Evaluation of Poly(N-Ethyl Pyrrolidine Methacrylamide) (EPA) and Derivatives as Polymeric Vehicles for miRNA Delivery to Neural Cells
Abstract
:1. Introduction
2. Materials and Methods
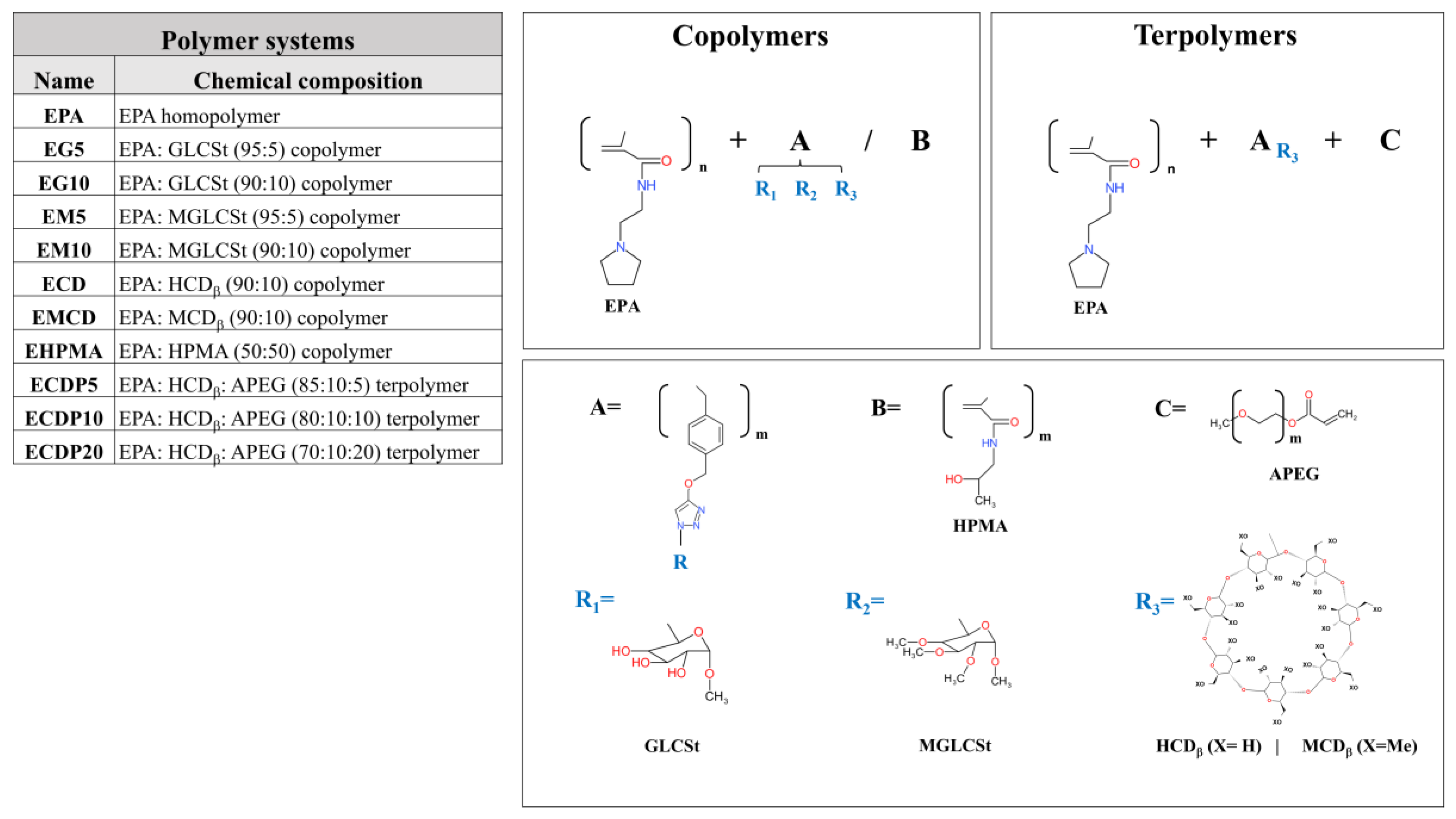
2.1. Synthesis of Homopolymers, Copolymers, and Terpolymers
2.2. miRNA Mimics
- (i)
- A synthetic miR-138 mimic (miR-138 mimic), a duplex RNA derivative composed of an antisense, and a sense strand. The antisense strand is a 5′-phosphate (P) modified RNA strand (5′-P-AGCUGGUGUUGUGAAUCAGGCCG-3′) corresponding to the human miR-138-5p sequence obtained from commercial sources (Merck-Sigma-Aldrich España, Madrid, Spain). The sense RNA strand was designed as the complementary sequence (5′-CGGCCUGAUUCACAACACCAGCU-3′) and was synthesized at a 1 μmol scale using 2’-O-TBDMS (t-butyldimethylsilyl) protected phosphoramidites (A, G, C and U) using a 3400 ABI DNA/RNA synthesizer. Synthesis was performed following standard protocols on DMT-ON mode. After the ammonia treatment, the solvent was evaporated to dryness, and 0.15 mL of triethylamine hydrofluoride/triethylamine /N-methylpyrrolidone (4:3:6) was added to remove t-butyldimethylsilyl groups. TBDMS-protected RNAs were incubated 2.5 h at 65 °C as described by Wincott et al. [23]. The deprotection reaction was quenched, and the crude product was purified using OPC cartridges (Glen Research). Oligoribonucleotides were quantified via UV absorption and characterized via MALDI-TOF mass spectrometry. The miR-138 mimic was obtained by annealing equimolar amounts of sense and antisense strands in 100 μL of 10 mM TRIS and 50 mM NaCl buffer. The resulting solution was heated at 94°C for 2 min, and then allowed to cool until it reached room temperature (RT). Then, 3 M sodium acetate pH = 5.2 was added (10 μL) along with EtOH (96%) (275 μL). Samples were stirred and precipitated at −20 °C. Finally, samples were centrifuged at 4 °C (15 min, 12000 rpm), and the supernatant was removed. The resulting pellets were dried using N2.
- (ii)
- A commercial cel-miR-67 mimic with a minimal amount of shared sequence identity with vertebrate miRNAs and no identified effects in vertebrate cells (miRIDIAN miRNA mimic negative control#1; cat#CN-001000-01; DharmaconTM) was employed as a negative control for functional assays (Neg. Ctrl mimic).
- (iii)
- Commercial Cy3-labeled cel-miR-67 (miRIDIAN miRNA Mimic Red Transfection Control; cat#CP-004500-01-05; DharmaconTM) was employed as a negative control in flow cytometry and immunofluorescence experiments (Cy3 Neg. Ctrl mimic).
- (iv)
- A DNA oligonucleotide with sequence equivalence to miR-138 mimic was employed for DLS and ζ-potential analyses to reduce the costs and degradation issues.
2.3. Characterization of Polymer Systems
- (i)
- Spectroscopic techniques: Polymer systems were characterized via 1H nuclear magnetic resonance spectroscopy (1H-NMR). Spectra were recorded in 5% deuterated chloroform (CDCl3) solution, deuterated methanol (CD3OD), or deuterated DMSO (DMSO-d6) with a Varian XLR-300 spectrometer using trimethylsilane (TMS) as the internal standard.
- (ii)
- Chromatographic techniques: The number average molecular weight (Mn) and polydispersity index (Ð) of the polymers were measured via gel permeation chromatography (GPC) with a Perkin Elmer (Waltham, MA, USA) chromatographic system equipped with a Waters (Milford, MA, USA) model 2414 refractive index detector using Styragel (300 mm × 7.8 mm and 5 μm nominal particle size) HR3 and HR5 water columns. Dimethylformamide (DMF) with 1 wt% LiBr was used as an eluent. Measurements were performed at 70 °C at a flow rate of 0.7 mL/min using a polymer concentration of 4 mg/mL. Calibration was performed with monodispersed polystyrene standards in the range between 2.0 and 9000.0 kDa.
2.4. Polyplexes Formation
2.5. Condensation Efficiency Assays
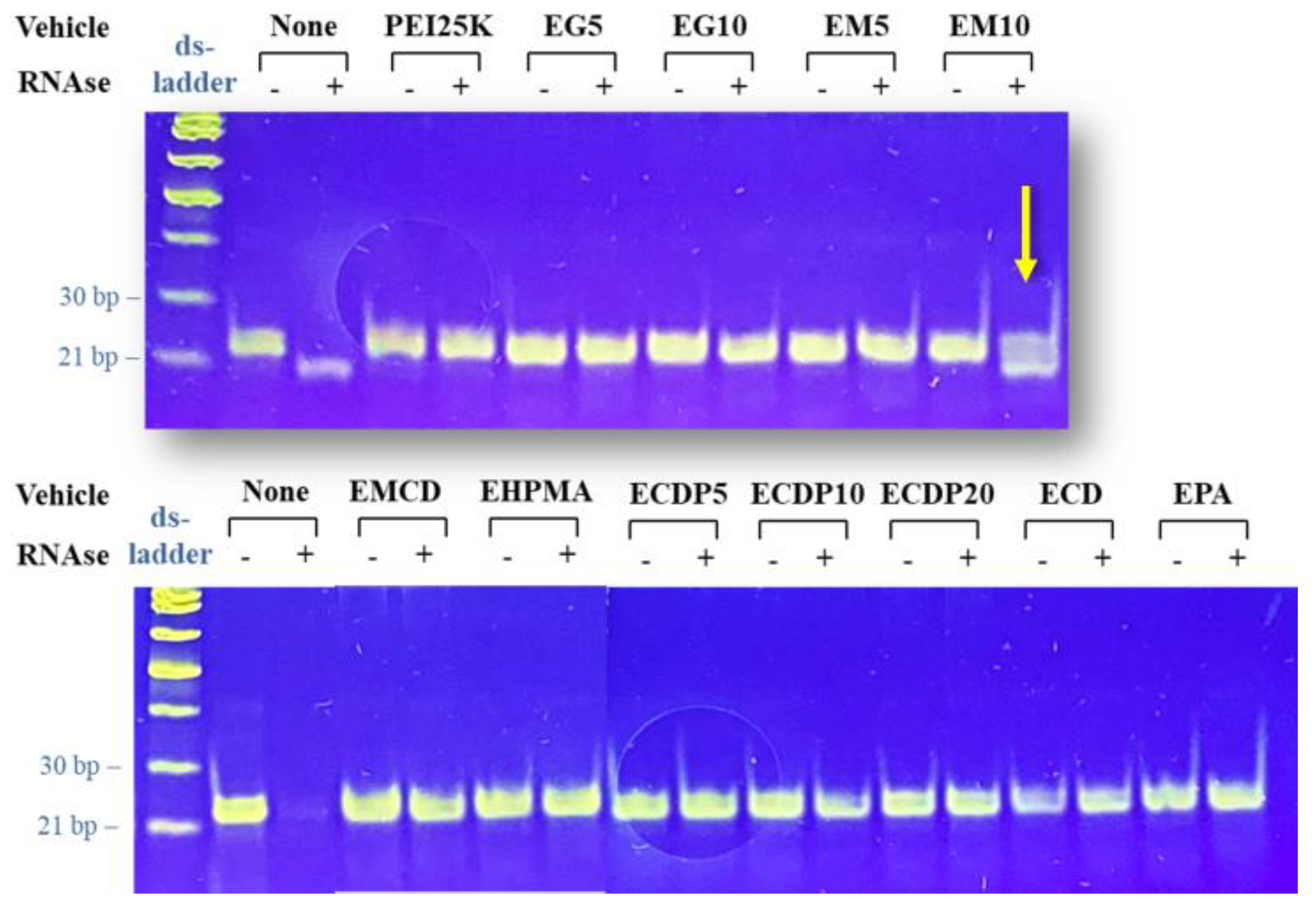
2.6. RNA Protection Assays
2.7. Cell Culture
2.8. Cell Transfection
2.9. Flow Cytometry
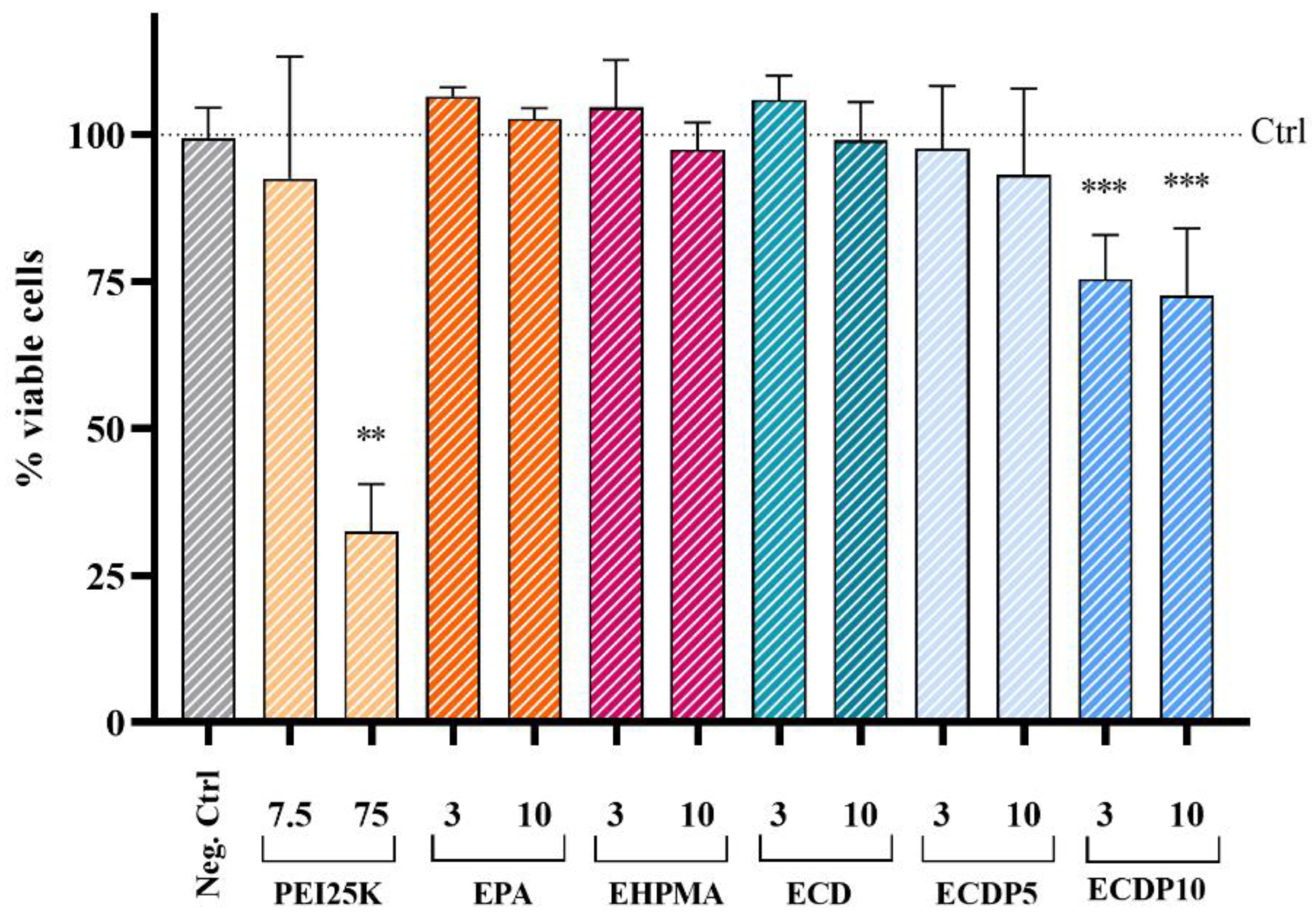
2.10. Cell Viability Assays
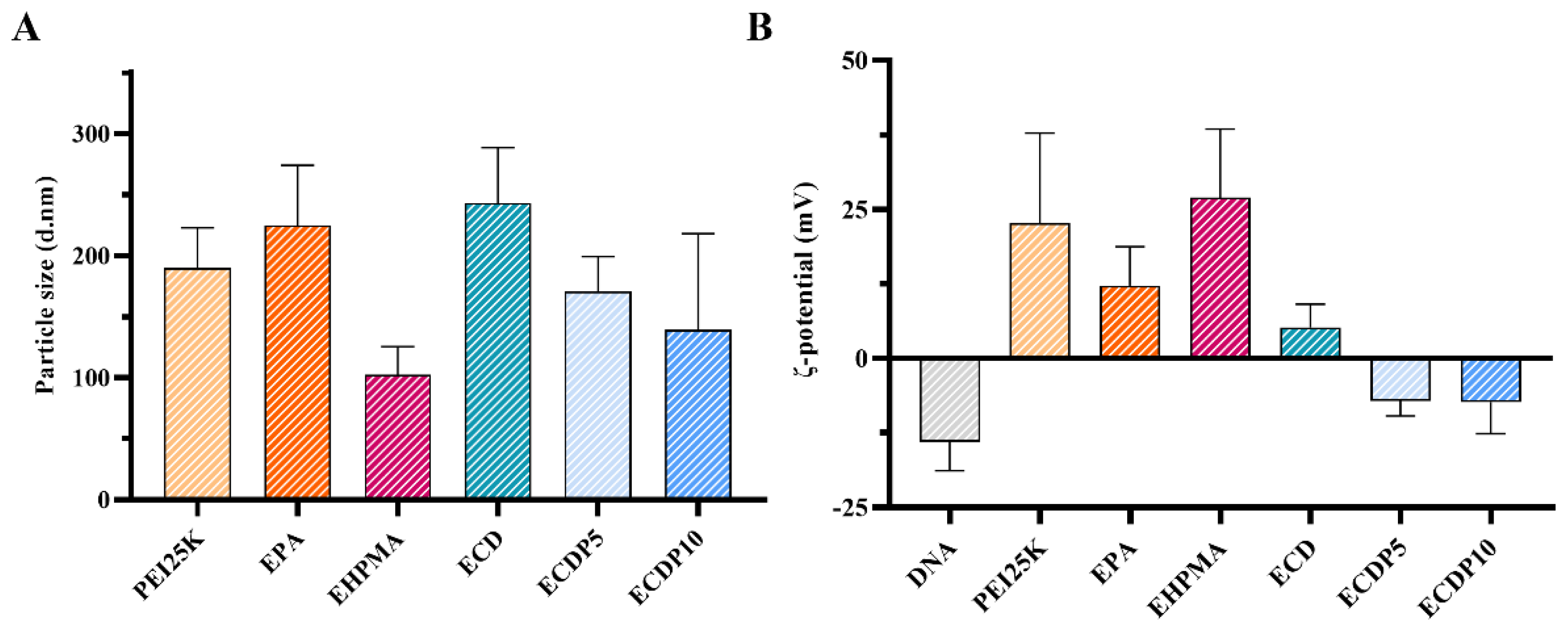
2.11. Particle Size and Zeta Potential
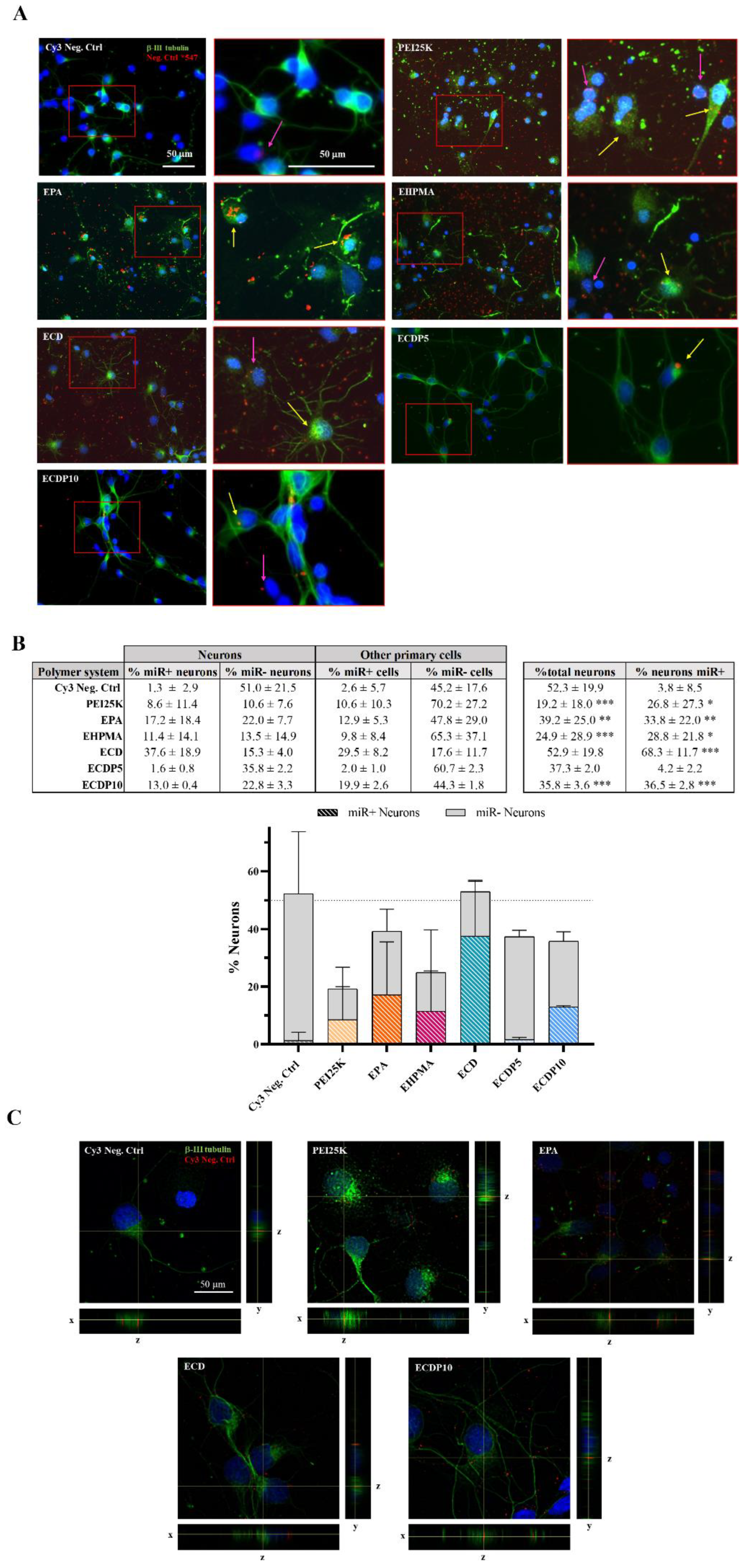
2.12. Immunofluorescence and Confocal Analysis
2.13. Endosomal Escape Assays
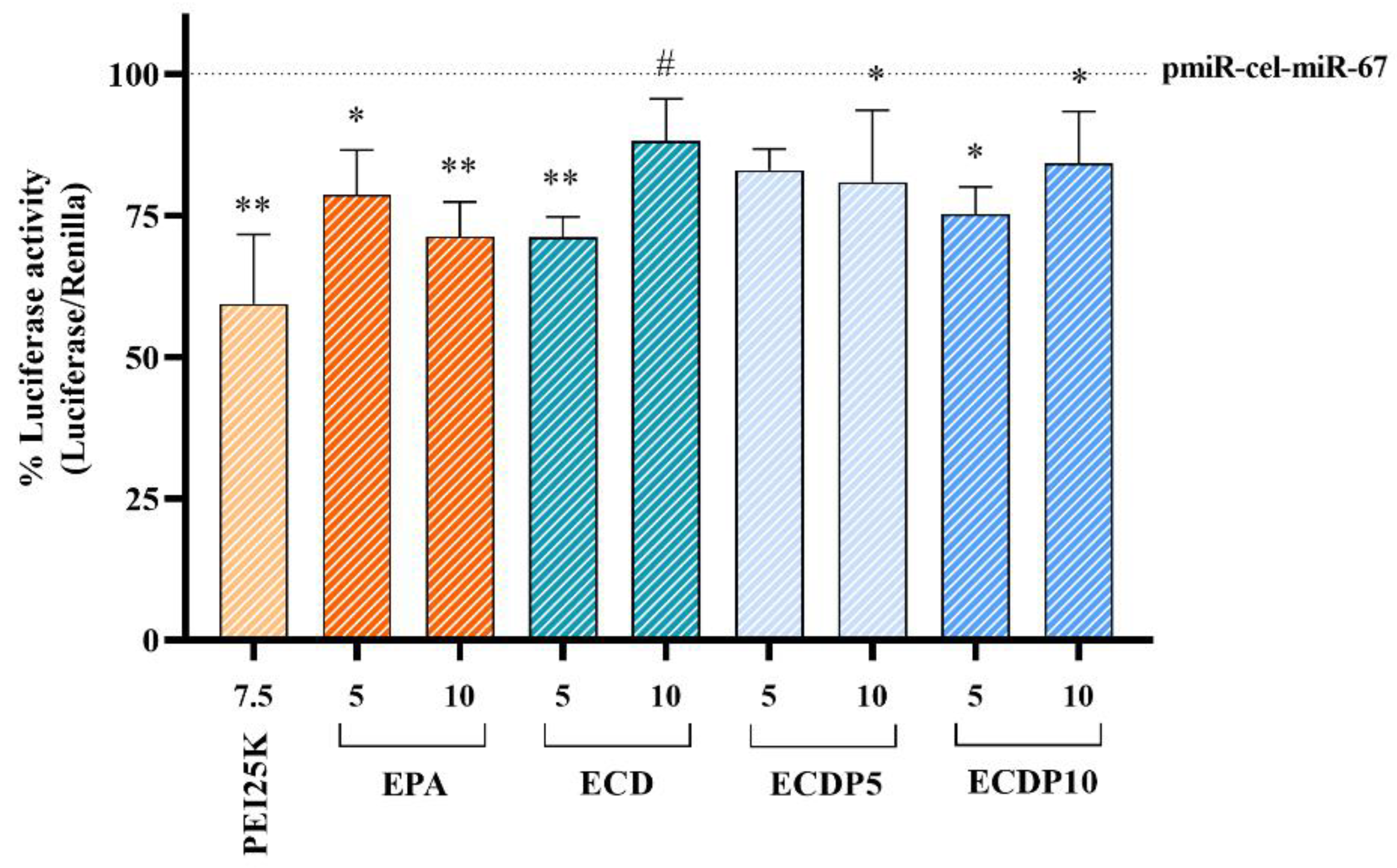
2.14. Dual-Luciferase Reporter Assay
2.15. Data Analysis
3. Results and Discussion
3.1. Synthesis and Characterization of Terpolymers
3.2. miRNA Condensation Efficiency
3.3. Protection against RNAses
3.4. Cytotoxicity and Cell Binding of the Polyplexes
3.5. Particle Size and ζ-Potential
3.6. Microscopy Analysis of miRNA Cell Binding and Internalization into Neural and Neuronal Cells
3.7. Endosomal Escape
3.8. miRNA Transfection Efficiency
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hu, B.; Zhong, L.; Weng, Y.; Peng, L.; Huang, Y.; Zhao, Y.; Liang, X.-J. Therapeutic siRNA: State of the art. Signal Transduct. Target. Ther. 2020, 5, 101. [Google Scholar] [CrossRef] [PubMed]
- Tan, J.-K.Y.; Sellers, D.L.; Pham, B.; Pun, S.H.; Horner, P.J. Non-Viral Nucleic Acid Delivery Strategies to the Central Nervous System. Front. Mol. Neurosci. 2016, 9, 108. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Huang, R. Non-viral nucleic acid delivery to the central nervous system and brain tumors. J. Gene Med. 2019, 21, e3091. [Google Scholar] [CrossRef]
- Mendonça, M.C.P.; Kont, A.; Aburto, M.R.; Cryan, J.F.; O’driscoll, C.M. Advances in the Design of (Nano)Formulations for Delivery of Antisense Oligonucleotides and Small Interfering RNA: Focus on the Central Nervous System. Mol. Pharm. 2021, 18, 1491–1506. [Google Scholar] [CrossRef] [PubMed]
- Liang, Y.; Liu, Z.; Shuai, X.; Wang, W.; Liu, J.; Bi, W.; Wang, C.; Jing, X.; Liu, Y.; Tao, E. Delivery of cationic polymer-siRNA nanoparticles for gene therapies in neural regeneration. Biochem. Biophys. Res. Commun. 2012, 421, 690–695. [Google Scholar] [CrossRef]
- Sun, P.; Liu, D.Z.; Jickling, G.C.; Sharp, F.R.; Yin, K.-J. MicroRNA-based therapeutics in central nervous system injuries. J. Cereb. Blood Flow Metab. 2018, 38, 1125–1148. [Google Scholar] [CrossRef]
- Liu, Z.; Wang, S.; Tapeinos, C.; Torrieri, G.; Känkänen, V.; El-Sayed, N.; Python, A.; Hirvonen, J.T.; Santos, H.A. Non-viral nanoparticles for RNA interference: Principles of design and practical guidelines. Adv. Drug Deliv. Rev. 2021, 174, 576–612. [Google Scholar] [CrossRef]
- Olden, B.R.; Cheng, Y.; Yu, J.L.; Pun, S.H. Cationic polymers for non-viral gene delivery to human T cells. J. Control. Release 2018, 282, 140–147. [Google Scholar] [CrossRef]
- Piotrowski-Daspit, A.S.; Kauffman, A.C.; Bracaglia, L.G.; Saltzman, W.M. Polymeric vehicles for nucleic acid delivery. Adv. Drug Deliv. Rev. 2020, 156, 119–132. [Google Scholar] [CrossRef]
- Singha, K.; Namgung, R.; Kim, W.J. Polymers in Small-Interfering RNA Delivery. Nucleic Acid Ther. 2011, 21, 133–147. [Google Scholar] [CrossRef]
- Lächelt, U.; Wagner, E. Nucleic Acid Therapeutics Using Polyplexes: A Journey of 50 Years (and Beyond). Chem. Rev. 2015, 115, 11043–11078. [Google Scholar] [CrossRef]
- Puhl, D.L.; D’amato, A.R.; Gilbert, R.J. Challenges of gene delivery to the central nervous system and the growing use of biomaterial vectors. Brain Res. Bull. 2019, 150, 216–230. [Google Scholar] [CrossRef] [PubMed]
- Zahir-Jouzdani, F.; Mottaghitalab, F.; Dinarvand, M.; Atyabi, F. siRNA delivery for treatment of degenerative diseases, new hopes and challenges. J. Drug Deliv. Sci. Technol. 2018, 45, 428–441. [Google Scholar] [CrossRef]
- Lai, W.-F.; Wong, W.-T. Design of Polymeric Gene Carriers for Effective Intracellular Delivery. Trends Biotechnol. 2018, 36, 713–728. [Google Scholar] [CrossRef]
- Velasco, D.; Elvira, C.; Román, J.S. New stimuli-responsive polymers derived from morpholine and pyrrolidine. J. Mater. Sci. Mater. Med. 2008, 19, 1453–1458. [Google Scholar] [CrossRef]
- Redondo, J.; Velasco, D.; Pérez-Perrino, M.; Reinecke, H.; Gallardo, A.; Pandit, A.; Elvira, C. Synergistic effect of pendant hydroxypropyl and pyrrolidine moieties randomly distributed along polymethacrylamide backbones on in vitro DNA-transfection. Eur. J. Pharm. Biopharm. 2015, 90, 38–43. [Google Scholar] [CrossRef]
- Velasco, D.; Collin, E.; Roman, J.S.; Pandit, A.; Elvira, C. End functionalized polymeric system derived from pyrrolidine provide high transfection efficiency. Eur. J. Pharm. Biopharm. 2011, 79, 485–494. [Google Scholar] [CrossRef] [PubMed]
- Velasco, D.; Réthoré, G.; Newland, B.; Parra, J.; Elvira, C.; Pandit, A.; Rojo, L.; Román, J.S. Low polydispersity (N-ethyl pyrrolidine methacrylamide-co-1-vinylimidazole) linear oligomers for gene therapy applications. Eur. J. Pharm. Biopharm. 2012, 82, 465–474. [Google Scholar] [CrossRef] [PubMed]
- Redondo, J.A.; Martínez-Campos, E.; Navarro, R.; Pérez-Perrino, M.; Reinecke, H.; Gallardo, A.; Corrales, G.; Fernández-Mayoralas, A.; Elvira, C. Hydroxyl versus permethylated glycopolymers as gene carriers. Eur. J. Pharm. Biopharm. 2017, 117, 68–76. [Google Scholar] [CrossRef]
- Maza, R.M.; Barreda-Manso, M.A.; Reigada, D.; Silván, Á.; Muñoz-Galdeano, T.; Soto, A.; Del Águila, Á.; Nieto-Díaz, M. MicroRNA-138-5p Targets Pro-Apoptotic Factors and Favors Neural Cell Survival: Analysis in the Injured Spinal Cord. Biomedicines 2022, 10, 1559. [Google Scholar] [CrossRef]
- Katakowski, M.; Buller, B.; Wang, X.; Rogers, T.; Chopp, M. Functional MicroRNA Is Transferred between Glioma Cells. Cancer Res. 2010, 70, 8259–8263. [Google Scholar] [CrossRef]
- Redondo, J.A.; Martínez-Campos, E.; Plet, L.; Pérez-Perrino, M.; Navarro, R.; Corrales, G.; Pandit, A.; Reinecke, H.; Gallardo, A.; López-Lacomba, J.L.; et al. Polymeric Gene Carriers Bearing Pendant β-Cyclodextrin: The Relevance of Glycoside Permethylation on the “In Vitro” Cell Response. Macromol. Rapid Commun. 2016, 37, 575–583. [Google Scholar] [CrossRef] [PubMed]
- Wincott, F.; DiRenzo, A.; Shaffer, C.; Grimm, S.; Tracz, D.; Workman, C.; Sweedler, D.; Gonzalez, C.; Scaringe, S.; Usman, N. Synthesis, deprotection, analysis and purification of RNA and ribosomes. Nucleic Acids Res. 1995, 23, 2677–2684. [Google Scholar] [CrossRef] [PubMed]
- Xie, Y.; Killinger, B.; Moszczynska, A.; Merkel, O.M. Targeted Delivery of siRNA to Transferrin Receptor Overexpressing Tumor Cells via Peptide Modified Polyethylenimine. Molecules 2016, 21, 1334. [Google Scholar] [CrossRef]
- Nakanishi, M.; Patil, R.; Ren, Y.; Shyam, R.; Wong, P.; Mao, H.-Q. Enhanced Stability and Knockdown Efficiency of Poly(ethylene glycol)-b-polyphosphoramidate/siRNA Micellar Nanoparticles by Co-condensation with Sodium Triphosphate. Pharm. Res. 2011, 28, 1723–1732. [Google Scholar] [CrossRef]
- Sage, D.; Donati, L.; Soulez, F.; Fortun, D.; Schmit, G.; Seitz, A.; Guiet, R.; Vonesch, C.; Unser, M. DeconvolutionLab2: An open-source software for deconvolution microscopy. Methods 2017, 115, 28–41. [Google Scholar] [CrossRef]
- Kongkatigumjorn, N.; Cortez-Jugo, C.; Czuba, E.; Wong, A.S.M.; Hodgetts, R.Y.; Johnston, A.P.R.; Such, G.K. Probing Endosomal Escape Using pHlexi Nanoparticles. Macromol. Biosci. 2017, 17, 1600248. [Google Scholar] [CrossRef]
- Pieczara, A.; Borek-Dorosz, A.; Buda, S.; Tipping, W.; Graham, D.; Pawlowski, R.; Mlynarski, J.; Baranska, M. Modified glucose as a sensor to track the metabolism of individual living endothelial cells—Observation of the 1602 cm−1 band called “Raman spectroscopic signature of life”. Biosens. Bioelectron. 2023, 230, 115234. [Google Scholar] [CrossRef]
- Liu, Z.; Ye, L.; Xi, J.; Wang, J.; Feng, Z.-G. Cyclodextrin polymers: Structure, synthesis, and use as drug carriers. Prog. Polym. Sci. 2021, 118, 101408. [Google Scholar] [CrossRef]
- Peng, L.; Wagner, E. Polymeric Carriers for Nucleic Acid Delivery: Current Designs and Future Directions. Biomacromolecules 2019, 20, 3613–3626. [Google Scholar] [CrossRef] [PubMed]
- Takeda, K.M.; Osada, K.; Tockary, T.A.; Dirisala, A.; Chen, Q.; Kataoka, K. Poly(ethylene glycol) Crowding as Critical Factor To Determine pDNA Packaging Scheme into Polyplex Micelles for Enhanced Gene Expression. Biomacromolecules 2017, 18, 36–43. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.; Zhou, S.; Ge, L.; Wu, W.; Jiang, X. Translatable High Drug Loading Drug Delivery Systems Based on Biocompatible Polymer Nanocarriers. Biomacromolecules 2018, 19, 1732–1745. [Google Scholar] [CrossRef]
- Breunig, M.; Lungwitz, U.; Liebl, R.; Goepferich, A. Breaking up the correlation between efficacy and toxicity for nonviral gene delivery. Proc. Natl. Acad. Sci. USA 2007, 104, 14454–14459. [Google Scholar] [CrossRef] [PubMed]
- Ma, D. Enhancing endosomal escape for nanoparticle mediated siRNA delivery. Nanoscale 2014, 6, 6415–6425. [Google Scholar] [CrossRef] [PubMed]
- Pei, M.; Xu, R.; Zhang, C.; Wang, X.; Li, C.; Hu, Y. Mannose-functionalized antigen nanoparticles for targeted dendritic cells, accelerated endosomal escape and enhanced MHC-I antigen presentation. Colloids Surf. B Biointerfaces 2021, 197, 111378. [Google Scholar] [CrossRef]
- Mahajan, S.; Tang, T. Polyethylenimine–DNA Nanoparticles under Endosomal Acidification and Implication to Gene Delivery. Langmuir 2022, 38, 8382–8397. [Google Scholar] [CrossRef]
- Varkouhi, A.K.; Scholte, M.; Storm, G.; Haisma, H.J. Endosomal escape pathways for delivery of biologicals. J. Control. Release 2011, 151, 220–228. [Google Scholar] [CrossRef]
- Chan, C.-L.; Majzoub, R.N.; Shirazi, R.S.; Ewert, K.K.; Chen, Y.-J.; Liang, K.S.; Safinya, C.R. Endosomal escape and transfection efficiency of PEGylated cationic liposome–DNA complexes prepared with an acid-labile PEG-lipid. Biomaterials 2012, 33, 4928–4935. [Google Scholar] [CrossRef]
- Moore, N.M.; Sheppard, C.L.; Sakiyama-Elbert, S.E. Characterization of a multifunctional PEG-based gene delivery system containing nuclear localization signals and endosomal escape peptides. Acta Biomater. 2009, 5, 854–864. [Google Scholar] [CrossRef]
- Martens, T.F.; Remaut, K.; Demeester, J.; De Smedt, S.C.; Braeckmans, K.; Martens, T.F.; Remaut, K.; Demeester, J.; De Smedt, S.C.; Braeckmans, K. Intracellular delivery of nanomaterials: How to catch endosomal escape in the act. Nano Today 2014, 9, 344–364. [Google Scholar] [CrossRef]
- Thomas, M.; Lange-Grünweller, K.; Dayyoub, E.; Bakowsky, U.; Weirauch, U.; Aigner, A.; Hartmann, R.K.; Grünweller, A. PEI-complexed LNA antiseeds as miRNA inhibitors. RNA Biol. 2012, 9, 1088–1098. [Google Scholar] [CrossRef] [PubMed]
- Hama, S.; Akita, H.; Ito, R.; Mizuguchi, H.; Hayakawa, T.; Harashima, H. Quantitative Comparison of Intracellular Trafficking and Nuclear Transcription between Adenoviral and Lipoplex Systems. Mol. Ther. 2006, 13, 786–794. [Google Scholar] [CrossRef] [PubMed]





Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Soto, A.; Nieto-Díaz, M.; Martínez-Campos, E.; Noalles-Dols, A.; Barreda-Manso, M.A.; Reviriego, F.; Reinecke, H.; Reigada, D.; Muñoz-Galdeano, T.; Novillo, I.; et al. Evaluation of Poly(N-Ethyl Pyrrolidine Methacrylamide) (EPA) and Derivatives as Polymeric Vehicles for miRNA Delivery to Neural Cells. Pharmaceutics 2023, 15, 1451. https://doi.org/10.3390/pharmaceutics15051451
Soto A, Nieto-Díaz M, Martínez-Campos E, Noalles-Dols A, Barreda-Manso MA, Reviriego F, Reinecke H, Reigada D, Muñoz-Galdeano T, Novillo I, et al. Evaluation of Poly(N-Ethyl Pyrrolidine Methacrylamide) (EPA) and Derivatives as Polymeric Vehicles for miRNA Delivery to Neural Cells. Pharmaceutics. 2023; 15(5):1451. https://doi.org/10.3390/pharmaceutics15051451
Chicago/Turabian StyleSoto, Altea, Manuel Nieto-Díaz, Enrique Martínez-Campos, Ana Noalles-Dols, María Asunción Barreda-Manso, Felipe Reviriego, Helmut Reinecke, David Reigada, Teresa Muñoz-Galdeano, Irene Novillo, and et al. 2023. "Evaluation of Poly(N-Ethyl Pyrrolidine Methacrylamide) (EPA) and Derivatives as Polymeric Vehicles for miRNA Delivery to Neural Cells" Pharmaceutics 15, no. 5: 1451. https://doi.org/10.3390/pharmaceutics15051451
APA StyleSoto, A., Nieto-Díaz, M., Martínez-Campos, E., Noalles-Dols, A., Barreda-Manso, M. A., Reviriego, F., Reinecke, H., Reigada, D., Muñoz-Galdeano, T., Novillo, I., Gallardo, A., Rodríguez-Hernández, J., Eritja, R., Aviñó, A., Elvira, C., & M. Maza, R. (2023). Evaluation of Poly(N-Ethyl Pyrrolidine Methacrylamide) (EPA) and Derivatives as Polymeric Vehicles for miRNA Delivery to Neural Cells. Pharmaceutics, 15(5), 1451. https://doi.org/10.3390/pharmaceutics15051451









