scholar] [CrossRef] [PubMed]
- Yáñez-Mó, M.; Siljander, P.R.M.; Andreu, Z.; Zavec, A.B.; Borràs, F.E.; Buzas, E.I.; Buzas, K.; Casal, E.; Cappello, F.; Carvalho, J.; et al. Biological properties of extracellular vesicles and their physiological functions. J. Extracell. Vesicles 2015, 4, 27066. [Google Scholar] [CrossRef] [PubMed]
- Doyle, L.M.; Wang, M.Z. Overview of Extracellular Vesicles, Their Origin, Composition, Purpose, and Methods for Exosome Isolation and Analysis. Cells 2019, 8, 727. [Google Scholar] [CrossRef] [PubMed]
- Zaborowski, M.P.; Balaj, L.; Breakefield, X.O.; Lai, C.P. Extracellular Vesicles: Composition, Biological Relevance, and Methods of Study. BioScience 2015, 65, 783–797. [Google Scholar] [CrossRef] [PubMed]
- van Niel, G.; D’Angelo, G.; Raposo, G. Shedding light on the cell biology of extracellular vesicles. Nat. Rev. Mol. Cell Biol. 2018, 19, 213–228. [Google Scholar] [CrossRef] [PubMed]
- De Toro, J.; Herschlik, L.; Waldner, C.; Mongini, C. Emerging Roles of Exosomes in Normal and Pathological Conditions: New Insights for Diagnosis and Therapeutic Applications. Front. Immunol. 2015, 6, 203. [Google Scholar] [CrossRef]
- McConnell, M.J. Extracellular vesicles and immune modulation. Immunol. Cell Biol. 2018, 96, 681–682. [Google Scholar] [CrossRef]
- Rak, J.; Guha, A. Extracellular vesicles–vehicles that spread cancer genes. BioEssays 2012, 34, 489–497. [Google Scholar] [CrossRef]
- Lai, R.C.; Arslan, F.; Lee, M.M.; Sze, N.S.K.; Choo, A.; Chen, T.S.; Salto-Tellez, M.; Timmers, L.; Lee, C.N.; El Oakley, R.M.; et al. Exosome secreted by MSC reduces myocardial ischemia/reperfusion injury. Stem Cell Res. 2010, 4, 214–222. [Google Scholar] [CrossRef]
- Lai, R.C.; Chen, T.S.; Lim, S.K. Mesenchymal stem cell exosome: A novel stem cell-based therapy for cardiovascular disease. Regen. Med. 2011, 6, 481–492. [Google Scholar] [CrossRef]
- Vu, N.B.; Nguyen, H.T.; Palumbo, R.; Pellicano, R.; Fagoonee, S.; Pham, P.V. Stem cell-derived exosomes for wound healing: Current status and promising directions. Minerva Med. 2021, 112, 384–400. [Google Scholar] [CrossRef] [PubMed]
- Ahn, S.-H.; Ryu, S.-W.; Choi, H.; You, S.; Park, J.; Choi, C. Manufacturing Therapeutic Exosomes: From Bench to Industry. Mol. Cells 2022, 45, 284–290. [Google Scholar] [CrossRef] [PubMed]
- Théry, C.; Amigorena, S.; Raposo, G.; Clayton, A. Isolation and Characterization of Exosomes from Cell Culture Supernatants and Biological Fluids. Curr. Protoc. Cell Biol. 2006, 30, 3.22.1–3.22.29. [Google Scholar] [CrossRef] [PubMed]
- Jang, S.C.; Kim, O.Y.; Yoon, C.M.; Choi, D.-S.; Roh, T.-Y.; Park, J.; Nilsson, J.; Lötvall, J.; Kim, Y.-K.; Gho, Y.S. Bioinspired Exosome-Mimetic Nanovesicles for Targeted Delivery of Chemotherapeutics to Malignant Tumors. ACS Nano 2013, 7, 7698–7710. [Google Scholar] [CrossRef] [PubMed]
- Jo, W.; Jeong, D.; Kim, J.; Cho, S.; Jang, S.C.; Han, C.; Kang, J.Y.; Gho, Y.S.; Park, J. Microfluidic fabrication of cell-derived nanovesicles as endogenous RNA carriers. Lab A Chip 2014, 14, 1261–1269. [Google Scholar] [CrossRef]
- Jo, W.; Kim, J.; Yoon, J.; Jeong, D.; Cho, S.; Jeong, H.; Yoon, Y.J.; Kim, S.C.; Gho, Y.S.; Park, J. Large-scale generation of cell-derived nanovesicles. Nanoscale 2014, 6, 12056–12064. [Google Scholar] [CrossRef]
- Yoon, J.; Jo, W.; Jeong, D.; Kim, J.; Jeong, H.; Park, J. Generation of nanovesicles with sliced cellular membrane fragments for exogenous material delivery. Biomaterials 2015, 59, 12–20. [Google Scholar] [CrossRef]
- Goh, W.J.; Zou, S.; Ong, W.Y.; Torta, F.; Alexandra, A.F.; Schiffelers, R.M.; Storm, G.; Wang, J.-W.; Czarny, B.; Pastorin, G. Bioinspired Cell-Derived Nanovesicles versus Exosomes as Drug Delivery Systems: A Cost-Effective Alternative. Sci. Rep. 2017, 7, 14322. [Google Scholar] [CrossRef]
- Neupane, Y.R.; Handral, H.K.; Alkaff, S.A.; Chng, W.H.; Venkatesan, G.; Huang, C.; Lee, C.K.; Wang, J.-W.; Sriram, G.; Dienzo, R.A.; et al. Cell-derived nanovesicles from mesenchymal stem cells as extracellular vesicle-mimetics in wound healing. Acta Pharm. Sin. B, 2022; in press. [Google Scholar]
- Rock, K.L.; Kono, H. The inflammatory response to cell death. Annu. Rev. Pathol. 2008, 3, 99–126. [Google Scholar] [CrossRef]
- Hwang, H.S.; Kim, H.; Han, G.; Lee, J.W.; Kim, K.; Kwon, I.C.; Yang, Y.; Kim, S.H. Extracellular Vesicles as Potential Therapeutics for Inflammatory Diseases. Int. J. Mol. Sci. 2021, 22, 5487. [Google Scholar]
- Teng, X.; Chen, L.; Chen, W.; Yang, J.; Yang, Z.; Shen, Z. Mesenchymal Stem Cell-Derived Exosomes Improve the Microenvironment of Infarcted Myocardium Contributing to Angiogenesis and Anti-Inflammation. Cell. Physiol. Biochem. 2015, 37, 2415–2424. [Google Scholar] [CrossRef] [PubMed]
- Hezel, M.E.V.; Nieuwland, R.; Bruggen, R.V.; Juffermans, N.P. The Ability of Extracellular Vesicles to Induce a Pro-Inflammatory Host Response. Int. J. Mol. Sci. 2017, 18, 1285. [Google Scholar] [CrossRef] [PubMed]
- Arslan, F.; Lai, R.C.; Smeets, M.B.; Akeroyd, L.; Choo, A.; Aguor, E.N.E.; Timmers, L.; van Rijen, H.V.; Doevendans, P.A.; Pasterkamp, G.; et al. Mesenchymal stem cell-derived exosomes increase ATP levels, decrease oxidative stress and activate PI3K/Akt pathway to enhance myocardial viability and prevent adverse remodeling after myocardial ischemia/reperfusion injury. Stem Cell Res. 2013, 10, 301–312. [Google Scholar] [CrossRef]
- Ugel, S.; Canè, S.; De Sanctis, F.; Bronte, V. Monocytes in the Tumor Microenvironment. Annu. Rev. Pathol. Mech. Dis. 2021, 16, 93–122. [Google Scholar] [CrossRef] [PubMed]
- Mentkowski, K.I.; Euscher, L.M.; Patel, A.; Alevriadou, B.R.; Lang, J.K. Monocyte recruitment and fate specification after myocardial infarction. Am. J. Physiol. Cell Physiol. 2020, 319, C797–C806. [Google Scholar] [CrossRef]
- Parihar, A.; Eubank, T.D.; Doseff, A.I. Monocytes and macrophages regulate immunity through dynamic networks of survival and cell death. J. Innate Immun. 2010, 2, 204–215. [Google Scholar] [CrossRef]
- Ogle, M.E.; Segar, C.E.; Sridhar, S.; Botchwey, E.A. Monocytes and macrophages in tissue repair: Implications for immunoregenerative biomaterial design. Exp. Biol. Med. 2016, 241, 1084–1097. [Google Scholar] [CrossRef]
- Haque, S.; Sinha, N.; Ranjit, S.; Midde, N.M.; Kashanchi, F.; Kumar, S. Monocyte-derived exosomes upon exposure to cigarette smoke condensate alter their characteristics and show protective effect against cytotoxicity and HIV-1 replication. Sci. Rep. 2017, 7, 16120. [Google Scholar] [CrossRef]
- Keerthikumar, S.; Chisanga, D.; Ariyaratne, D.; Al Saffar, H.; Anand, S.; Zhao, K.; Samuel, M.; Pathan, M.; Jois, M.; Chilamkurti, N.; et al. ExoCarta: A Web-Based Compendium of Exosomal Cargo. J. Mol. Biol. 2016, 428, 688–692. [Google Scholar] [CrossRef]
- Haraszti, R.A.; Didiot, M.-C.; Sapp, E.; Leszyk, J.; Shaffer, S.A.; Rockwell, H.E.; Gao, F.; Narain, N.R.; DiFiglia, M.; Kiebish, M.A.; et al. High-resolution proteomic and lipidomic analysis of exosomes and microvesicles from different cell sources. J. Extracell. Vesicles 2016, 5, 32570. [Google Scholar] [CrossRef]
- Kooijmans, S.A.A.; Vader, P.; van Dommelen, S.M.; van Solinge, W.W.; Schiffelers, R.M. Exosome mimetics: A novel class of drug delivery systems. Int. J. Nanomed. 2012, 7, 1525–1541. [Google Scholar]
- Verderio, C.; Gabrielli, M.; Giussani, P. Role of sphingolipids in the biogenesis and biological activity of extracellular vesicles. J. Lipid Res. 2018, 59, 1325–1340. [Google Scholar] [CrossRef] [PubMed]
- Fadok, V.A.; Voelker, D.R.; Campbell, P.A.; Cohen, J.J.; Bratton, D.L.; Henson, P.M. Exposure of phosphatidylserine on the surface of apoptotic lymphocytes triggers specific recognition and removal by macrophages. J. Immunol. 1992, 148, 2207. [Google Scholar] [CrossRef] [PubMed]
- Fadok, V.A.; Bratton, D.L.; Frasch, S.C.; Warner, M.L.; Henson, P.M. The role of phosphatidylserine in recognition of apoptotic cells by phagocytes. Cell Death Differ. 1998, 5, 551–562. [Google Scholar] [CrossRef]
- Tuteja, N.; Chandra, M.; Tuteja, R.; Misra, M.K. Nitric Oxide as a Unique Bioactive Signaling Messenger in Physiology and Pathophysiology. J. Biomed. Biotechnol. 2004, 2004, 227–237. [Google Scholar] [CrossRef] [PubMed]
- Farah, C.; Michel, L.Y.M.; Balligand, J.-L. Nitric oxide signalling in cardiovascular health and disease. Nat. Rev. Cardiol. 2018, 15, 292–316. [Google Scholar] [CrossRef]
- Green, S.J.; Mellouk, S.; Hoffman, S.L.; Meltzer, M.S.; Nacy, C.A. Cellular mechanisms of nonspecific immunity to intracellular infection: Cytokine-induced synthesis of toxic nitrogen oxides from l-arginine by macrophages and hepatocytes. Immunol. Lett. 1990, 25, 15–19. [Google Scholar] [CrossRef]
- Bogdan, C. Nitric oxide and the immune response. Nat. Immunol. 2001, 2, 907–916. [Google Scholar] [CrossRef]
- Bryan, N.S.; Grisham, M.B. Methods to detect nitric oxide and its metabolites in biological samples. Free. Radic. Biol. Med. 2007, 43, 645–657. [Google Scholar] [CrossRef]
- Halim, A.T.A.; Ariffin, N.A.F.M.; Azlan, M. Review: The Multiple Roles of Monocytic Microparticles. Inflammation 2016, 39, 1277–1284. [Google Scholar] [CrossRef]
- Niki, E. Antioxidant Defenses in Eukariotic Cells: An Overview. In Free Radicals: From Basic Science to Medicine; Poli, G., Albano, E., Dianzani, M.U., Eds.; Birkhäuser: Basel, Switzerland, 1993; pp. 365–373. [Google Scholar]
- Ighodaro, O.M.; Akinloye, O.A. First line defence antioxidants-superoxide dismutase (SOD), catalase (CAT) and glutathione peroxidase (GPX): Their fundamental role in the entire antioxidant defence grid. Alex. J. Med. 2018, 54, 287–293. [Google Scholar] [CrossRef]
- Ayala, A.; Muñoz, M.F.; Argüelles, S. Lipid peroxidation: Production, metabolism, and signaling mechanisms of malondialdehyde and 4-hydroxy-2-nonenal. Oxidative Med. Cell. Longev. 2014, 2014, 360438. [Google Scholar] [CrossRef] [PubMed]
- Lewis, S.M.; Williams, A.; Eisenbarth, S.C. Structure and function of the immune system in the spleen. Sci. Immunol. 2019, 4, eaau6085. [Google Scholar] [CrossRef]
- Kauffman, K.J.; Mir, F.F.; Jhunjhunwala, S.; Kaczmarek, J.C.; Hurtado, J.E.; Yang, J.H.; Webber, M.J.; Kowalski, P.S.; Heartlein, M.W.; DeRosa, F.; et al. Efficacy and immunogenicity of unmodified and pseudouridine-modified mRNA delivered systemically with lipid nanoparticles in vivo. Biomaterials 2016, 109, 78–87. [Google Scholar] [CrossRef]
- Ou, Y.-H.; Liang, J.; Chng, W.H.; Muthuramalingam, R.P.K.; Ng, Z.X.; Lee, C.K.; Neupane, Y.R.; Yau, J.N.N.; Zhang, S.; Lou, C.K.L.; et al. Investigations on Cellular Uptake Mechanisms and Immunogenicity Profile of Novel Bio-Hybrid Nanovesicles. Pharmaceutics 2022, 14, 1738. [Google Scholar] [CrossRef]
- Marzio, R.; Jirillo, E.; Ransijn, A.; Mauël, J.; Corradin, S.B. Expression and function of the early activation antigen CD69 in murine macrophages. J. Leukoc. Biol. 1997, 62, 349–355. [Google Scholar] [CrossRef] [PubMed]
- Cibrián, D.; Sánchez-Madrid, F. CD69: From activation marker to metabolic gatekeeper. Eur. J. Immunol. 2017, 47, 946–953. [Google Scholar] [CrossRef]











| Resting Macrophages | Activated Macrophages | |||
|---|---|---|---|---|
| U937 EVs | U937 CDNs | U937 EVs | U937 CDNs | |
| Nitrite (proxy for NO) | ↑ | ↑ | ↑ | n.s. |
| TNF-α | ↑ | ↑ | ↑ | n.s. |
| IL-6 | ↑ | ↑ | ↑ | n.s. |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chng, W.H.; Muthuramalingam, R.P.K.; Lou, C.K.L.; New, S.; Neupane, Y.R.; Lee, C.K.; Altay Benetti, A.; Huang, C.; Thoniyot, P.; Toh, W.S.; et al. Extracellular Vesicles and Their Mimetics: A Comparative Study of Their Pharmacological Activities and Immunogenicity Profiles. Pharmaceutics 2023, 15, 1290. https://doi.org/10.3390/pharmaceutics15041290
Chng WH, Muthuramalingam RPK, Lou CKL, New S, Neupane YR, Lee CK, Altay Benetti A, Huang C, Thoniyot P, Toh WS, et al. Extracellular Vesicles and Their Mimetics: A Comparative Study of Their Pharmacological Activities and Immunogenicity Profiles. Pharmaceutics. 2023; 15(4):1290. https://doi.org/10.3390/pharmaceutics15041290
Chicago/Turabian StyleChng, Wei Heng, Ram Pravin Kumar Muthuramalingam, Charles Kang Liang Lou, Silas New, Yub Raj Neupane, Choon Keong Lee, Ayca Altay Benetti, Chenyuan Huang, Praveen Thoniyot, Wei Seong Toh, and et al. 2023. "Extracellular Vesicles and Their Mimetics: A Comparative Study of Their Pharmacological Activities and Immunogenicity Profiles" Pharmaceutics 15, no. 4: 1290. https://doi.org/10.3390/pharmaceutics15041290
APA StyleChng, W. H., Muthuramalingam, R. P. K., Lou, C. K. L., New, S., Neupane, Y. R., Lee, C. K., Altay Benetti, A., Huang, C., Thoniyot, P., Toh, W. S., Wang, J.-W., & Pastorin, G. (2023). Extracellular Vesicles and Their Mimetics: A Comparative Study of Their Pharmacological Activities and Immunogenicity Profiles. Pharmaceutics, 15(4), 1290. https://doi.org/10.3390/pharmaceutics15041290