Postbiotic Metabolite of Lactiplantibacillus plantarum PD18 against Periodontal Pathogens and Their Virulence Markers in Biofilm Formation
Abstract
:1. Introduction
2. Materials and Methods
2.1. Bacterial Strains and Growth Conditions
2.2. Lactobacilli Isolated from Fermented Foods
2.3. Antimicrobial Activity of PM from Isolated Lactobacillus sp.
2.4. Characterizing and Determining Minimum Inhibitory Concentration (MIC) of Lactobacillus PM
2.5. Determining Minimum Biofilm Inhibitory Concentration (MBIC)
2.6. Molecular Identification
2.7. Effect of Lactobacillus PM on Biofilm Formation and the Viability of Biofilm Cells
2.8. Effect of Contact Time of Lactobacillus PM on Oral Bacterial Biofilm
2.9. CLSM Analysis of Bacterial Biofilm
2.10. Measuring Biofilm Formation and Residual Cells of Bacteria Using Scanning Electron Microscopy (SEM)
2.11. Evaluating the Capability of Reducing Biofilms from Typodont Teeth
2.12. Statistical Analysis
3. Results
3.1. Antimicrobial Activity of PM of Isolated Lactobacillus Strains
3.2. Characterizing and Determining the Minimum Inhibitory Concentration (MIC) of Lactobacillus PM
3.3. Determining Minimum Biofilm Inhibitory Concentration (MBIC)
3.4. Molecular Identification
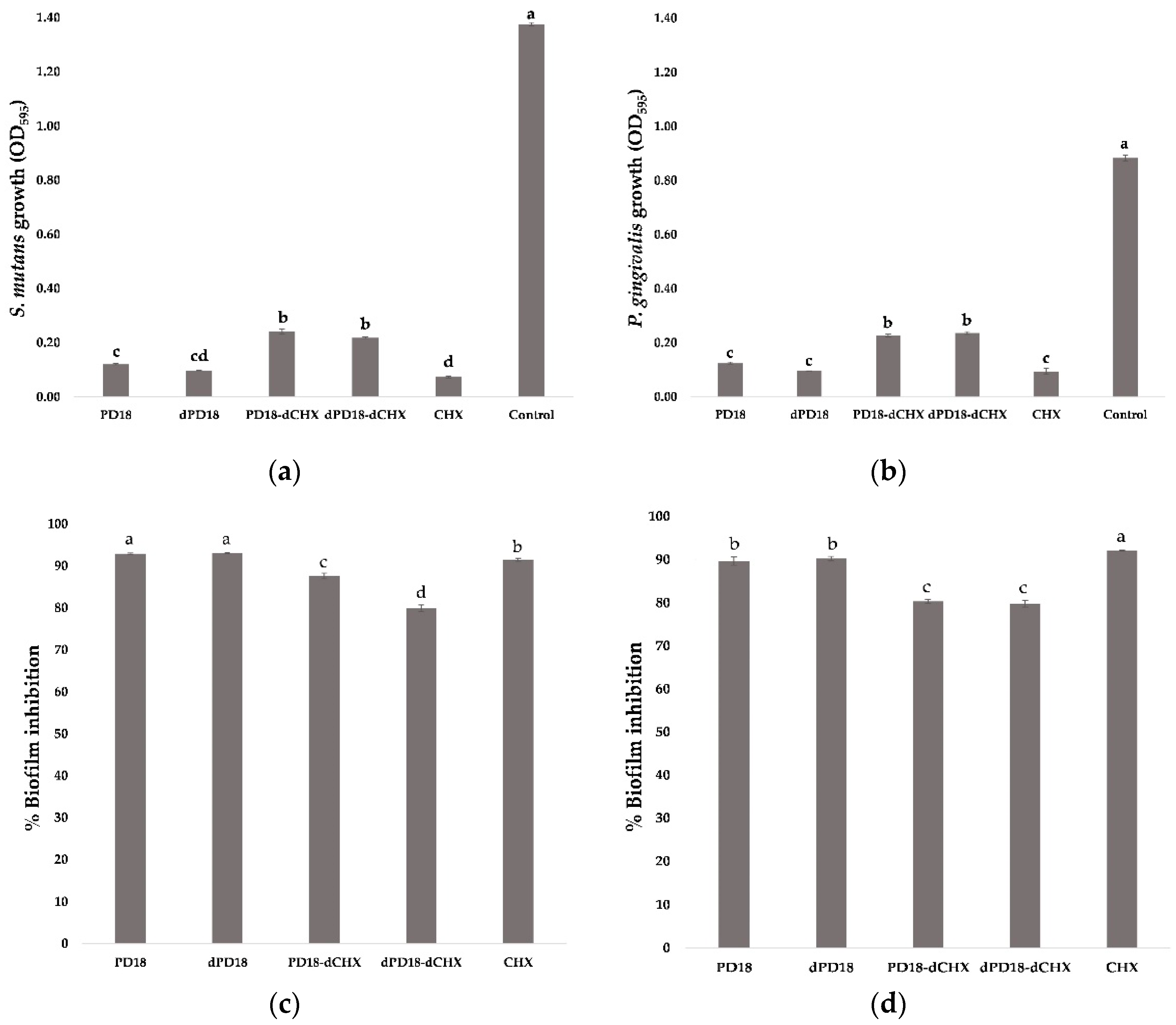
3.5. Effect of Lactobacillus PM on Biofilm Formation and Viability of Biofilm Cells
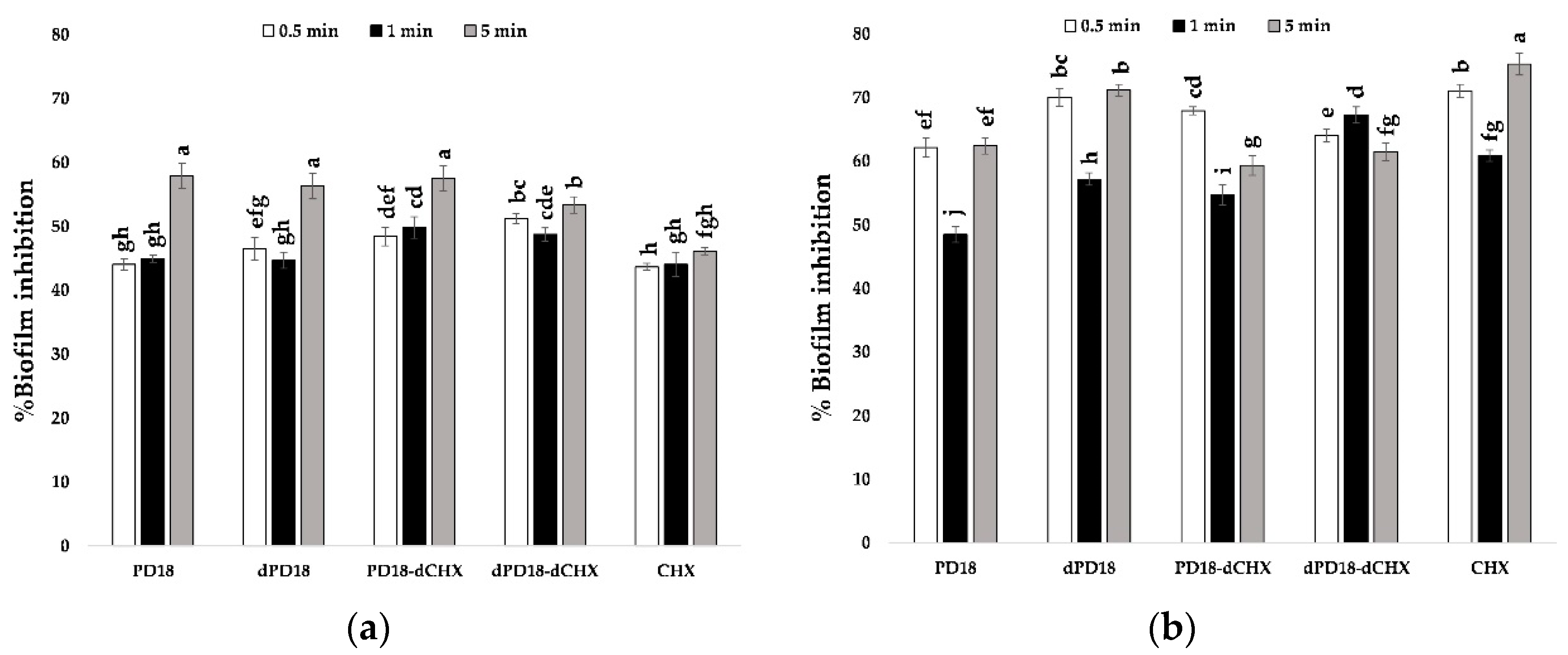
3.6. Effect of Contact Time of Lactobacillus PM on Oral Bacterial Biofilm
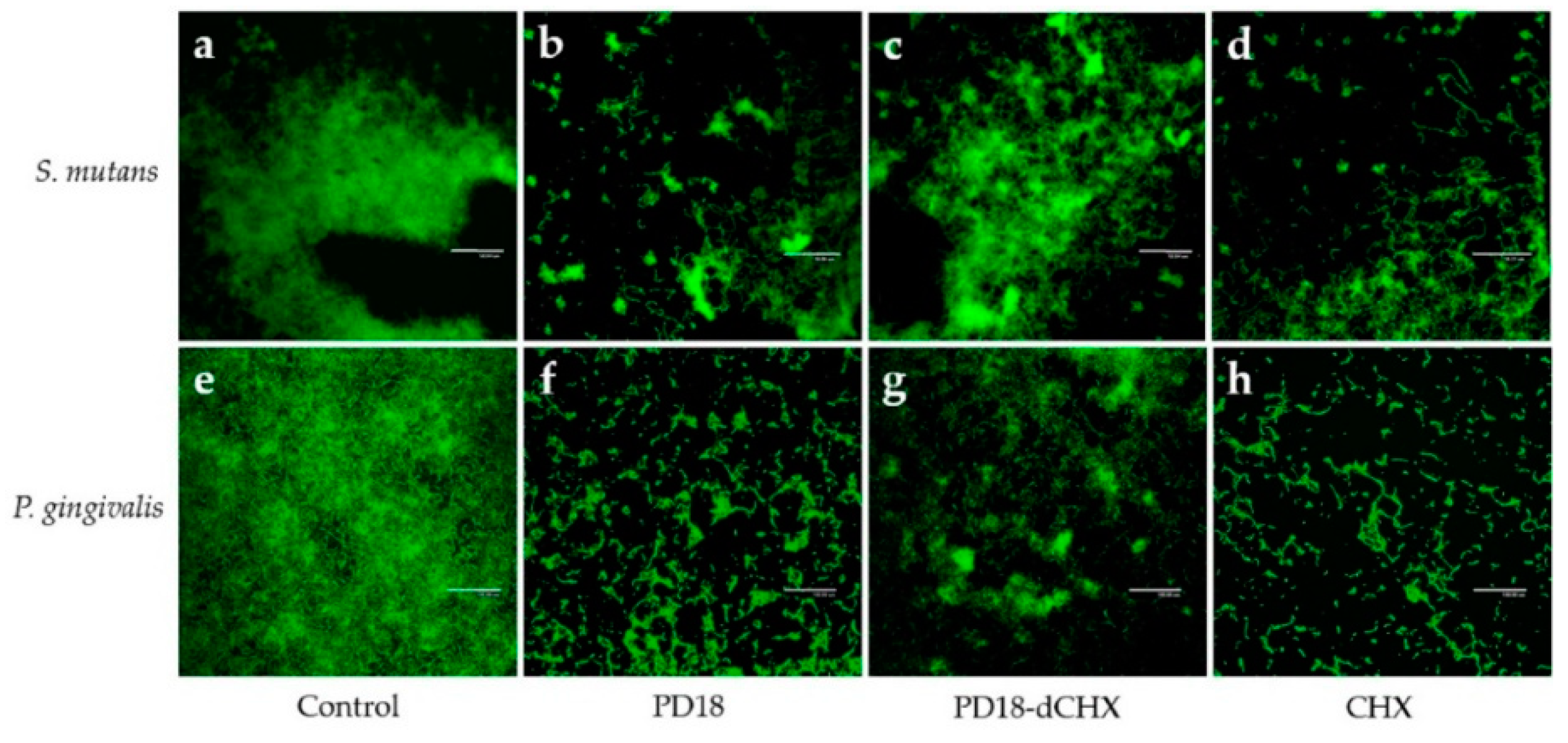
3.7. Biofilm Observation by CLSM
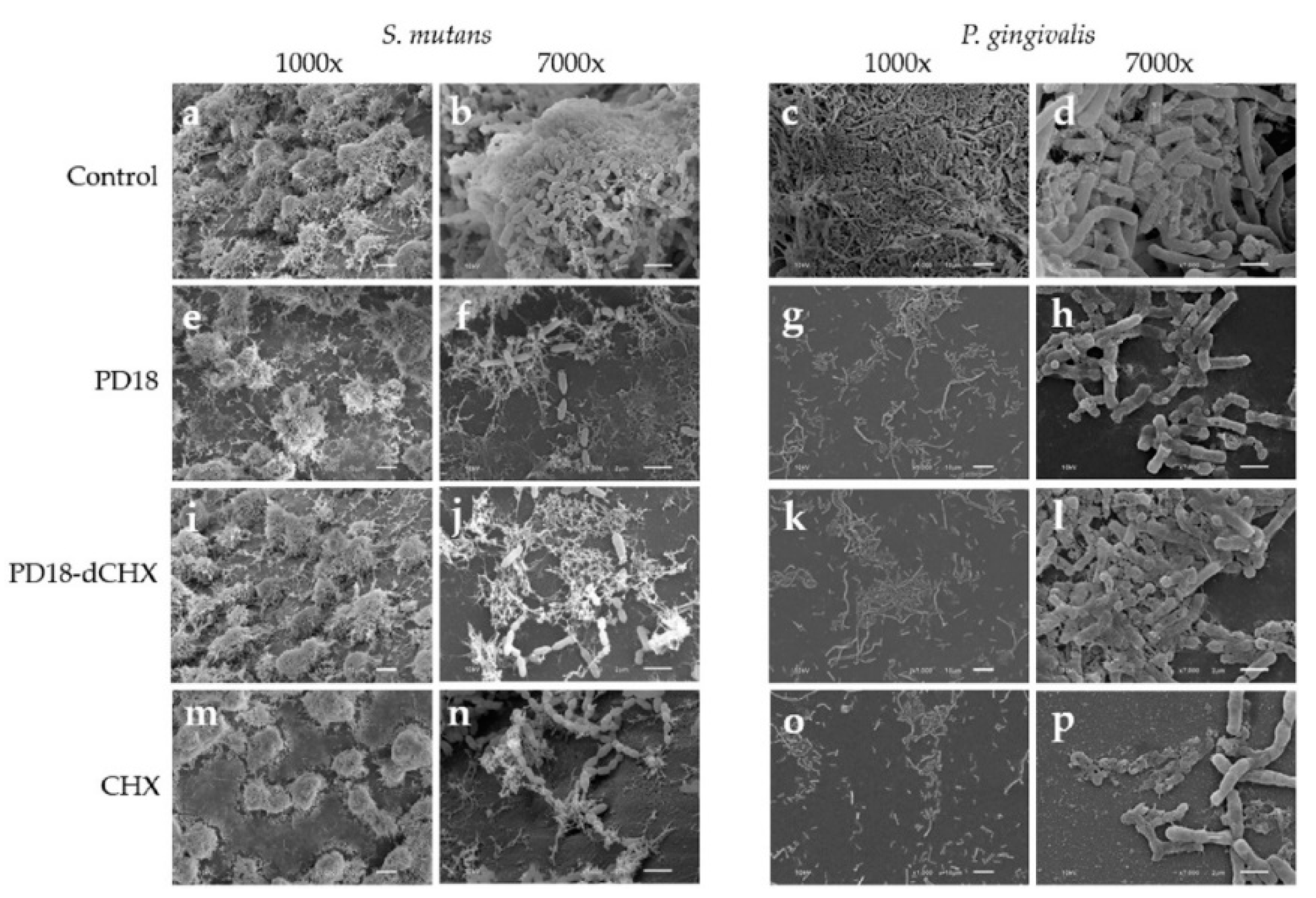
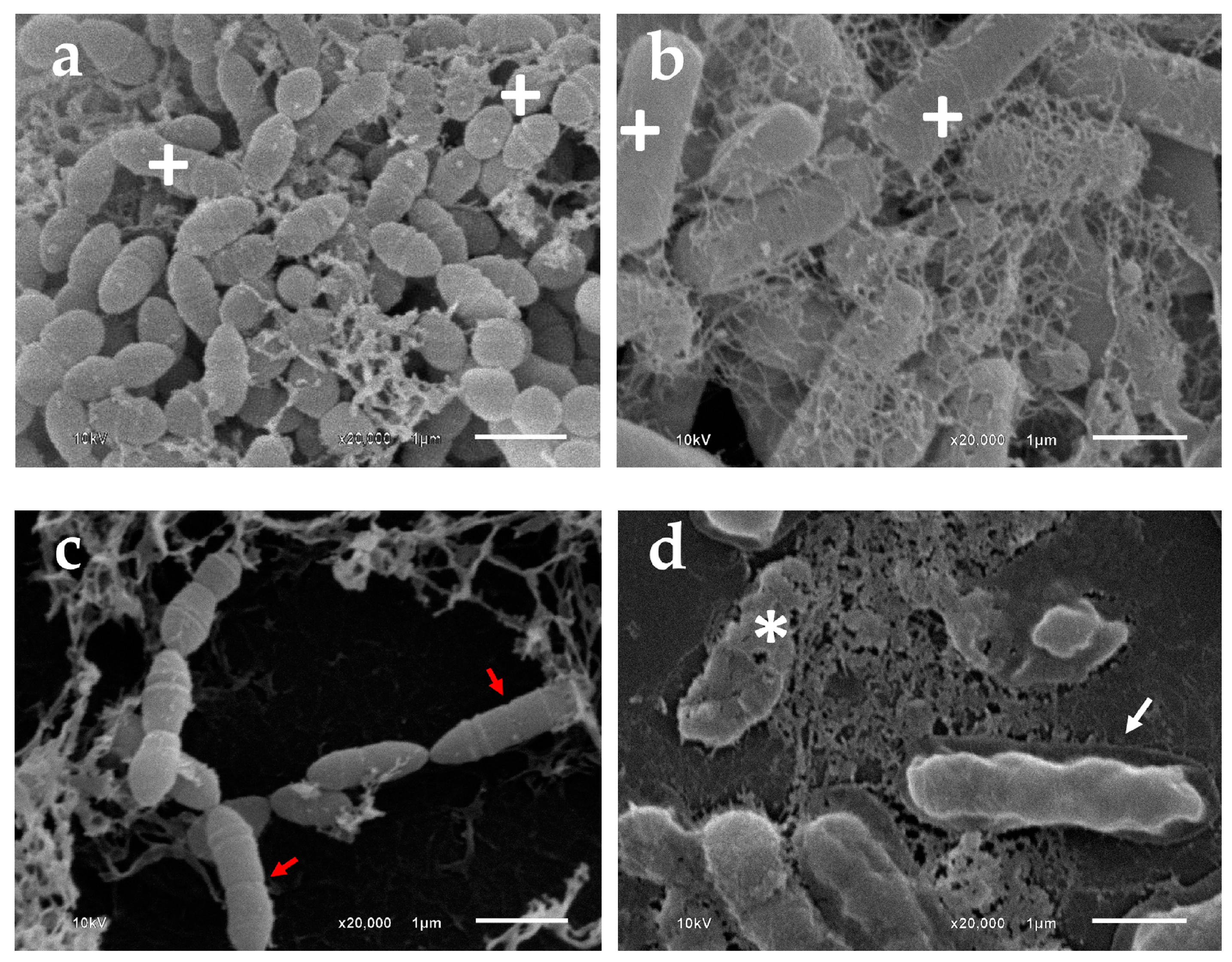
3.8. Morphologic Analysis Using SEM
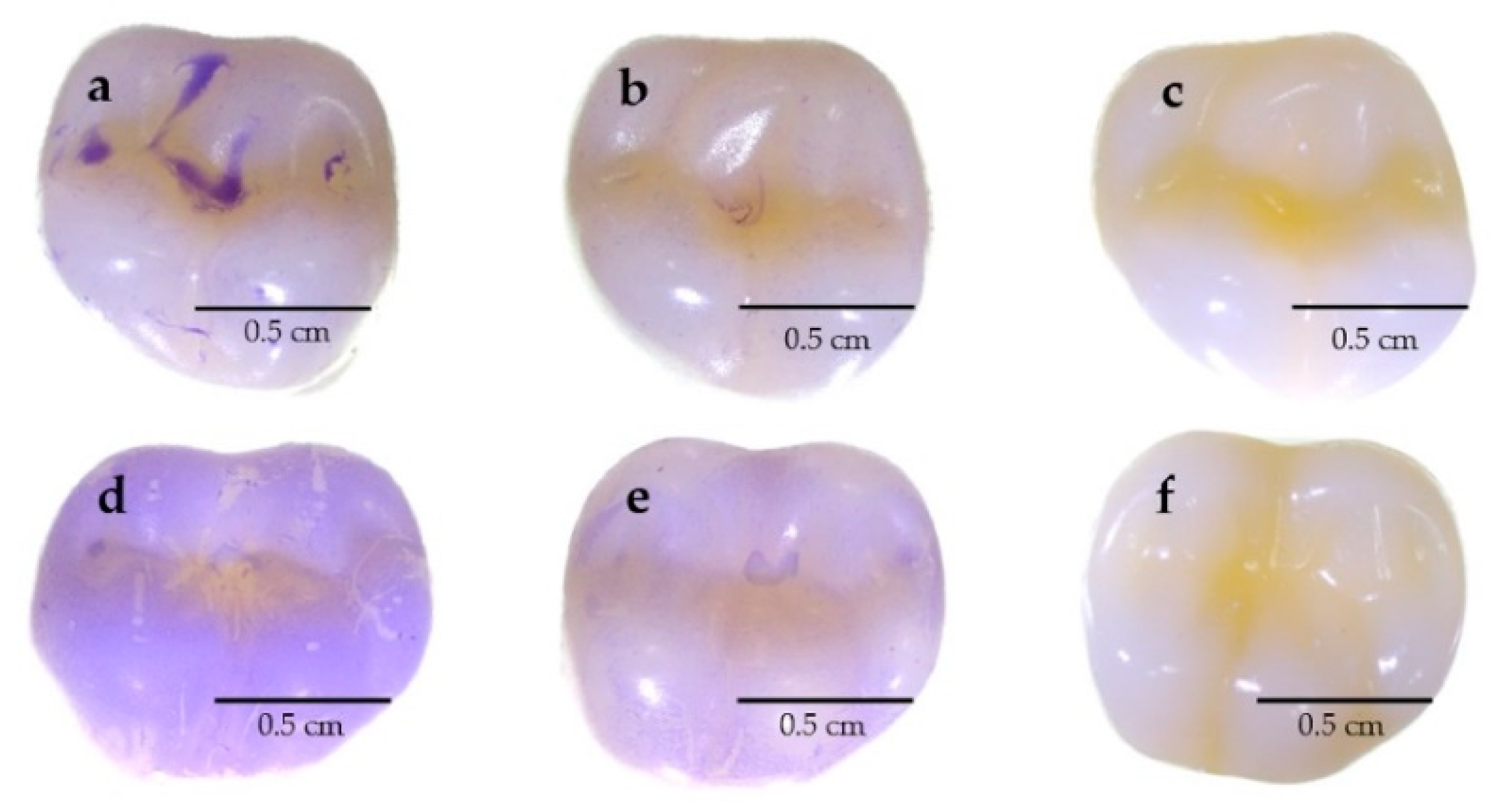
3.9. Evaluating the Capability to Reduce Biofilms on Typodont Teeth
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Haque, M.; Sartelli, M.; Haque, S.Z. Dental infection and resistance—Global health consequences. Dent. J. 2019, 7, 22. [Google Scholar] [CrossRef] [PubMed]
- Dembowska, E.; Jaroń, A.; Gabrysz-Trybek, E.; Bladowska, J.; Trybek, G. Evaluation of Common Factors of Periodontitis and Cardiovascular Disease in Patients with the Acute Coronary Syndrome. Int. J. Environ. Res. Public Health 2022, 19, 8139. [Google Scholar] [CrossRef] [PubMed]
- Sedghi, L.M.; Bacino, M.; Kapila, Y.L. Periodontal disease: The good, the bad, and the unknown. Front. Cell. Infect. Microbiol. 2021, 11, 1210. [Google Scholar] [CrossRef]
- Kõll-Klais, P.; Mändar, R.; Leibur, E.; Marcotte, H.; Hammarström, L.; Mikelsaar, M. Oral lactobacilli in chronic periodontitis and periodontal health: Species composition and antimicrobial activity. Oral Microbiol. Immunol. 2005, 20, 354–361. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Zheng, C.; Yang, J.; Li, B. Intersection between macrophages and periodontal pathogens in periodontitis. J. Leukoc. Biol. 2021, 110, 577–583. [Google Scholar] [CrossRef] [PubMed]
- Albaghdadi, S.Z.; Altaher, J.B.; Drobiova, H.; Bhardwaj, R.G.; Karched, M. In vitro characterization of biofilm formation in Prevotella species. Front. Oral Health 2021, 2, 724194. [Google Scholar] [CrossRef]
- Lai, S.; Lingström, P.; Cagetti, M.G.; Cocco, F.; Meloni, G.; Arrica, M.A.; Campus, G. Effect of Lactobacillus brevis CD2 containing lozenges and plaque pH and cariogenic bacteria in diabetic children: A randomised clinical trial. Clin. Oral Investig. 2021, 25, 115–123. [Google Scholar] [CrossRef]
- Yang, H.; Bi, Y.; Shang, X.; Wang, M.; Linden, S.B.; Li, Y.; Li, Y.; Nelson, D.C.; Wei, H. Antibiofilm activities of a novel chimeolysin against Streptococcus mutans under physiological and cariogenic conditions. Antimicrob. Agents Chemother. 2016, 60, 7436–7443. [Google Scholar] [CrossRef]
- Giordani, B.; Parolin, C.; Vitali, B. Lactobacilli as anti-biofilm strategy in oral infectious diseases: A mini-review. Front. Med. Technol. 2021, 3, 769172. [Google Scholar] [CrossRef]
- Hajishengallis, G.; Lamont, R.J. Beyond the red complex and into more complexity: The polymicrobial synergy and dysbiosis (PSD) model of periodontal disease etiology. Mol. Oral Microbiol. 2012, 27, 409–419. [Google Scholar] [CrossRef]
- Xu, W.; Zhou, W.; Wang, H.; Liang, S. Roles of Porphyromonas gingivalis and its virulence factors in periodontitis. Adv. Protein Chem. Struct. Biol. 2020, 120, 45–84. [Google Scholar]
- Smalley, J.W.; Olczak, T. Heme acquisition mechanisms of Porphyromonas gingivalis–strategies used in a polymicrobial community in a heme-limited host environment. Mol. Oral Microbiol. 2017, 32, 1–23. [Google Scholar] [CrossRef]
- Stobernack, T.; du Teil Espina, M.; Mulder, L.M.; Palma Medina, L.M.; Piebenga, D.R.; Gabarrini, G.; Zhao, X.; Janssen, K.M.; Hulzebos, J.; Brouwer, E. A secreted bacterial peptidylarginine deiminase can neutralize human innate immune defenses. MBio 2018, 9, e01704–e01718. [Google Scholar] [CrossRef] [PubMed]
- Aleksijević, L.H.; Aleksijević, M.; Škrlec, I.; Šram, M.; Šram, M.; Talapko, J. Porphyromonas gingivalis Virulence Factors and Clinical Significance in Periodontal Disease and Coronary Artery Diseases. Pathogens 2022, 11, 1173. [Google Scholar] [CrossRef]
- Sharma, A. Virulence mechanisms of Tannerella forsythia. Periodontology 2000 2010, 54, 106. [Google Scholar] [CrossRef]
- Bravo-Lopez, M.; Villa-Islas, V.; Rocha Arriaga, C.; Villaseñor-Altamirano, A.B.; Guzmán-Solís, A.; Sandoval-Velasco, M.; Wesp, J.K.; Alcantara, K.; López-Corral, A.; Gómez-Valdés, J. Paleogenomic insights into the red complex bacteria Tannerella forsythia in Pre-Hispanic and Colonial individuals from Mexico. Philos. Trans. R. Soc. B 2020, 375, 20190580. [Google Scholar] [CrossRef]
- Sharma, G.; Garg, N.; Hasan, S.; Shirodkar, S. Prevotella: An insight into its characteristics and associated virulence factors. Microb. Pathog. 2022, 169, 105673. [Google Scholar] [CrossRef]
- Banas, J.A. Virulence properties of Streptococcus mutans. Front. Biosci. Landmark 2004, 9, 1267–1277. [Google Scholar] [CrossRef]
- Bedoya-Correa, C.M.; Rodríguez, R.J.R.; Parada-Sanchez, M.T. Genomic and phenotypic diversity of Streptococcus mutans. J. Oral Biosci. 2019, 61, 22–31. [Google Scholar] [CrossRef]
- Zhang, Q.; Ma, Q.; Wang, Y.; Wu, H.; Zou, J. Molecular mechanisms of inhibiting glucosyltransferases for biofilm formation in Streptococcus mutans. Int. J. Oral Sci. 2021, 13, 30. [Google Scholar] [CrossRef]
- Habash, M.; Reid, G. Microbial biofilms: Their development and significance for medical device—Related infections. J. Clin. Pharmacol. 1999, 39, 887–898. [Google Scholar] [CrossRef]
- Sun, X.; Wang, L.; Lynch, C.D.; Sun, X.; Li, X.; Qi, M.; Ma, C.; Li, C.; Dong, B.; Zhou, Y. Nanoparticles having amphiphilic silane containing Chlorin e6 with strong anti-biofilm activity against periodontitis-related pathogens. J. Dent. 2019, 81, 70–84. [Google Scholar] [CrossRef]
- Saïz, P.; Taveira, N.; Alves, R. Probiotics in oral health and disease: A systematic review. Appl. Sci. 2021, 11, 8070. [Google Scholar] [CrossRef]
- Lorusso, F.; Tartaglia, G.; Inchingolo, F.; Scarano, A. Early Response and Clinical Efficacy of a Mouthwash Containing Chlorhexidine, Anti Discoloration System, Polyvinylpyrrolidone/Vinyl Acetate and Sodium DNA in Periodontitis Model: A Triple-Blind Randomized Controlled Clinical Trial. Dent. J. 2022, 10, 101. [Google Scholar] [CrossRef]
- Makino-Oi, A.; Ishii, Y.; Hoshino, T.; Okubo, N.; Sugito, H.; Hosaka, Y.; Fukaya, C.; Nakagawa, T.; Saito, A. Effect of periodontal surgery on oral health-related quality of life in patients who have completed initial periodontal therapy. J. Periodontal Res. 2016, 51, 212–220. [Google Scholar] [CrossRef]
- Huang, M.-C.J.; Tang, J. Probiotics in personal care products. Microbiol. Discov. 2015, 3, 5. [Google Scholar] [CrossRef]
- Jung, J.-I.; Baek, S.-M.; Nguyen, T.H.; Kim, J.W.; Kang, C.-H.; Kim, S.; Imm, J.-Y. Effects of probiotic culture supernatant on cariogenic biofilm formation and RANKL-induced osteoclastogenesis in RAW 264.7 macrophages. Molecules 2021, 26, 733. [Google Scholar] [CrossRef] [PubMed]
- Inchingolo, A.D.; Malcangi, G.; Semjonova, A.; Inchingolo, A.M.; Patano, A.; Coloccia, G.; Ceci, S.; Marinelli, G.; Di Pede, C.; Ciocia, A.M. Oralbiotica/Oralbiotics: The Impact of Oral Microbiota on Dental Health and Demineralization: A Systematic Review of the Literature. Children 2022, 9, 1014. [Google Scholar] [CrossRef] [PubMed]
- FAO. Guidelines for the Evaluation of Probiotics in Food, Report of a Joint FAO/WHO Working Group on Drafting Guideline for the Evaluation of Probiotic in Food; World Health Organization: Geneva, Switzerland, 2002. [Google Scholar]
- Butera, A.; Gallo, S.; Pascadopoli, M.; Taccardi, D.; Scribante, A. Home oral care of periodontal patients using antimicrobial gel with postbiotics, lactoferrin, and aloe barbadensis leaf juice powder vs. conventional chlorhexidine gel: A split-mouth randomized clinical trial. Antibiotics 2022, 11, 118. [Google Scholar] [CrossRef]
- Gueimonde, M.; Ouwehand, A.C.; Salminen, S. Safety of probiotics. Scand. J. Nutr. 2004, 48, 42–48. [Google Scholar] [CrossRef]
- Seddik, H.A.; Bendali, F.; Gancel, F.; Fliss, I.; Spano, G.; Drider, D. Lactobacillus plantarum and its probiotic and food potentialities. Probiotics Antimicrob. Proteins 2017, 9, 111–122. [Google Scholar] [CrossRef]
- Endo, A.; Maeno, S.; Tanizawa, Y.; Kneifel, W.; Arita, M.; Dicks, L.; Salminen, S. Fructophilic lactic acid bacteria, a unique group of fructose-fermenting microbes. Appl. Environ. Microbiol. 2018, 84, e01290-18. [Google Scholar] [CrossRef] [PubMed]
- Ciandrini, E.; Campana, R.; Casettari, L.; Perinelli, D.R.; Fagioli, L.; Manti, A.; Palmieri, G.F.; Papa, S.; Baffone, W. Characterization of biosurfactants produced by Lactobacillus spp. and their activity against oral streptococci biofilm. Appl. Microbiol. Biotechnol. 2016, 100, 6767–6777. [Google Scholar] [CrossRef] [PubMed]
- Aguilar-Toalá, J.; Garcia-Varela, R.; Garcia, H.; Mata-Haro, V.; González-Córdova, A.; Vallejo-Cordoba, B.; Hernández-Mendoza, A. Postbiotics: An evolving term within the functional foods field. Trends Food Sci. Technol. 2018, 75, 105–114. [Google Scholar] [CrossRef]
- Moradi, M.; Kousheh, S.A.; Almasi, H.; Alizadeh, A.; Guimarães, J.T.; Yılmaz, N.; Lotfi, A. Postbiotics produced by lactic acid bacteria: The next frontier in food safety. Compr. Rev. Food Sci. Food Saf. 2020, 19, 3390–3415. [Google Scholar] [CrossRef]
- Zheng, J.; Wittouck, S.; Salvetti, E.; Franz, C.M.; Harris, H.M.; Mattarelli, P.; O’toole, P.W.; Pot, B.; Vandamme, P.; Walter, J. A taxonomic note on the genus Lactobacillus: Description of 23 novel genera, emended description of the genus Lactobacillus Beijerinck 1901, and union of Lactobacillaceae and Leuconostocaceae. Int. J. Syst. Evol. Microbiol. 2020, 70, 2782–2858. [Google Scholar] [CrossRef]
- Humam, A.M.; Loh, T.C.; Foo, H.L.; Samsudin, A.A.; Mustapha, N.M.; Zulkifli, I.; Izuddin, W.I. Effects of feeding different postbiotics produced by Lactobacillus plantarum on growth performance, carcass yield, intestinal morphology, gut microbiota composition, immune status, and growth gene expression in broilers under heat stress. Animals 2019, 9, 644. [Google Scholar] [CrossRef]
- Lin, X.; Chen, X.; Chen, Y.; Jiang, W.; Chen, H. The effect of five probiotic lactobacilli strains on the growth and biofilm formation of Streptococcus mutans. Oral Dis. 2015, 21, e128–e134. [Google Scholar] [CrossRef]
- Jaffar, N.; Ishikawa, Y.; Mizuno, K.; Okinaga, T.; Maeda, T. Mature biofilm degradation by potential probiotics: Aggregatibacter actinomycetemcomitans versus Lactobacillus spp. PLoS ONE 2016, 11, e0159466. [Google Scholar] [CrossRef]
- Jeong, D.; Kim, D.-H.; Song, K.-Y.; Seo, K.-H. Antimicrobial and anti-biofilm activities of Lactobacillus kefiranofaciens DD2 against oral pathogens. J. Oral Microbiol. 2018, 10, 1472985. [Google Scholar] [CrossRef]
- Rossoni, R.D.; dos Santos Velloso, M.; de Barros, P.P.; de Alvarenga, J.A.; Dos Santos, J.D.; dos Santos Prado, A.C.C.; de Camargo Ribeiro, F.; Anbinder, A.L.; Junqueira, J.C. Inhibitory effect of probiotic Lactobacillus supernatants from the oral cavity on Streptococcus mutans biofilms. Microb. Pathog. 2018, 123, 361–367. [Google Scholar] [CrossRef]
- Srivastava, N.; Ellepola, K.; Venkiteswaran, N.; Chai, L.Y.A.; Ohshima, T.; Seneviratne, C.J. Lactobacillus Plantarum 108 inhibits Streptococcus mutans and Candida albicans mixed-species biofilm formation. Antibiotics 2020, 9, 478. [Google Scholar] [CrossRef]
- Mohd-Zubri, N.S.; Ramasamy, K.; Abdul-Rahman, N.Z. Characterization and potential oral probiotic properties of Lactobacillus plantarum FT 12 and Lactobacillus brevis FT 6 isolated from Malaysian fermented food. Arch. Oral Biol. 2022, 143, 105515. [Google Scholar] [CrossRef]
- Söderling, E.M.; Marttinen, A.M.; Haukioja, A.L. Probiotic lactobacilli interfere with Streptococcus mutans biofilm formation in vitro. Curr. Microbiol. 2011, 62, 618–622. [Google Scholar] [CrossRef]
- Marttinen, A.M.; Haukioja, A.L.; Keskin, M.; Söderling, E.M. Effects of Lactobacillus reuteri PTA 5289 and L. paracasei DSMZ16671 on the adhesion and biofilm formation of Streptococcus mutans. Curr. Microbiol. 2013, 67, 193–199. [Google Scholar] [CrossRef]
- Prabhurajeshwar, C.; Chandrakanth, R.K. Probiotic potential of Lactobacilli with antagonistic activity against pathogenic strains: An in vitro validation for the production of inhibitory substances. Biomed. J. 2017, 40, 270–283. [Google Scholar] [CrossRef]
- Missaoui, J.; Saidane, D.; Mzoughi, R.; Minervini, F. Fermented seeds (“Zgougou”) from aleppo pine as a novel source of potentially probiotic lactic acid bacteria. Microorganisms 2019, 7, 709. [Google Scholar] [CrossRef]
- Thieme, L.; Hartung, A.; Tramm, K.; Klinger-Strobel, M.; Jandt, K.D.; Makarewicz, O.; Pletz, M.W. MBEC versus MBIC: The lack of differentiation between biofilm reducing and inhibitory effects as a current problem in biofilm methodology. Biol. Proced. Online 2019, 21, 1–5. [Google Scholar] [CrossRef]
- Haney, E.F.; Trimble, M.J.; Cheng, J.T.; Vallé, Q.; Hancock, R.E. Critical assessment of methods to quantify biofilm growth and evaluate antibiofilm activity of host defence peptides. Biomolecules 2018, 8, 29. [Google Scholar] [CrossRef]
- Ben Slama, R.; Kouidhi, B.; Zmantar, T.; Chaieb, K.; Bakhrouf, A. Anti-listerial and anti-biofilm activities of potential Probiotic Lactobacillus strains isolated from Tunisian traditional fermented food. J. Food Saf. 2013, 33, 8–16. [Google Scholar] [CrossRef]
- Lin, X.; Xu, J.; Shi, Z.; Xu, Y.; Fu, T.; Zhang, L.; He, F. Evaluation of the antibacterial effects and mechanism of Plantaricin 149 from Lactobacillus plantarum NRIC 149 on the peri-implantitis pathogens. Sci. Rep. 2021, 11, 21022. [Google Scholar] [CrossRef]
- Moraes, R.M.; Schlagenhauf, U.; Anbinder, A.L. Outside the limits of bacterial viability: Postbiotics in the management of periodontitis. Biochem. Pharmacol. 2022, 2022, 115072. [Google Scholar] [CrossRef]
- Koll, P.; Mändar, R.; Marcotte, H.; Leibur, E.; Mikelsaar, M.; Hammarström, L. Characterization of oral lactobacilli as potential probiotics for oral health. Oral Microbiol. Immunol. 2008, 23, 139–147. [Google Scholar] [CrossRef] [PubMed]
- Bustamante, M.; Oomah, B.D.; Mosi-Roa, Y.; Rubilar, M.; Burgos-Díaz, C. Probiotics as an adjunct therapy for the treatment of halitosis, dental caries and periodontitis. Probiotics Antimicrob. Proteins 2020, 12, 325–334. [Google Scholar] [CrossRef]
- Kareem, K.Y.; Hooi Ling, F.; Teck Chwen, L.; May Foong, O.; Anjas Asmara, S. Inhibitory activity of postbiotic produced by strains of Lactobacillus plantarum using reconstituted media supplemented with inulin. Gut Pathog. 2014, 6, 1–7. [Google Scholar] [CrossRef]
- Wang, Y.; Pei, H.; Liu, Y.; Huang, X.; Deng, L.; Lan, Q.; Chen, S.; He, L.; Liu, A.; Ao, X. Inhibitory mechanism of cell-free supernatants of Lactobacillus plantarum on Proteus mirabilis and influence of the expression of histamine synthesis-related genes. Food Control 2021, 125, 107982. [Google Scholar] [CrossRef]
- Todorov, S.D.; Franco, B.D.G.D.M. Lactobacillus plantarum: Characterization of the species and application in food production. Food Rev. Int. 2010, 26, 205–229. [Google Scholar] [CrossRef]
- Dani, S.; Prabhu, A.; Chaitra, K.; Desai, N.; Patil, S.R.; Rajeev, R. Assessment of Streptococcus mutans in healthy versus gingivitis and chronic periodontitis: A clinico-microbiological study. Contemp. Clin. Dent. 2016, 7, 529. [Google Scholar] [CrossRef]
- Gerits, E.; Verstraeten, N.; Michiels, J. New approaches to combat Porphyromonas gingivalis biofilms. J. Oral Microbiol. 2017, 9, 1300366. [Google Scholar] [CrossRef]
- Prado, M.M.; Kovalski, D.J.; Torrez, W.B.; Bueno-Silva, B.; Feres, M.; de Almeida, J.; Porto, L.M. Development of a multispecies periodontal biofilm model within a stirred bioreactor. Biofouling 2020, 36, 725–735. [Google Scholar] [CrossRef]
- Preza, D.; Olsen, I.; Aas, J.A.; Willumsen, T.; Grinde, B.; Paster, B.J. Bacterial profiles of root caries in elderly patients. J. Clin. Microbiol. 2008, 46, 2015–2021. [Google Scholar] [CrossRef] [PubMed]
- Yip, S.; Dehcheshmeh, M.M.; McLelland, D.J.; Boardman, W.S.; Saputra, S.; Ebrahimie, E.; Weyrich, L.S.; Bird, P.S.; Trott, D.J. Porphyromonas spp., Fusobacterium spp., and Bacteroides spp. dominate microbiota in the course of macropod progressive periodontal disease. Sci. Rep. 2021, 11, 17775. [Google Scholar] [CrossRef]
- Diaz, P.I.; Slakeski, N.; Reynolds, E.C.; Morona, R.; Rogers, A.H.; Kolenbrander, P.E. Role of oxyR in the oral anaerobe Porphyromonas gingivalis. J. Bacteriol. 2006, 188, 2454–2462. [Google Scholar] [CrossRef]
- He, J.; Miyazaki, H.; Anaya, C.; Yu, F.; Yeudall, W.A.; Lewis, J.P. Role of Porphyromonas gingivalis FeoB2 in metal uptake and oxidative stress protection. Infect. Immun. 2006, 74, 4214–4223. [Google Scholar] [CrossRef]
- Wang, Z.; Shen, Y.; Haapasalo, M. Dynamics of dissolution, killing, and inhibition of dental plaque biofilm. Front. Microbiol. 2020, 11, 964. [Google Scholar] [CrossRef]
- Prasanth, M. Antimicrobial efficacy of different toothpastes and mouthrinses: An in vitro study. Dent. Res. J. 2011, 8, 85. [Google Scholar]
- Schestakow, A.; Guth, M.S.; Eisenmenger, T.A.; Hannig, M. Evaluation of anti-biofilm activity of mouthrinses containing tannic acid or chitosan on dentin in situ. Molecules 2021, 26, 1351. [Google Scholar] [CrossRef]
- Guo, Y.; Wang, Y.; Wang, Y.; Jin, Y.; Wang, C. Heme competition triggers an increase in the pathogenic potential of Porphyromonas gingivalis in Porphyromonas gingivalis-Candida albicans mixed biofilm. Front. Microbiol. 2020, 11, 596459. [Google Scholar] [CrossRef] [PubMed]
- Tahmourespour, A.; Salehi, R.; Kermanshahi, R.; Eslami, G. The anti-biofouling effect of Lactobacillus fermentum-derived biosurfactant against Streptococcus mutans. Biofouling 2011, 27, 385–392. [Google Scholar] [CrossRef] [PubMed]
- Savabi, O.; Kazemi, M.; Kamali, S.; Salehi, A.R.; Eslami, G.; Tahmourespour, A.; Salehi, R. Effects of biosurfactant produced by Lactobacillus casei on gtfB, gtfC, and ftf gene expression level in S. mutans by real-time RT-PCR. Adv. Biomed. Res. 2014, 3, 231. [Google Scholar]
- Yan, X.; Gu, S.; Cui, X.; Shi, Y.; Wen, S.; Chen, H.; Ge, J. Antimicrobial, anti-adhesive and anti-biofilm potential of biosurfactants isolated from Pediococcus acidilactici and Lactobacillus plantarum against Staphylococcus aureus CMCC26003. Microb. Pathog. 2019, 127, 12–20. [Google Scholar] [CrossRef] [PubMed]

| Lactobacillus PM | Inhibition Zone (mm) | |||
|---|---|---|---|---|
| P. gingivalis | T. forsythia | P. loescheii | S. mutans | |
| PD01 | NI | 7.58 ± 0.04 c | NI | NI |
| PD02 | NI | 7.61 ± 0.11 c | NI | NI |
| PD03 | NI | 7.04 ± 0.08 b | NI | NI |
| PD04 | NI | 7.15 ± 0.06 b | NI | NI |
| PD05 | NI | 7.10 ± 0.09 b | NI | NI |
| PD06 | NI | 6.19 ± 0.21 a | NI | NI |
| PD07 | 7.21 ± 0.10 c | 7.19 ± 0.12 b | 7.59 ± 0.06 e | 6.38 ± 0.04 b |
| PD08 | 6.24 ± 0.12 a | 7.68 ± 0.24 c | 7.62 ± 0.15 e | 6.18 ± 0.06 c |
| PD09 | 6.64 ± 0.12 b | 7.14 ± 0.14 b | 6.23 ± 0.06 b | 5.73 ± 0.10 d |
| PD10 | 6.07 ± 0.05 a | 7.16 ± 0.28 b | 5.57 ± 0.12 a | 5.84 ± 0.16 d |
| PD11 | 6.11 ± 0.07 a | 6.29 ± 0.17 a | 7.12 ± 0.09 d | 6.17 ± 0.06 c |
| PD12 | NI | NI | 7.08 ± 0.14 d | NI |
| PD13 | NI | NI | 7.14 ± 0.09 d | NI |
| PD14 | 6.06 ± 0.06 a | 6.22 ± 0.16 a | 6.60 ± 0.13 c | 6.13 ± 0.11 c |
| PD15 | NI | NI | 7.08 ± 0.20 d | NI |
| PD16 | NI | NI | 7.18 ± 0.15 d | NI |
| PD17 | 7.10 ± 0.16 c | 6.07 ± 0.27 a | 6.58 ± 0.08 c | 6.12 ± 0.07 c |
| PD18 | 7.78 ± 0.19 d | 7.69 ± 0.11 c | 7.52 ± 0.07 e | 10.63 ± 0.05 a |
| PD19 | NI | NI | 7.18 ± 0.04 d | NI |
| PD20 | 6.17 ± 0.04 a | NI | 7.54 ± 0.10 e | NI |
| PD21 | NI | 7.51 ± 0.11 c | 7.17 ± 0.10 d | NI |
| Lactobacillus PM | Treatment | P. gingivalis | T. forsythia | P. loescheii | S. mutans |
|---|---|---|---|---|---|
| PD7 | Crude | 1:4 | 1:4 | 1:4 | 1:4 |
| Neutralized pH | ND | ND | ND | ND | |
| Proteinase K | ND | 1:2 | ND | ND | |
| Catalase | ND | ND | ND | ND | |
| PD8 | Crude | 1:2 | 1:4 | 1:4 | 1:2 |
| Neutralized pH | ND | 1:4 | 1:2 | ND | |
| Proteinase K | ND | ND | ND | ND | |
| Catalase | ND | ND | ND | ND | |
| PD9 | Crude | 1:2 | 1:4 | 1:4 | 1:4 |
| Neutralized pH | ND | 1:4 | ND | ND | |
| Proteinase K | 1:2 | ND | ND | ND | |
| Catalase | ND | ND | 1:2 | ND | |
| PD10 | Crude | 1:2 | 1:4 | 1:4 | 1:2 |
| Neutralized pH | ND | 1:4 | ND | ND | |
| Proteinase K | ND | ND | ND | ND | |
| Catalase | ND | ND | 1:2 | ND | |
| PD11 | Crude | 1:2 | 1:4 | 1:4 | 1:2 |
| Neutralized pH | ND | 1:4 | ND | ND | |
| Proteinase K | ND | ND | ND | ND | |
| Catalase | ND | ND | 1:2 | ND | |
| PD14 | Crude | 1:2 | 1:4 | 1:4 | 1:2 |
| Neutralized pH | ND | ND | ND | ND | |
| Proteinase K | ND | ND | ND | ND | |
| Catalase | ND | ND | 1:2 | ND | |
| PD17 | Crude | 1:4 | 1:2 | 1:4 | 1:2 |
| Neutralized pH | ND | ND | ND | ND | |
| Proteinase K | ND | ND | ND | ND | |
| Catalase | ND | ND | ND | ND | |
| PD18 | Crude | 1:2 | 1:4 | 1:4 | 1:2 |
| Neutralized pH | ND | 1:2 | 1:2 | ND | |
| Proteinase K | ND | ND | 1:2 | ND | |
| Catalase | ND | ND | 1:2 | ND |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Butrungrod, W.; Chaiyasut, C.; Makhamrueang, N.; Peerajan, S.; Chaiyana, W.; Sirilun, S. Postbiotic Metabolite of Lactiplantibacillus plantarum PD18 against Periodontal Pathogens and Their Virulence Markers in Biofilm Formation. Pharmaceutics 2023, 15, 1419. https://doi.org/10.3390/pharmaceutics15051419
Butrungrod W, Chaiyasut C, Makhamrueang N, Peerajan S, Chaiyana W, Sirilun S. Postbiotic Metabolite of Lactiplantibacillus plantarum PD18 against Periodontal Pathogens and Their Virulence Markers in Biofilm Formation. Pharmaceutics. 2023; 15(5):1419. https://doi.org/10.3390/pharmaceutics15051419
Chicago/Turabian StyleButrungrod, Widawal, Chaiyavat Chaiyasut, Netnapa Makhamrueang, Sartjin Peerajan, Wantida Chaiyana, and Sasithorn Sirilun. 2023. "Postbiotic Metabolite of Lactiplantibacillus plantarum PD18 against Periodontal Pathogens and Their Virulence Markers in Biofilm Formation" Pharmaceutics 15, no. 5: 1419. https://doi.org/10.3390/pharmaceutics15051419
APA StyleButrungrod, W., Chaiyasut, C., Makhamrueang, N., Peerajan, S., Chaiyana, W., & Sirilun, S. (2023). Postbiotic Metabolite of Lactiplantibacillus plantarum PD18 against Periodontal Pathogens and Their Virulence Markers in Biofilm Formation. Pharmaceutics, 15(5), 1419. https://doi.org/10.3390/pharmaceutics15051419







