Screening Libraries to Discover Molecular Design Principles for the Targeted Delivery of mRNA with One-Component Ionizable Amphiphilic Janus Dendrimers Derived from Plant Phenolic Acids †
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Characterization and Methods
2.2.1. 1H and 13C NMR
2.2.2. Thin-Layer Chromatography (TLC)
2.2.3. High-Pressure Liquid Chromatography (HPLC)
2.2.4. Matrix-Assisted Laser Desorption Ionization–Time-of-Flight (MALDI-TOF) Mass Spectrometry
2.2.5. Dynamic Light Scattering (DLS)
2.2.6. pKa Measurements of Individual IAJDs
2.2.7. Production of Nucleoside-Modified Luc-mRNA
2.2.8. Formulation of DNPs Co-Assembled from IAJDs and Luc-mRNA
2.2.9. Luminescence Characterization for In Vivo Transfection Experiments
2.2.10. In Vivo mRNA Delivery in Mice with DNPs
2.2.11. Molecular Modelling
2.3. Synthesis of IAJDs
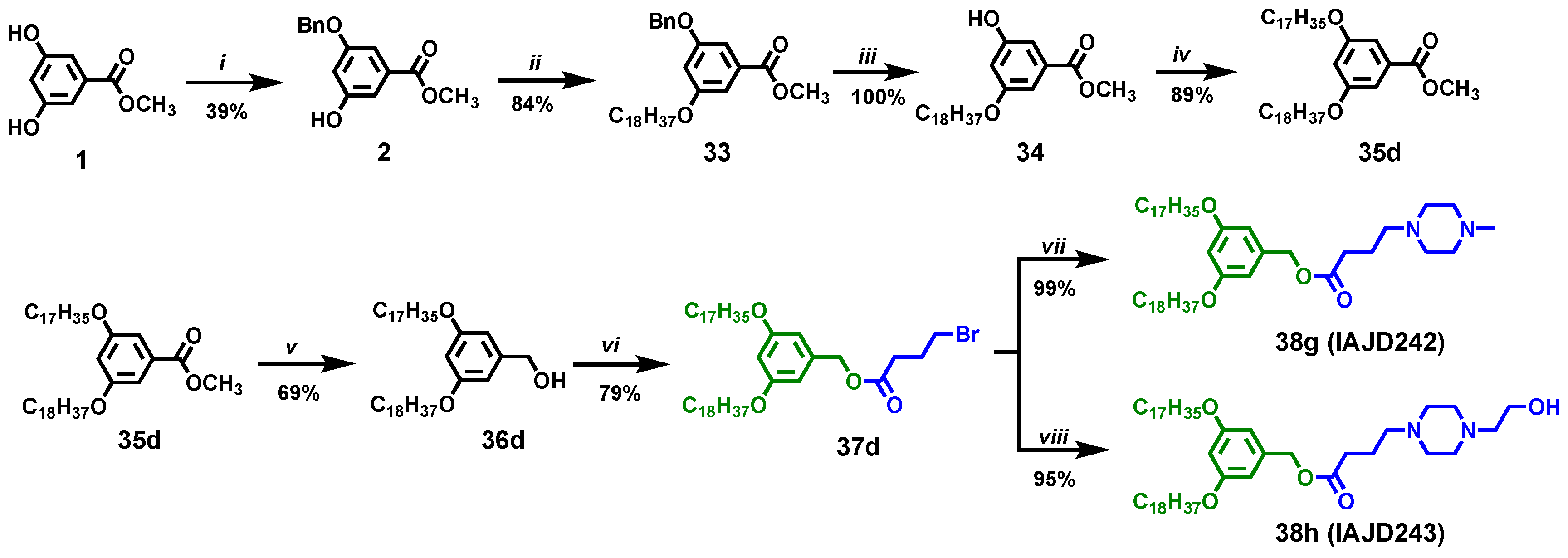
2.3.1. Synthesis of the Monoprotected Benzyl Ether of Methyl 3,5-Dihydroxybenzoate, 2
2.3.2. Synthesis of Methyl 3-(benzyloxy)-5-(octadecyloxy)benzoate, 33
2.3.3. Synthesis of the Methyl 3-hydroxy-5-(octadecyloxy)benzoate, 34
2.3.4. Synthesis of Methyl 3-(heptadecyloxy)-5-(octadecyloxy)benzoate (35d)
2.3.5. Synthesis of (3-(Heptadecyloxy)-5-(octadecyloxy)phenyl)methanol (36d)
2.3.6. Synthesis of 3-(Heptadecyloxy)-5-(octadecyloxy)benzyl 4-bromobutanoate, 37d
2.3.7. Synthesis of 3-(Heptadecyloxy)-5-(octadecyloxy)benzyl 4-(4-methylpiperazin-1-yl)butanoate, 38g, IAJD242
2.3.8. Synthesis of 3-(Heptadecyloxy)-5-(octadecyloxy)benzyl 4-(4-(2-hydroxyethyl)piperazin-1-yl)butanoate, 38h, IAJD243
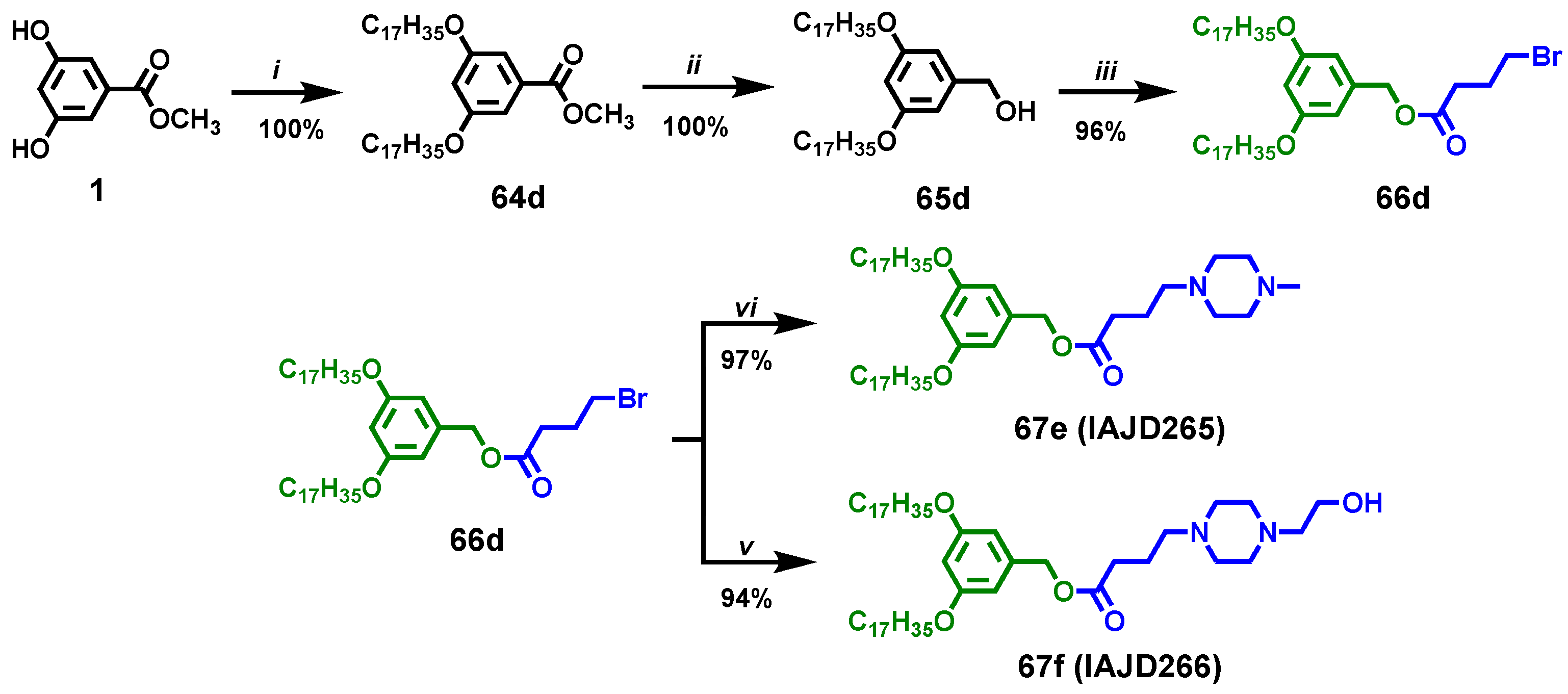
2.3.9. Synthesis of Methyl 3,5-bis(heptadecyloxy)benzoate), 64d
2.3.10. Synthesis of (3,5-Bis(heptadecyloxy)phenyl)methanol, 65d
2.3.11. Synthesis of 3,5-Bis(heptadecyloxy)benzyl 4-bromobutanoate, 66d
2.3.12. Synthesis of 3,5-Bis(pentadecyloxy)benzyl 4-(4-methylpiperazin-1-yl)butanoate. 67e, IAJD265
2.3.13. Synthesis of 3,5-Bis(pentadecyloxy)benzyl 4-(4-(2-hydroxyethyl)piperazin-1-yl)butanoate, 67f, IAJD266
3. Results and Discussion
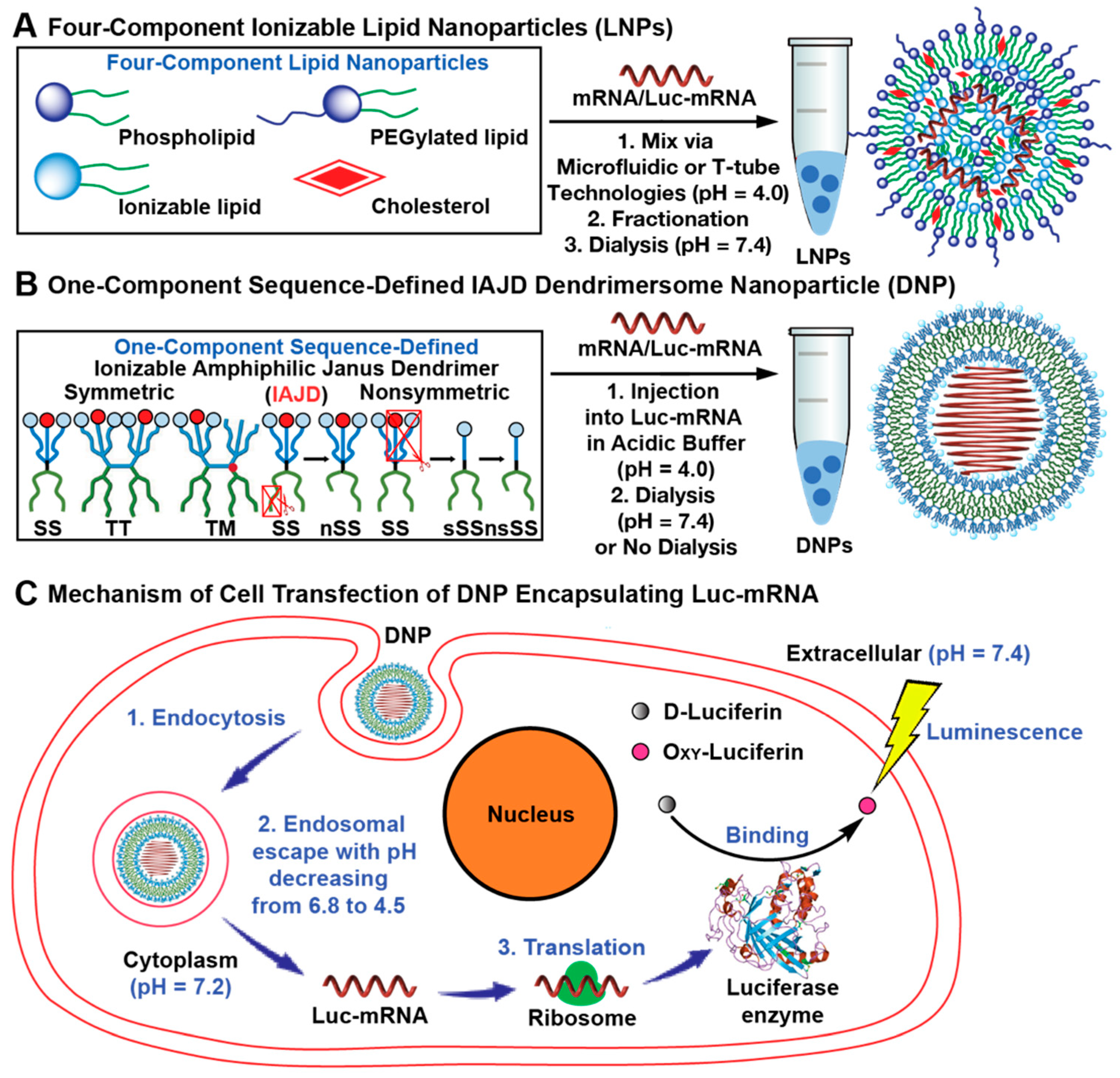
3.1. Concept, Strategy, and Methodology
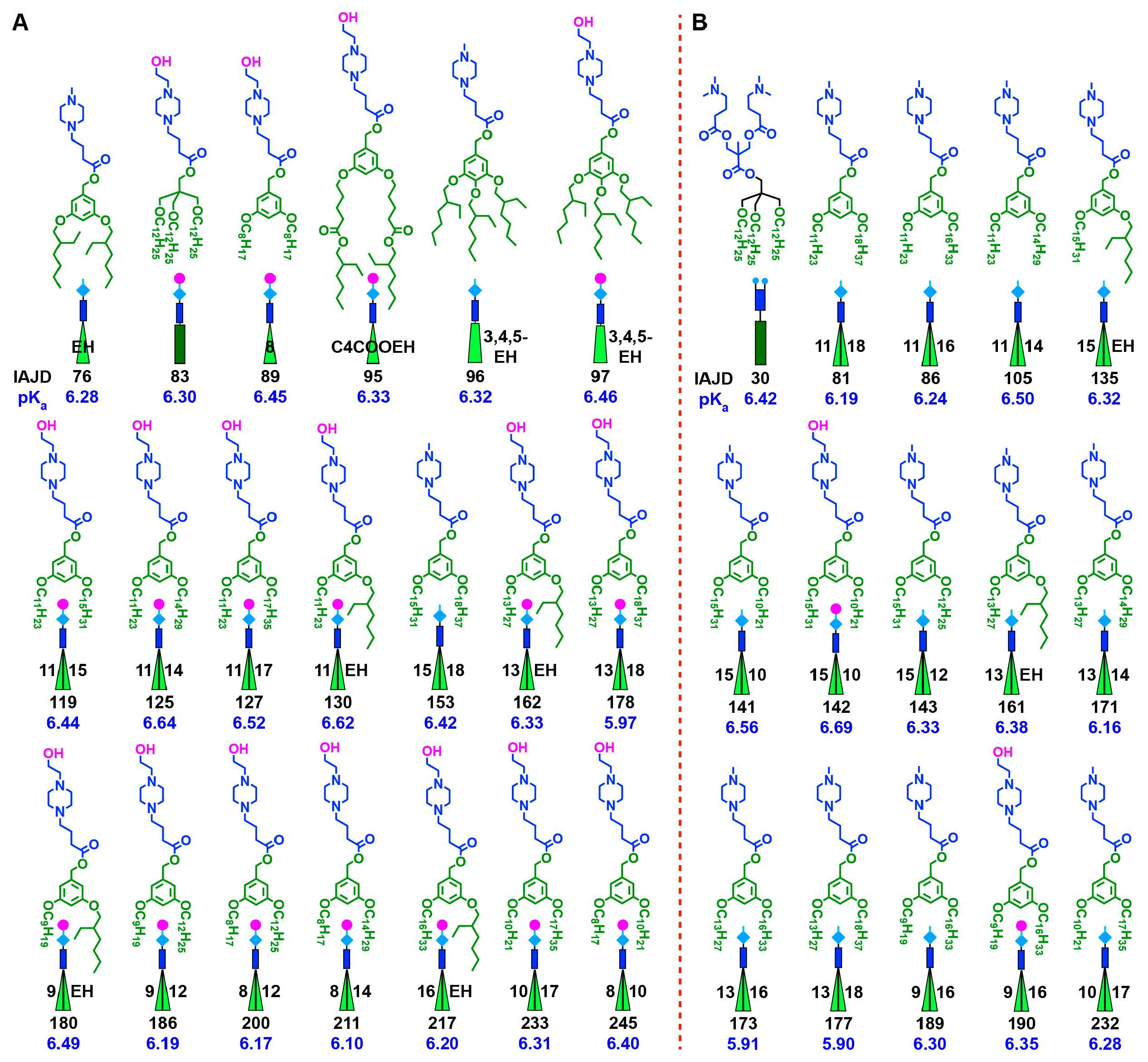
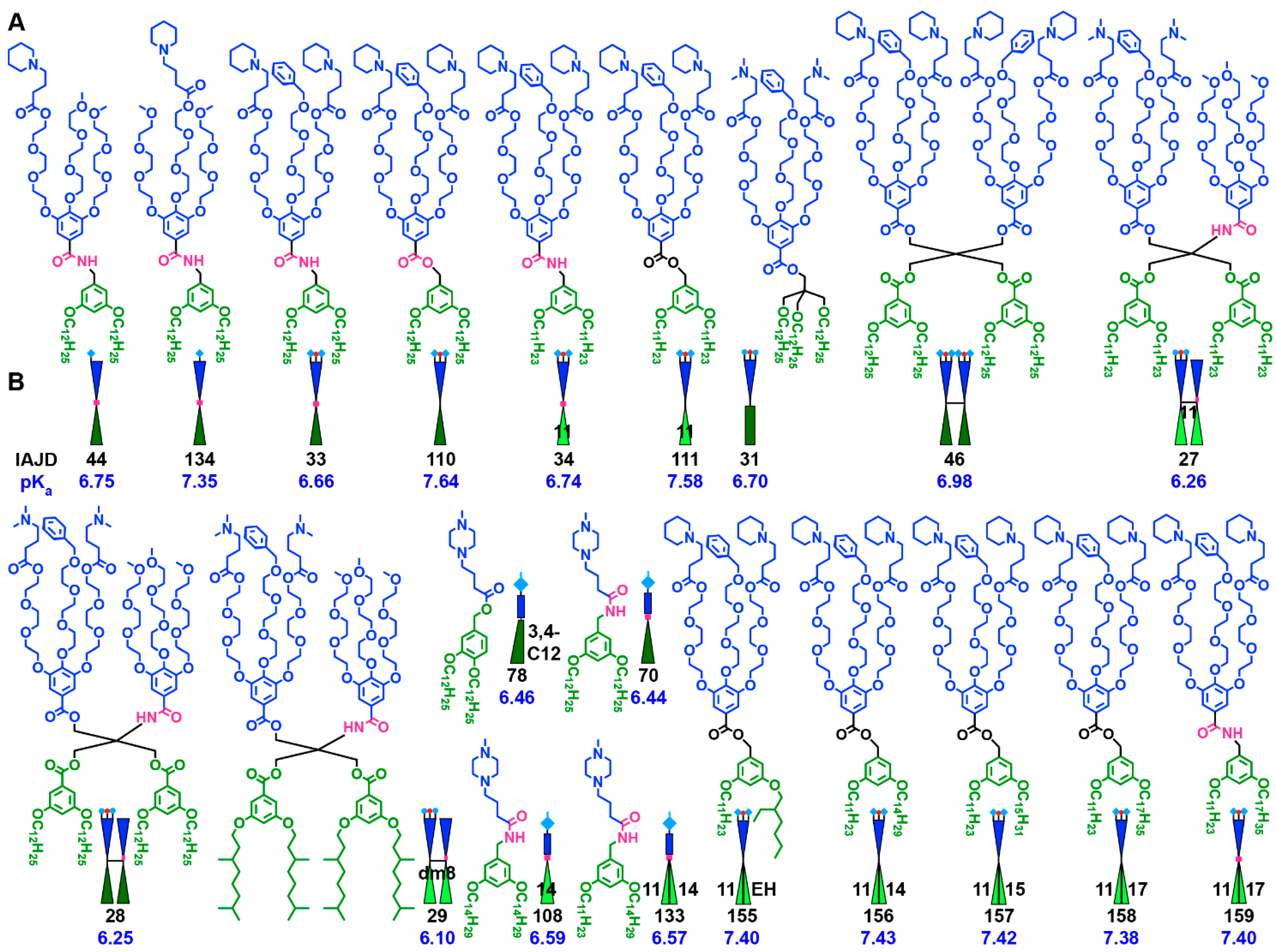
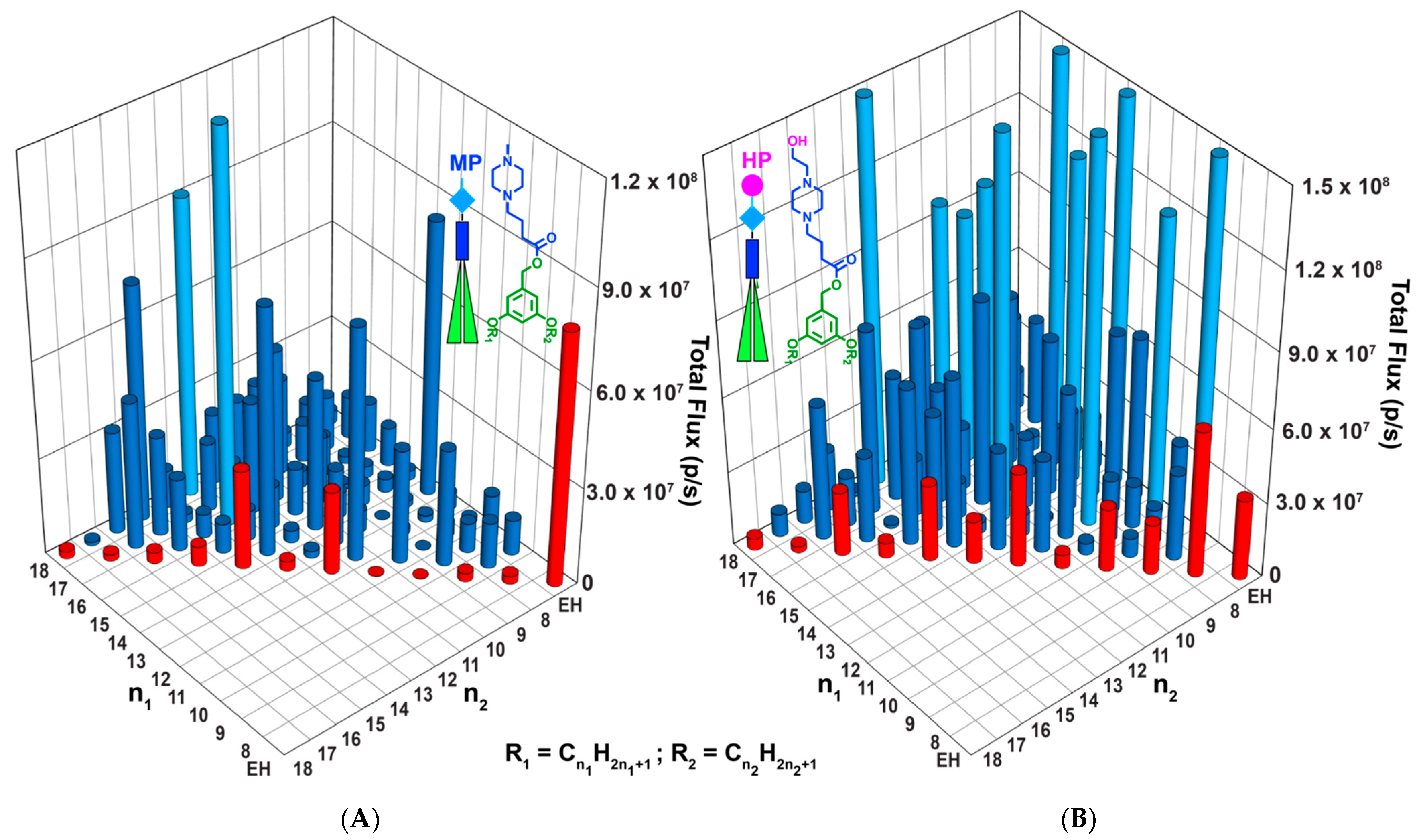
3.2. Accelerated Modular–Orthogonal Synthesis of Two Libraries of nsSS IAJDs
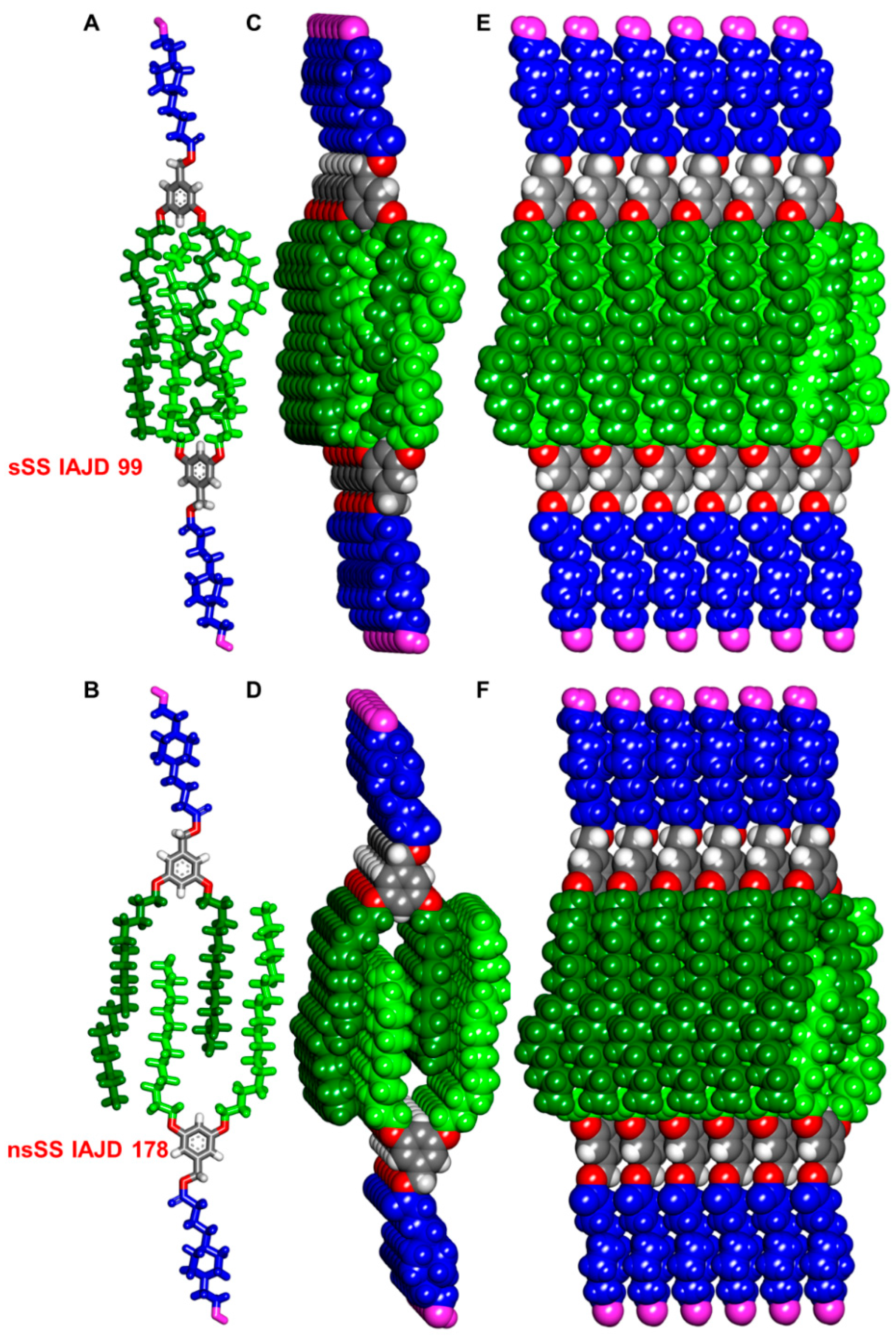
3.3. Accelerated Modular–Orthogonal Synthesis of a Complete Library of sSS IAJDs
3.4. Co-Assembly of IAJDs with Luc-mRNA into Dendrimersome Nanoparticles (DNPs) via Simple Injection
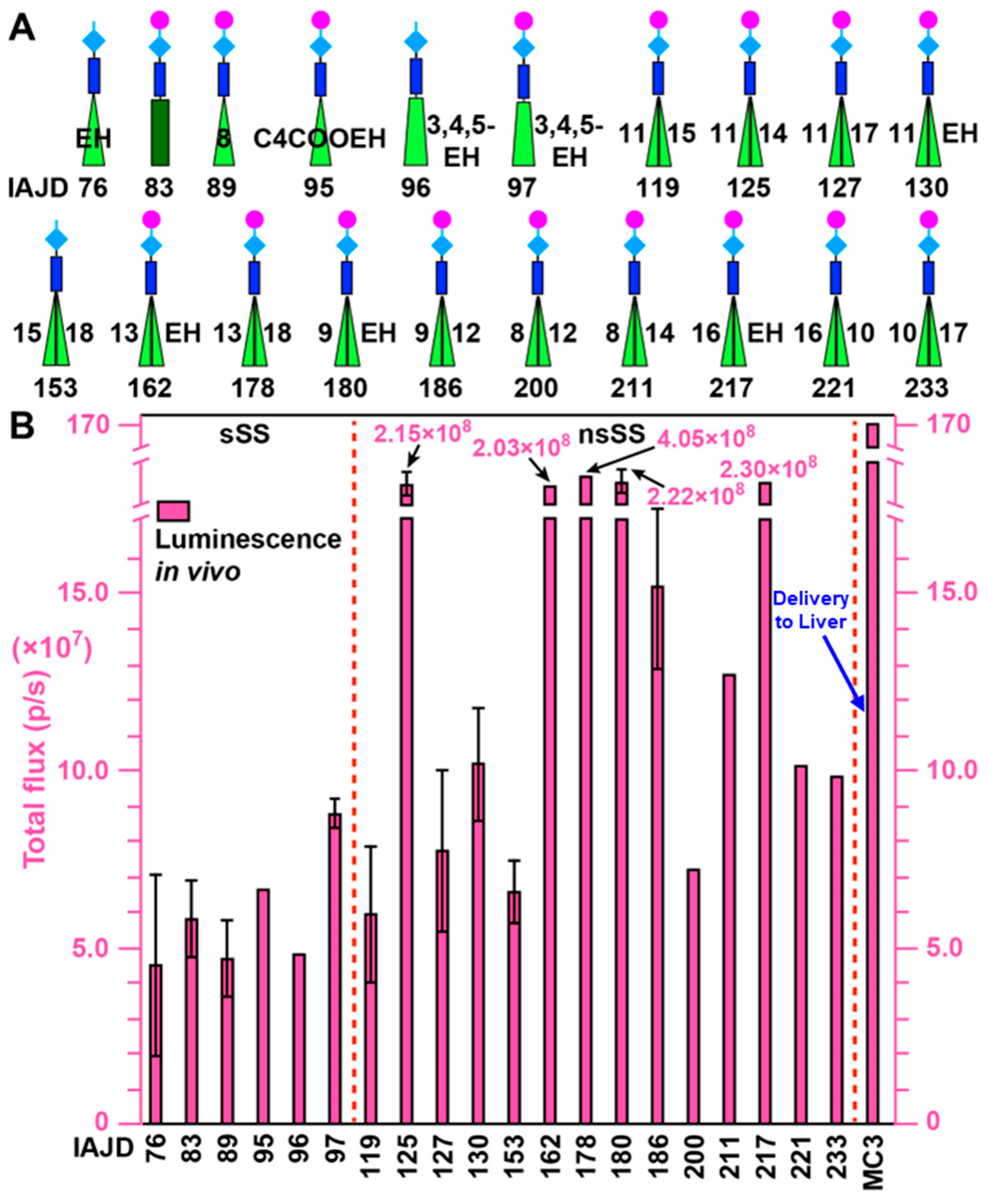
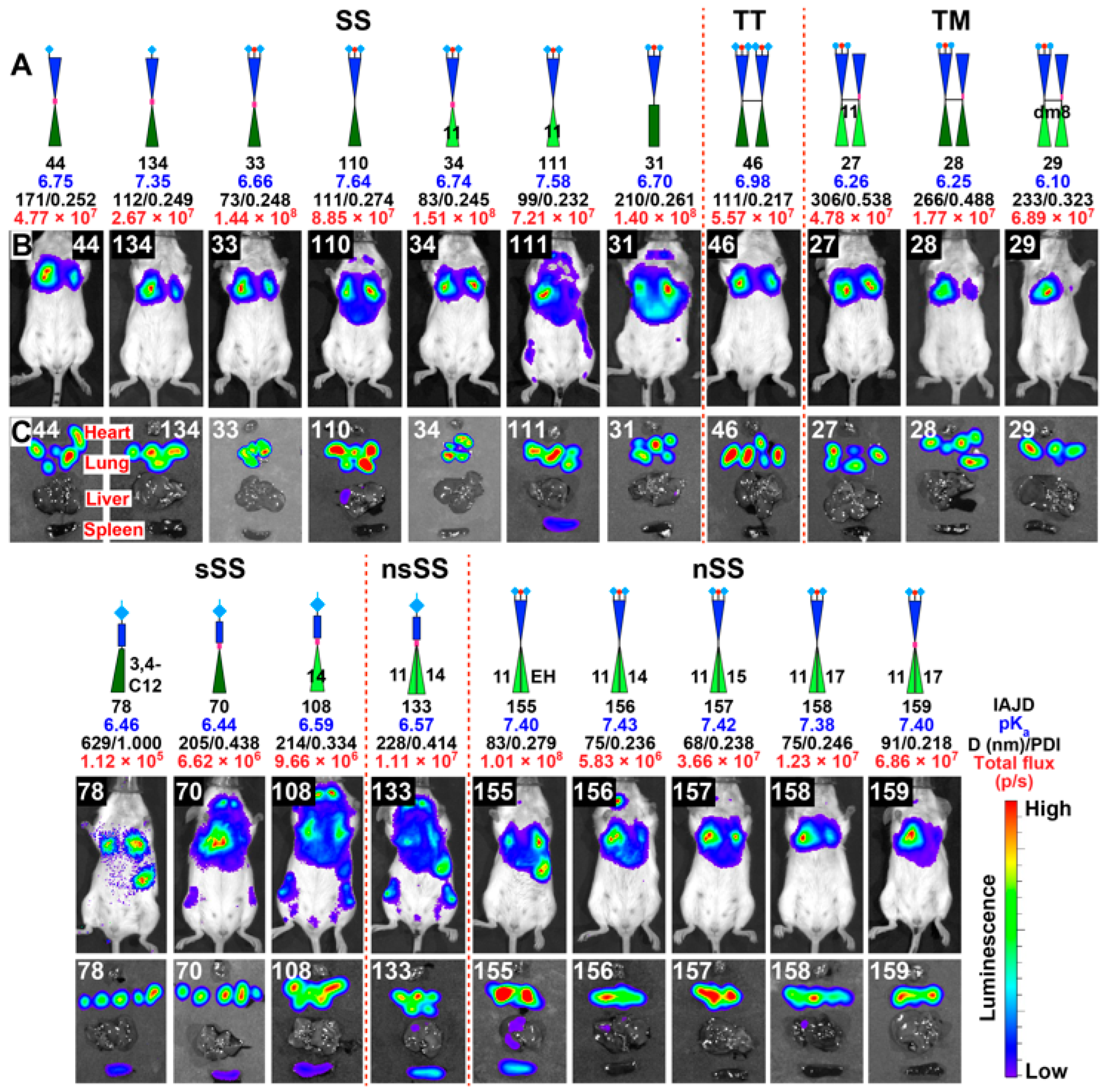
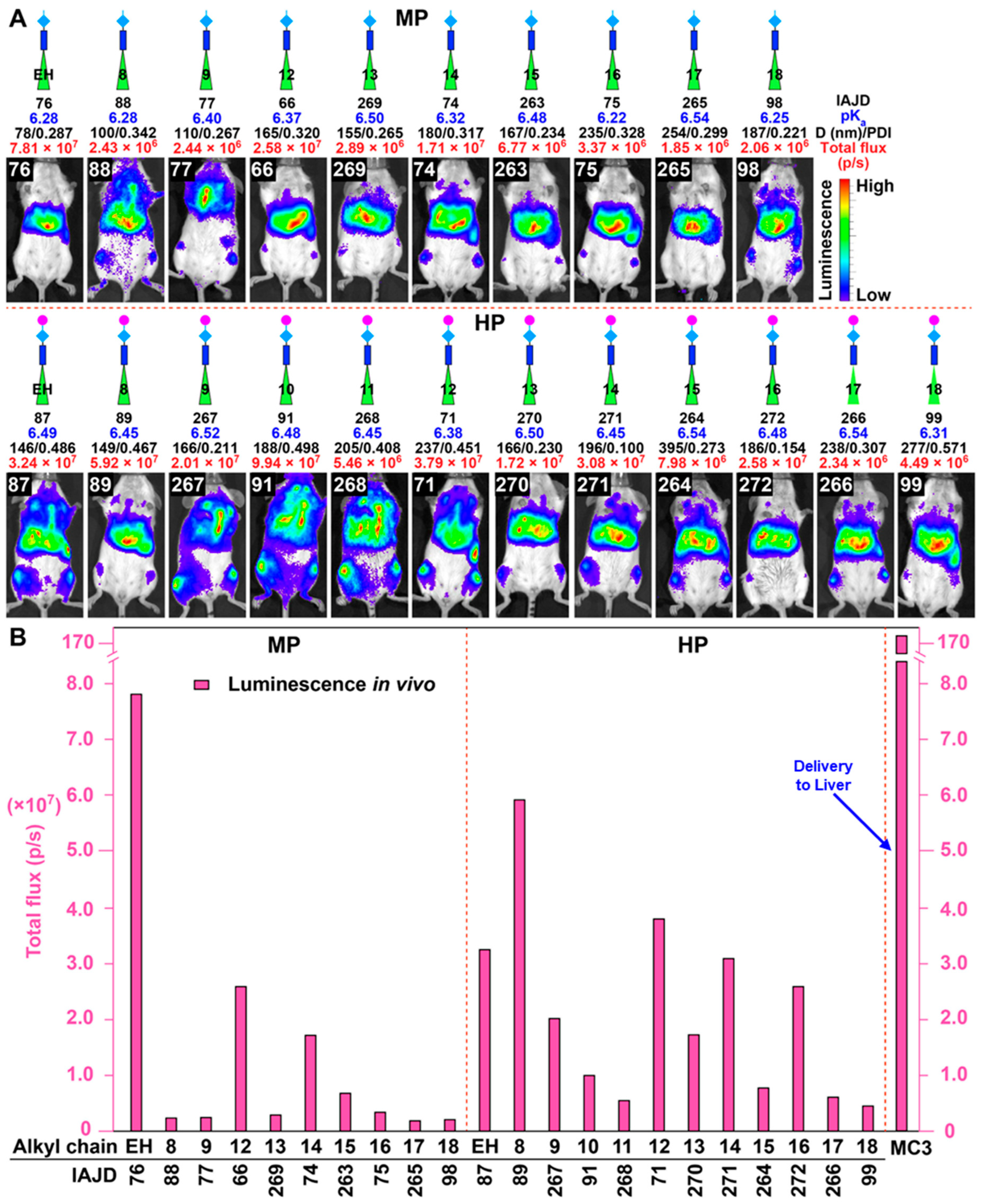
3.5. In Vivo Delivery of Mice with DNPs Containing Luc-mRNA
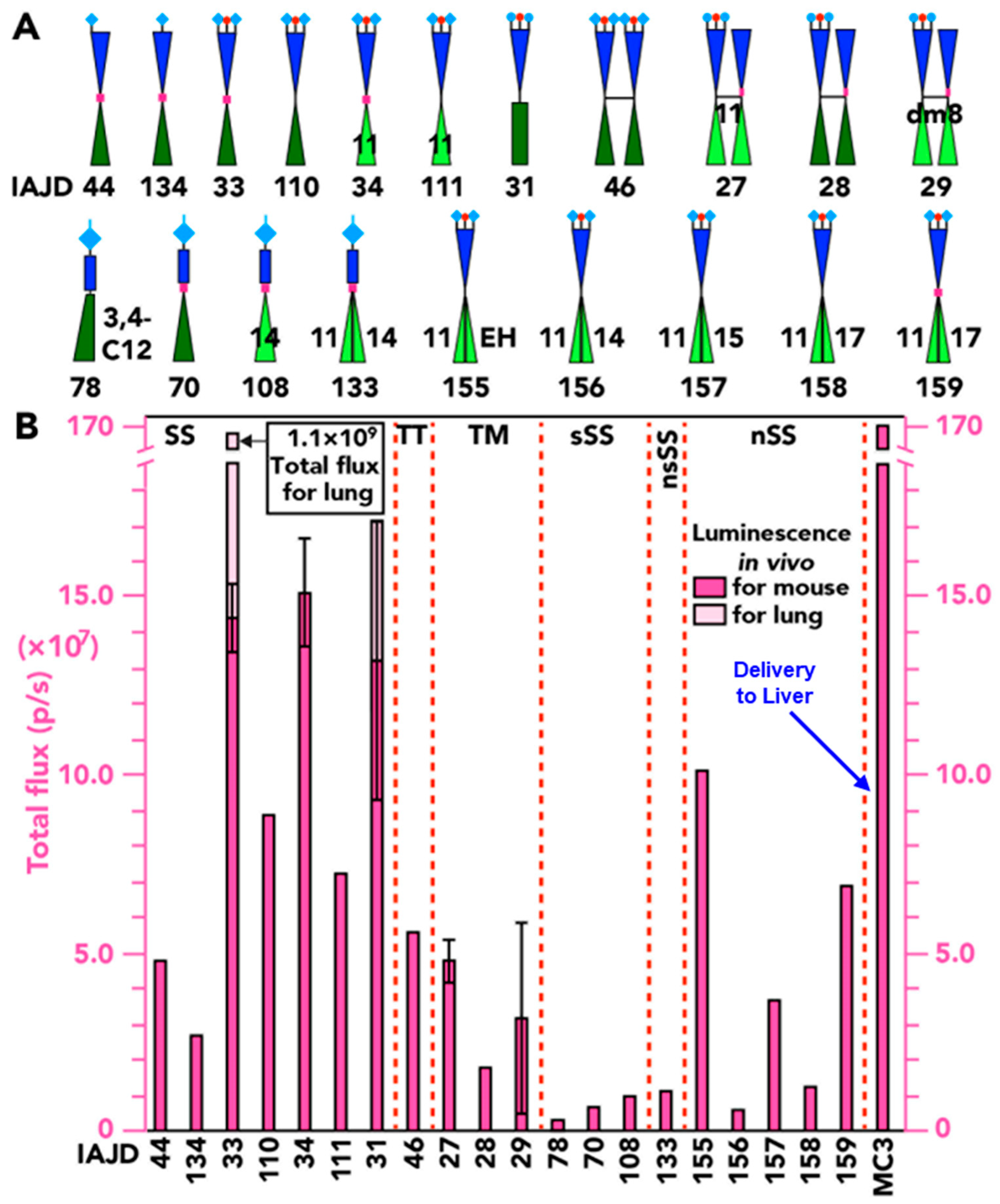
3.6. In Vivo Delivery of Luc-mRNA with nsSS DNPs to Spleen and Liver
3.7. In Vivo Delivery of Luc-mRNA with DNPs to Lung
3.8. In Vivo Delivery of Luc-mRNA with sSS DNPs
3.9. A Hypothetic Mechanism Explaining the Activity of mRNA Delivery via the Hydrophobic Part of IAJDs
3.10. The Current Status of the Molecular Design Principles to Target the Delivery of mRNA Mediated via IAJDs
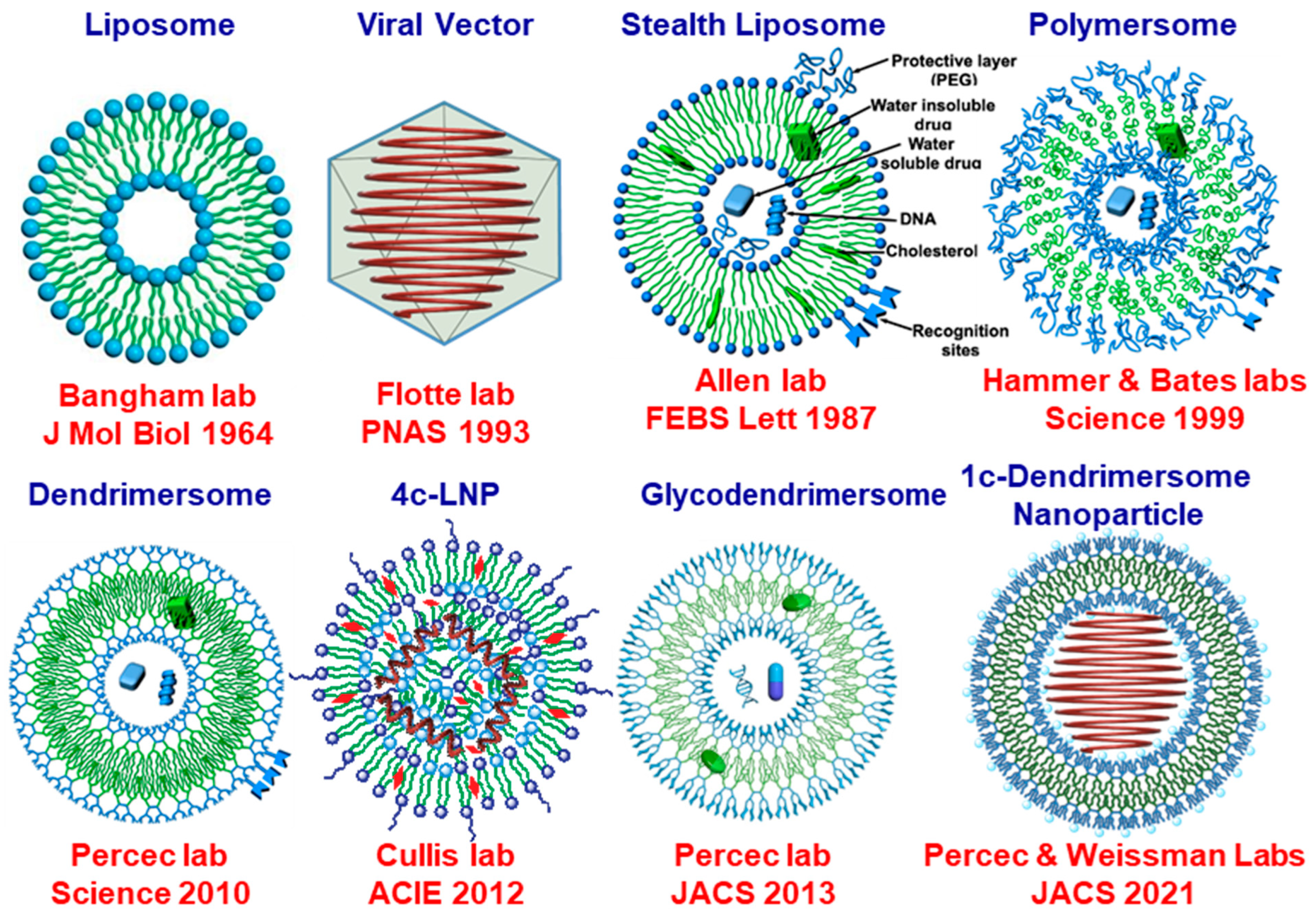
4. The Early Days of Nucleic Acid Delivery and Their Evolution to the Current Methodologies
5. Inspiration from and Collaboration with Donald A. Tomalia
6. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Waehler, R.; Russell, S.J.; Curiel, D.T. Engineering Targeted Viral Vectors for Gene Therapy. Nat. Rev. Genet. 2007, 8, 573–587. [Google Scholar] [CrossRef] [PubMed]
- Thomas, C.E.; Ehrhardt, A.; Kay, M.A. Progress and Problems with the Use of Viral Vectors for Gene Therapy. Nat. Rev. Genet. 2003, 4, 346–358. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.; Kong, N.; Zhang, X.; Cao, Y.; Langer, R.; Tao, W. The Landscape of mRNA Nanomedicine. Nat. Med. 2022, 28, 2273–2287. [Google Scholar] [CrossRef] [PubMed]
- Yin, H.; Kanasty, R.L.; Eltoukhy, A.A.; Vegas, A.J.; Dorkin, J.R.; Anderson, D.G. Non-Viral Vectors for Gene-Based Therapy. Nat. Rev. Genet. 2014, 15, 541–555. [Google Scholar] [CrossRef] [PubMed]
- Hajj, K.A.; Whitehead, K.A. Tools for Translation: Non-Viral Materials for Therapeutic mRNA Delivery. Nat. Rev. Mater. 2017, 2, 17056. [Google Scholar] [CrossRef]
- Zhang, Y.; Sun, C.; Wang, C.; Jankovic, K.E.; Dong, Y. Lipids and Lipid Derivatives for RNA Delivery. Chem. Rev. 2021, 121, 12181–12277. [Google Scholar] [CrossRef]
- Mintzer, M.A.; Simanek, E.E. Nonviral Vectors for Gene Delivery. Chem. Rev. 2009, 109, 259–302. [Google Scholar] [CrossRef]
- Mendes, L.P.; Pan, J.; Torchilin, V.P. Dendrimers as Nanocarriers for Nucleic Acid and Drug Delivery in Cancer Therapy. Molecules 2017, 22, 1401. [Google Scholar] [CrossRef]
- Benner, N.L.; McClellan, R.L.; Turlington, C.R.; Haabeth, O.A.W.; Waymouth, R.M.; Wender, P.A. Oligo (Serine Ester) Charge-Altering Releasable Transporters: Organocatalytic Ring-Opening Polymerization and Their Use for in Vitro and in Vivo mRNA Delivery. J. Am. Chem. Soc. 2019, 141, 8416–8421. [Google Scholar] [CrossRef]
- Haabeth, O.A.W.; Lohmeyer, J.J.K.; Sallets, A.; Blake, T.R.; Sagiv-Barfi, I.; Czerwinski, D.K.; McCarthy, B.; Powell, A.E.; Wender, P.A.; Waymouth, R.M.; et al. An mRNA SARS-CoV-2 Vaccine Employing Charge-Altering Releasable Transporters with a TLR-9 Agonist Induces Neutralizing Antibodies and T Cell Memory. ACS Cent. Sci. 2021, 7, 1191–1204. [Google Scholar] [CrossRef]
- Liu, S.; Wang, X.; Yu, X.; Cheng, Q.; Johnson, L.T.; Chatterjee, S.; Zhang, D.; Lee, S.M.; Sun, Y.; Lin, T.-C.; et al. Zwitterionic Phospholipidation of Cationic Polymers Facilitates Systemic mRNA Delivery to Spleen and Lymph Nodes. J. Am. Chem. Soc. 2021, 143, 21321–21330. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Q.; Wei, T.; Fabiak, L.; Johnson, L.T.; Dilliard, S.A.; Siegwart, D.J. Selective Organ Targeting (SORT) Nanoparticles for Tissue-Specific mRNA Delivery and CRISPR-Cas Gene Editing. Nat. Nanotechnol. 2020, 15, 313–320. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Zhou, Q.; Li, Y.; Garner, L.V.; Watkins, S.P.; Carter, L.J.; Smoot, J.; Gregg, A.C.; Daniels, A.D.; Jervey, S.; et al. Research and Development on Therapeutic Agents and Vaccines for COVID-19 and Related Human Coronavirus Diseases. ACS Cent. Sci. 2020, 6, 315–331. [Google Scholar] [CrossRef] [PubMed]
- Corbett, K.S.; Edwards, D.K.; Leist, S.R.; Abiona, O.M.; Boyoglu-Barnum, S.; Gillespie, R.A.; Himansu, S.; Schäfer, A.; Ziwawo, C.T.; DiPiazza, A.T.; et al. SARS-CoV-2 mRNA Vaccine Design Enabled by Prototype Pathogen Preparedness. Nature 2020, 586, 567–571. [Google Scholar] [CrossRef]
- Mulligan, M.J.; Lyke, K.E.; Kitchin, N.; Absalon, J.; Gurtman, A.; Lockhart, S.; Neuzil, K.; Raabe, V.; Bailey, R.; Swanson, K.A.; et al. Phase I/II Study of COVID-19 RNA Vaccine BNT162b1 in Adults. Nature 2020, 586, 589–593. [Google Scholar] [CrossRef]
- Ansell, S.M.; Du, X. Preparation of Novel Lipids and Lipid Nanoparticle Formulations for Delivery of Nucleic Acids. WO 2017/075531 A1, 4 May 2017. [Google Scholar]
- Sabnis, S.; Kumarasinghe, E.S.; Salerno, T.; Mihai, C.; Ketova, T.; Senn, J.J.; Lynn, A.; Bulychev, A.; McFadyen, I.; Chan, J.; et al. A Novel Amino Lipid Series for mRNA Delivery: Improved Endosomal Escape and Sustained Pharmacology and Safety in Non-human Primates. Mol. Ther. 2018, 26, 1509–1519. [Google Scholar] [CrossRef]
- Ambegia, E.; Ansell, S.; Cullis, P.; Heyes, J.; Palmer, L.; MacLachlan, I. Stabilized Plasmid–Lipid Particles Containing PEG-Diacylglycerols Exhibit Extended Circulation Lifetimes and Tumor Selective Gene Expression. Biochim. Biophys. Acta Biomembr. 2005, 1669, 155–163. [Google Scholar] [CrossRef]
- Chaudhary, N.; Weissman, D.; Whitehead, K.A. mRNA Vaccines for Infectious Diseases: Principles, Delivery and Clinical Translation. Nat. Rev. Drug Discov. 2021, 20, 817–838. [Google Scholar] [CrossRef]
- Kon, E.; Elia, U.; Peer, D. Principles for Designing an Optimal mRNA Lipid Nanoparticle Vaccine. Curr. Opin. Biotechnol. 2022, 73, 329–336. [Google Scholar] [CrossRef]
- Hou, X.; Zaks, T.; Langer, R.; Dong, Y. Lipid Nanoparticles for mRNA Delivery. Nat. Rev. Mater. 2021, 6, 1078–1094. [Google Scholar] [CrossRef]
- Han, X.; Zhang, H.; Butowska, K.; Swingle, K.L.; Alameh, M.-G.; Weissman, D.; Mitchell, M.J. An Ionizable Lipid Toolbox for RNA Delivery. Nat. Commun. 2021, 12, 7233. [Google Scholar] [CrossRef]
- Pardi, N.; Hogan, M.J.; Porter, F.W.; Weissman, D. mRNA Vaccines—A New Era in Vaccinology. Nat. Rev. Drug. Discov. 2018, 17, 261–279. [Google Scholar] [CrossRef]
- Zhang, D.; Atochina-Vasserman, E.N.; Maurya, D.S.; Huang, N.; Xiao, Q.; Ona, N.; Liu, M.; Shahnawaz, H.; Ni, H.; Kim, K.; et al. One-Component Multifunctional Sequence-Defined Ionizable Amphiphilic Janus Dendrimer Delivery Systems for mRNA. J. Am. Chem. Soc. 2021, 143, 12315–12327. [Google Scholar] [CrossRef]
- Zhang, D.; Atochina-Vasserman, E.N.; Maurya, D.S.; Liu, M.; Xiao, Q.; Lu, J.; Lauri, G.; Ona, N.; Reagan, E.K.; Ni, H.; et al. Targeted Delivery of mRNA with One-Component Ionizable Amphiphilic Janus Dendrimers. J. Am. Chem. Soc. 2021, 143, 17975–17982. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.; Atochina-Vasserman, E.N.; Lu, J.; Maurya, D.S.; Xiao, Q.; Liu, M.; Adamson, J.; Ona, N.; Reagan, E.K.; Ni, H.; et al. The Unexpected Importance of the Primary Structure of the Hydrophobic Part of One-Component Ionizable Amphiphilic Janus Dendrimers in Targeted mRNA Delivery Activity. J. Am. Chem. Soc. 2022, 144, 4746–4753. [Google Scholar] [CrossRef] [PubMed]
- Cullis, P.R.; Hope, M.J. Lipid Nanoparticle Systems for Enabling Gene Therapies. Mol. Ther. 2017, 25, 1467–1475. [Google Scholar] [CrossRef] [PubMed]
- Kulkarni, J.A.; Darjuan, M.M.; Mercer, J.E.; Chen, S.; van der Meel, R.; Thewalt, J.L.; Tam, Y.Y.C.; Cullis, P.R. On the Formation and Morphology of Lipid Nanoparticles Containing Ionizable Cationic Lipids and siRNA. ACS Nano 2018, 12, 4787–4795. [Google Scholar] [CrossRef]
- Jayaraman, M.; Ansell, S.M.; Mui, B.L.; Tam, Y.K.; Chen, J.; Du, X.; Butler, D.; Eltepu, L.; Matsuda, S.; Narayanannair, J.K.; et al. Maximizing the Potency of siRNA Lipid Nanoparticles for Hepatic Gene Silencing In Vivo. Angew. Chem. Int. Ed. 2012, 51, 8529–8533. [Google Scholar] [CrossRef]
- Ramezanpour, M.; Schmidt, M.L.; Bodnariuc, I.; Kulkarni, J.A.; Leung, S.S.W.; Cullis, P.R.; Thewalt, J.L.; Tieleman, D.P. Ionizable Amino Lipid Interactions with POPC: Implications for Lipid Nanoparticle Function. Nanoscale 2019, 11, 14141–14146. [Google Scholar] [CrossRef]
- Hatakeyama, H.; Akita, H.; Harashima, H. A Multifunctional Envelope Type Nano Device (MEND) for Gene Delivery to Tumours Based on the EPR Effect: A Strategy for Overcoming the PEG Dilemma. Adv. Drug Deliv. Rev. 2011, 63, 152–160. [Google Scholar] [CrossRef]
- Sato, Y.; Hatakeyama, H.; Sakurai, Y.; Hyodo, M.; Akita, H.; Harashima, H. A pH-Sensitive Cationic Lipid Facilitates the Delivery of Liposomal siRNA and Gene Silencing Activity in Vitro and in Vivo. J. Control. Release 2012, 163, 267–276. [Google Scholar] [CrossRef] [PubMed]
- Choi, J.S.; MacKay, J.A.; Szoka, F.C. Low-pH-Sensitive PEG-Stabilized Plasmid–Lipid Nanoparticles: Preparation and Characterization. Bioconjugate Chem. 2003, 14, 420–429. [Google Scholar] [CrossRef] [PubMed]
- Pelegri-O’Day, E.M.; Lin, E.-W.; Maynard, H.D. Therapeutic Protein–Polymer Conjugates: Advancing Beyond PEGylation. J. Am. Chem. Soc. 2014, 136, 14323–14332. [Google Scholar] [CrossRef] [PubMed]
- Moore, J.S.; Stupp, S.I. Room Temperature Polyesterification. Macromolecules 1990, 23, 65–70. [Google Scholar] [CrossRef]
- Pardi, N.; Muramatsu, H.; Weissman, D.; Kariko, K. In Vitro Transcription of Long RNA Containing Modified Nucleosides. Methods Mol. Biol. 2013, 969, 29–42. [Google Scholar] [CrossRef] [PubMed]
- Baiersdörfer, M.; Boros, G.; Muramatsu, H.; Mahiny, A.; Vlatkovic, I.; Sahin, U.; Karikó, K. A Facile Method for the Removal of DsRNA Contaminant from In Vitro-Transcribed MRNA. Mol. Ther. Nucleic Acids 2019, 15, 26–35. [Google Scholar] [CrossRef]
- May, J.P.; Undzys, E.; Roy, A.; Li, S.-D. Synthesis of a Gemcitabine Prodrug for Remote Loading into Liposomes and Improved Therapeutic Effect. Bioconjugate Chem. 2016, 27, 226–237. [Google Scholar] [CrossRef]
- Percec, V.; Wilson, D.A.; Leowanawat, P.; Wilson, C.J.; Hughes, A.D.; Kaucher, M.S.; Hammer, D.A.; Levine, D.H.; Kim, A.J.; Bates, F.S.; et al. Self-Assembly of Janus Dendrimers into Uniform Dendrimersomes and Other Complex Architectures. Science 2010, 328, 1009–1014. [Google Scholar] [CrossRef]
- Peterca, M.; Percec, V.; Leowanawat, P.; Bertin, A. Predicting the Size and Properties of Dendrimersomes from the Lamellar Structure of Their Amphiphilic Janus Dendrimers. J. Am. Chem. Soc. 2011, 133, 20507–20520. [Google Scholar] [CrossRef]
- Zhang, S.; Sun, H.-J.; Hughes, A.D.; Moussodia, R.-O.; Bertin, A.; Chen, Y.; Pochan, D.J.; Heiney, P.A.; Klein, M.L.; Percec, V. Self-Assembly of Amphiphilic Janus Dendrimers into Uniform Onion-Like Dendrimersomes with Predictable Size and Number of Bilayers. Proc. Natl. Acad. Sci. USA 2014, 111, 9058–9063. [Google Scholar] [CrossRef]
- Yadavalli, S.S.; Xiao, Q.; Sherman, S.E.; Hasley, W.D.; Klein, M.L.; Goulian, M.; Percec, V. Bioactive Cell-Like Hybrids from Dendrimersomes with a Human Cell Membrane and Its Components. Proc. Natl. Acad. Sci. USA 2019, 116, 744–752. [Google Scholar] [CrossRef]
- Xiao, Q.; Sherman, S.E.; Wilner, S.E.; Zhou, X.; Dazen, C.; Baumgart, T.; Reed, E.H.; Hammer, D.A.; Shinoda, W.; Klein, M.L.; et al. Janus Dendrimersomes Coassembled from Fluorinated, Hydrogenated, and Hybrid Janus Dendrimers as Models for Cell Fusion and Fission. Proc. Natl. Acad. Sci. USA 2017, 114, E7045–E7053. [Google Scholar] [CrossRef]
- Joseph, A.; Wagner, A.M.; Garay-Sarmiento, M.; Aleksanyan, M.; Haraszti, T.; Söder, D.; Georgiev, V.N.; Dimova, R.; Percec, V.; Rodriguez-Emmenegger, C. Zwitterionic Dendrimersomes: A Closer Xenobiotic Mimic of Cell Membranes. In Advanced Materials; Wiley Online Library: Hoboken, NJ, USA, 2022; p. 2206288. [Google Scholar] [CrossRef]
- Kostina, N.Y.; Rahimi, K.; Xiao, Q.; Haraszti, T.; Dedisch, S.; Spatz, J.P.; Schwaneberg, U.; Klein, M.L.; Percec, V.; Möller, M.; et al. Membrane-Mimetic Dendrimersomes Engulf Living Bacteria via Endocytosis. Nano Lett. 2019, 19, 5732–5738. [Google Scholar] [CrossRef]
- Kostina, N.Y.; Wagner, A.M.; Haraszti, T.; Rahimi, K.; Xiao, Q.; Klein, M.L.; Percec, V.; Rodriguez-Emmenegger, C. Unraveling Topology-Induced Shape Transformations in Dendrimersomes. Soft Matter 2021, 17, 254–267. [Google Scholar] [CrossRef]
- Buzzacchera, I.; Xiao, Q.; Han, H.; Rahimi, K.; Li, S.; Kostina, N.Y.; Toebes, B.J.; Wilner, S.E.; Möller, M.; Rodriguez-Emmenegger, C.; et al. Screening Libraries of Amphiphilic Janus Dendrimers Based on Natural Phenolic Acids to Discover Monodisperse Unilamellar Dendrimersomes. Biomacromolecules 2019, 20, 712–727. [Google Scholar] [CrossRef]
- Li, S.; Xia, B.; Javed, B.; Hasley, W.D.; Melendez-Davila, A.; Liu, M.; Kerzner, M.; Agarwal, S.; Xiao, Q.; Torre, P.; et al. Good. Direct Visualization of Vesicle Disassembly and Reassembly Using Photocleavable Dendrimers Elucidates Cargo Release Mechanisms. ACS Nano 2020, 14, 7398–7411. [Google Scholar] [CrossRef]
- Zhang, D.; Xiao, Q.; Rahimzadeh, M.; Liu, M.; Rodriguez-Emmenegger, C.; Miyazaki, Y.; Shinoda, W.; Percec, V. Self-Assembly of Glycerol-Amphiphilic Janus Dendrimers Amplifies and Indicates Principles for the Selection of Stereochemistry by Biological Membranes. J. Am. Chem. Soc. 2023, 145, 4311–4323. [Google Scholar] [CrossRef]
- Sun, H.-J.; Zhang, S.; Percec, V. From Structure to Function via Complex Supramolecular Dendrimer Systems. Chem. Soc. Rev. 2015, 44, 3900–3923. [Google Scholar] [CrossRef]
- Xiao, Q.; Rubien, J.; Wang, Z.; Reed, E.; Hammer, D.; Sahoo, D.; Heiney, P.; Yadavalli, S.; Goulian, M.; Wilner, S.; et al. Self-Sorting and Co-Assembly of Fluorinated, Hydrogenated, and Hybrid Janus Dendrimers into Dendrimersomes. J. Am. Chem. Soc. 2016, 138, 12655–12663. [Google Scholar] [CrossRef]
- Percec, V.; Leowanawat, P.; Sun, H.-J.; Kulikov, O.; Nusbaum, C.D.; Tran, T.M.; Bertin, A.; Wilson, D.A.; Peterca, M.; Zhang, S.; et al. Modular Synthesis of Amphiphilic Janus Glycodendrimers and Their Self-Assembly into Glycodendrimersomes and Other Complex Architectures with Bioactivity to Biomedically Relevant Lectins. J. Am. Chem. Soc. 2013, 135, 9055–9077. [Google Scholar] [CrossRef]
- Zhang, S.; Xiao, Q.; Sherman, S.E.; Muncan, A.; Ramos Vicente, A.D.M.; Wang, Z.; Hammer, D.A.; Williams, D.; Chen, Y.; Pochan, D.J.; et al. Glycodendrimersomes from Sequence-Defined Janus Glycodendrimers Reveal High Activity and Sensor Capacity for the Agglutination by Natural Variants of Human Lectins. J. Am. Chem. Soc. 2015, 137, 13334–13344. [Google Scholar] [CrossRef] [PubMed]
- Xiao, Q.; Zhang, S.; Wang, Z.; Sherman, S.E.; Moussodia, R.-O.; Peterca, M.; Muncan, A.; Williams, D.R.; Hammer, D.A.; Vértesy, S.; et al. Onion-like Glycodendrimersomes from Sequence-Defined Janus Glycodendrimers and Influence of Architecture on Reactivity to a Lectin. Proc. Natl. Acad. Sci. USA 2016, 113, 1162–1167. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez-Emmenegger, C.; Xiao, Q.; Kostina, N.Y.; Sherman, S.E.; Rahimi, K.; Partridge, B.E.; Li, S.; Sahoo, D.; Perez, A.M.R.; Buzzacchera, I.; et al. Encoding Biological Recognition in a Bicomponent Cell-Membrane Mimic. Proc. Natl. Acad. Sci. USA 2019, 116, 5376–5382. [Google Scholar] [CrossRef] [PubMed]
- Xiao, Q.; Delbianco, M.; Sherman, S.E.; Perez, A.M.R.; Bharate, P.; Pardo-Vargas, A.; Rodriguez-Emmenegger, C.; Kostina, N.Y.; Rahimi, K.; Söder, D.; et al. Nanovesicles Displaying Functional Linear and Branched Oligomannose Self-Assembled from Sequence-Defined Janus Glycodendrimers. Proc. Natl. Acad. Sci. USA 2020, 117, 11931–11939. [Google Scholar] [CrossRef]
- Kostina, N.Y.; Söder, D.; Haraszti, T.; Xiao, Q.; Rahimi, K.; Partridge, B.E.; Klein, M.L.; Percec, V.; Rodriguez-Emmenegger, C. Enhanced Concanavalin A Binding to Preorganized Mannose Nanoarrays in Glycodendrimersomes Revealed Multivalent Interactions. Angew. Chem. Int. Ed. 2021, 60, 8352–8360. [Google Scholar] [CrossRef]
- Kopitz, J.; Xiao, Q.; Ludwig, A.-K.; Romero, A.; Michalak, M.; Sherman, S.E.; Zhou, X.; Dazen, C.; Vértesy, S.; Kaltner, H.; et al. Reaction of a Programmable Glycan Presentation of Glycodendrimersomes and Cells with Engineered Human Lectins to Show the Sugar Functionality of the Cell Surface. Angew. Chem. 2017, 129, 14869–14873. [Google Scholar] [CrossRef]
- Kopitz, J.; Xiao, Q.; Ludwig, A.-K.; Romero, A.; Michalak, M.; Sherman, S.E.; Zhou, X.; Dazen, C.; Vértesy, S.; Kaltner, H.; et al. Reaction of a Programmable Glycan Presentation of Glycodendrimersomes and Cells with Engineered Human Lectins to Show the Sugar Functionality of the Cell Surface. Angew. Chem. Int. Ed. 2017, 56, 14677–14681. [Google Scholar] [CrossRef]
- Ludwig, A.-K.; Michalak, M.; Xiao, Q.; Gilles, U.; Medrano, F.J.; Ma, H.; FitzGerald, F.G.; Hasley, W.D.; Melendez-Davila, A.; Liu, M.; et al. Design–Functionality Relationships for Adhesion/Growth-Regulatory Galectins. Proc. Natl. Acad. Sci. USA 2019, 116, 2837–2842. [Google Scholar] [CrossRef]
- Murphy, P.V.; Romero, A.; Xiao, Q.; Ludwig, A.-K.; Jogula, S.; Shilova, N.V.; Singh, T.; Gabba, A.; Javed, B.; Zhang, D.; et al. Hans-Joachim Gabius Probing Sulfatide-Tissue Lectin Recognition with Functionalized Glycodendrimersomes. iScience 2021, 24, 101919. [Google Scholar] [CrossRef]
- Wilner, S.E.; Xiao, Q.; Graber, Z.T.; Sherman, S.E.; Percec, V.; Baumgart, T. Dendrimersomes Exhibit Lamellar-to-Sponge Phase Transitions. Langmuir 2018, 34, 5527–5534. [Google Scholar] [CrossRef]
- Torre, P.; Xiao, Q.; Buzzacchera, I.; Sherman, S.E.; Rahimi, K.; Kostina, N.Y.; Rodriguez-Emmenegger, C.; Möller, M.; Wilson, C.J.; Klein, M.L.; et al. Encapsulation of Hydrophobic Components in Dendrimersomes and Decoration of Their Surface with Proteins and Nucleic Acids. Proc. Natl. Acad. Sci. USA 2019, 116, 15378–15385. [Google Scholar] [CrossRef] [PubMed]
- Xiao, Q.; Ludwig, A.-K.; Romanò, C.; Buzzacchera, I.; Sherman, S.E.; Vetro, M.; Vértesy, S.; Kaltner, H.; Reed, E.H.; Möller, M.; et al. Exploring Functional Pairing between Surface Glycoconjugates and Human Galectins Using Programmable Glycodendrimersomes. Proc. Natl. Acad. Sci. USA 2018, 115, E2509–E2518. [Google Scholar] [CrossRef]
- Xiao, Q.; Wang, Z.; Williams, D.; Leowanawat, P.; Peterca, M.; Sherman, S.E.; Zhang, S.; Hammer, D.A.; Heiney, P.A.; King, S.R.; et al. Why Do Membranes of Some Unhealthy Cells Adopt a Cubic Architecture? ACS Cent. Sci. 2016, 2, 943–953. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Moussodia, R.-O.; Vértesy, S.; André, S.; Klein, M.L.; Gabius, H.-J.; Percec, V. Unraveling Functional Significance of Natural Variations of a Human Galectin by Glycodendrimersomes with Programmable Glycan Surface. Proc. Natl. Acad. Sci. USA 2015, 112, 5585–5590. [Google Scholar] [CrossRef]
- Zhang, S.; Moussodia, R.-O.; Murzeau, C.; Sun, H.-J.; Klein, M.L.; Vértesy, S.; André, S.; Roy, R.; Gabius, H.-J.; Percec, V. Dissecting Molecular Aspects of Cell Interactions Using Glycodendrimersomes with Programmable Glycan Presentation and Engineered Human Lectins. Angew. Chem. Int. Ed. 2015, 54, 4036–4040. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Moussodia, R.-O.; Sun, H.-J.; Leowanawat, P.; Muncan, A.; Nusbaum, C.D.; Chelling, K.M.; Heiney, P.A.; Klein, M.L.; André, S.; et al. Mimicking Biological Membranes with Programmable Glycan Ligands Self-Assembled from Amphiphilic Janus Glycodendrimers. Angew. Chem. 2014, 126, 11079–11083. [Google Scholar] [CrossRef]
- Zhang, S.; Moussodia, R.-O.; Sun, H.-J.; Leowanawat, P.; Muncan, A.; Nusbaum, C.D.; Chelling, K.M.; Heiney, P.A.; Klein, M.L.; André, S.; et al. Mimicking Biological Membranes with Programmable Glycan Ligands Self-Assembled from Amphiphilic Janus Glycodendrimers. Angew. Chem. Int. Ed. 2014, 53, 10899–10903. [Google Scholar] [CrossRef]
- Zhang, S.; Sun, H.-J.; Hughes, A.D.; Draghici, B.; Lejnieks, J.; Leowanawat, P.; Bertin, A.; De Leon, L.O.; Kulikov, O.V.; Chen, Y.; et al. “Single–Single” Amphiphilic Janus Dendrimers Self-Assemble into Uniform Dendrimersomes with Predictable Size. ACS Nano 2014, 8, 1554–1565. [Google Scholar] [CrossRef]
- Sherman, S.E.; Xiao, Q.; Percec, V. Mimicking Complex Biological Membranes and Their Programmable Glycan Ligands with Dendrimersomes and Glycodendrimersomes. Chem. Rev. 2017, 117, 6538–6631. [Google Scholar] [CrossRef]
- Xiao, Q.; Yadavalli, S.S.; Zhang, S.; Sherman, S.E.; Fiorin, E.; Silva, L.D.; Wilson, D.A.; Hammer, D.A.; André, S.; Gabius, H.-J.; et al. Bioactive Cell-Like Hybrids Coassembled from (Glyco)Dendrimersomes with Bacterial Membranes. Proc. Natl. Acad. Sci. USA 2016, 113, E1134–E1141. [Google Scholar] [CrossRef]
- Allen, T.M.; Chonn, A. Large Unilamellar Liposomes with Low Uptake into the Reticuloendothelial System. FEBS Lett. 1987, 223, 42–46. [Google Scholar] [CrossRef]
- Allen, T.M.; Cullis, P.R. Drug Delivery Systems: Entering the Mainstream. Science 2004, 303, 1818–1822. [Google Scholar] [CrossRef] [PubMed]
- Klibanov, A.L.; Maruyama, K.; Torchilin, V.P.; Huang, L. Amphipathic Polyethyleneglycols Effectively Prolong the Circulation Time of Liposomes. FEBS Lett. 1990, 268, 235–237. [Google Scholar] [CrossRef]
- Papahadjopoulos, D.; Allen, T.M.; Gabizon, A.; Mayhew, E.; Matthay, K.; Huang, S.K.; Lee, K.D.; Woodle, M.C.; Lasic, D.D.; Redemann, C. Sterically Stabilized Liposomes: Improvements in Pharmacokinetics and Antitumor Therapeutic Efficacy. Proc. Natl. Acad. Sci. USA 1991, 88, 11460–11464. [Google Scholar] [CrossRef] [PubMed]
- Olson, F.; Hunt, C.A.; Szoka, F.C.; Vail, W.J.; Papahadjopoulos, D. Preparation of Liposomes of Defined Size Distribution by Extrusion through Polycarbonate Membranes. Biochim. Biophys. Acta BBA Biomembr. 1979, 557, 9–23. [Google Scholar] [CrossRef]
- Roche, C.; Sun, H.-J.; Leowanawat, P.; Araoka, F.; Partridge, B.E.; Peterca, M.; Wilson, D.A.; Prendergast, M.E.; Heiney, P.A.; Graf, R.; et al. A Supramolecular Helix That Disregards Chirality. Nat. Chem. 2016, 8, 80–89. [Google Scholar] [CrossRef] [PubMed]
- Partridge, B.E.; Wang, L.; Sahoo, D.; Olsen, J.T.; Leowanawat, P.; Roche, C.; Ferreira, H.; Reilly, K.J.; Zeng, X.; Ungar, G.; et al. Sequence-Defined Dendrons Dictate Supramolecular Cogwheel Assembly of Dendronized Perylene Bisimides. J. Am. Chem. Soc. 2019, 141, 15761–15766. [Google Scholar] [CrossRef]
- Wang, L.; Partridge, B.E.; Huang, N.; Olsen, J.T.; Sahoo, D.; Zeng, X.; Ungar, G.; Graf, R.; Spiess, H.W.; Percec, V. Extraordinary Acceleration of Cogwheel Helical Self-Organization of Dendronized Perylene Bisimides by the Dendron Sequence Encoding Their Tertiary Structure. J. Am. Chem. Soc. 2020, 142, 9525–9536. [Google Scholar] [CrossRef] [PubMed]
- Darnell, J.; Lodish, H.; Baltimore, D. Molecular Cell Biology; Scientific American Books: New York, NY, USA, 1986; pp. 567–615. [Google Scholar]
- Garrido, J.; Garrido, E.M.; Borges, F. Studies on the Food Additive Propyl Gallate: Synthesis, Structural Characterization, and Evaluation of the Antioxidant Activity. J. Chem. Educ. 2012, 89, 130–133. [Google Scholar] [CrossRef]
- Percec, V.; Wang, S.; Huang, N.; Partridge, B.E.; Wang, X.; Sahoo, D.; Hoffman, D.J.; Malineni, J.; Peterca, M.; Jezorek, R.L.; et al. An Accelerated Modular-Orthogonal Ni-Catalyzed Methodology to Symmetric and Nonsymmetric Constitutional Isomeric AB2 to AB9 Dendrons Exhibiting Unprecedented Self-Organizing Principles. J. Am. Chem. Soc. 2021, 143, 17724–17743. [Google Scholar] [CrossRef] [PubMed]
- Helfrich, W. Elastic Properties of Lipid Bilayers: Theory and Possible Experiments. Z. Nat. C 1973, 28, 693–703. [Google Scholar] [CrossRef] [PubMed]
- Tanford, C. The Hydrophobic Effect and the Organization of Living Matter. Science 1978, 200, 1012–1018. [Google Scholar] [CrossRef] [PubMed]
- Szoka, F.; Papahadjopoulos, D. Comparative Properties and Methods of Preparation of Lipid Vesicles (Liposomes). Annu. Rev. Biophys. Bioeng. 1980, 9, 467–508. [Google Scholar] [CrossRef]
- Gabizon, A.; Papahadjopoulos, D. Liposome Formulations with Prolonged Circulation Time in Blood and Enhanced Uptake by Tumors. Proc. Natl. Acad. Sci. USA 1988, 85, 6949–6953. [Google Scholar] [CrossRef] [PubMed]
- Hamann, M.V.; Stanke, N.; Müllers, E.; Stirnnagel, K.; Hütter, S.; Artegiani, B.; Alonso, S.B.; Calegari, F.; Lindemann, D. Efficient Transient Genetic Manipulation In Vitro and In Vivo by Prototype Foamy Virus-Mediated Nonviral RNA Transfer. Mol. Ther. 2014, 22, 1460–1471. [Google Scholar] [CrossRef]
- Kay, M.A.; Holterman, A.-X.; Meuse, L.; Gown, A.; Ochs, H.D.; Linsley, P.S.; Wilson, C.B. Long–Term Hepatic Adenovirus–Mediated Gene Expression in Mice Following CTLA4Ig Administration. Nat. Genet. 1995, 11, 191–197. [Google Scholar] [CrossRef] [PubMed]
- Flotte, T.R.; Afione, S.A.; Conrad, C.; McGrath, S.A.; Solow, R.; Oka, H.; Zeitlin, P.L.; Guggino, W.B.; Carter, B.J. Stable in Vivo Expression of the Cystic Fibrosis Transmembrane Conductance Regulator with an Adeno-Associated Virus Vector. Proc. Natl. Acad. Sci. USA 1993, 90, 10613–10617. [Google Scholar] [CrossRef]
- Bukrinsky, M.I. HIV-1 Nuclear Import: In Search of a Leader. Front. Biosci. 1999, 4, 772–781. [Google Scholar] [CrossRef]
- Zhao, Z.; Anselmo, A.C.; Mitragotri, S. Viral vector-based Gene Therapies in the Clinic. Bioeng. Transl. Med. 2022, 7, E10258. [Google Scholar] [CrossRef]
- Lundstrom, K. Viral Vectors in Gene Therapy: Where Do We Stand in 2023? Viruses 2023, 15, 698. [Google Scholar] [CrossRef]
- Gao, G.-P.; Alvira, M.R.; Wang, L.; Calcedo, R.; Johnston, J.; Wilson, J.M. Novel Adeno-Associated Viruses from Rhesus Monkeys as Vectors for Human Gene Therapy. Proc. Natl. Acad. Sci. USA 2002, 99, 11854–11859. [Google Scholar] [CrossRef] [PubMed]
- Felgner, P.L.; Gadek, T.R.; Holm, M.; Roman, R.; Chan, H.W.; Wenz, M.; Northrop, J.P.; Ringold, G.M.; Danielsen, M. Lipofection: A Highly Efficient, Lipid-Mediated DNA-Transfection Procedure. Proc. Natl. Acad. Sci. USA 1987, 84, 7413–7417. [Google Scholar] [CrossRef] [PubMed]
- Felgner, J.; Martin, M.; Tsai, Y.; Felgner, P.L. Cationic Lipid-Mediated Transfection in Mammalian Cells: “Lipofection”. J. Tissue Cult. Methods 1993, 15, 63–68. [Google Scholar] [CrossRef]
- Semple, S.C.; Akinc, A.; Chen, J.; Sandhu, A.P.; Mui, B.L.; Cho, C.K.; Sah, D.W.Y.; Stebbing, D.; Crosley, E.J.; Yaworski, E.; et al. Rational Design of Cationic Lipids for SiRNA Delivery. Nat. Biotechnol. 2010, 28, 172–176. [Google Scholar] [CrossRef]
- Haensler, J.; Szoka, F.C. Polyamidoamine Cascade Polymers Mediate Efficient Transfection of Cells in Culture. Bioconjugate Chem. 1993, 4, 372–379. [Google Scholar] [CrossRef]
- Kukowska-Latallo, J.F.; Bielinska, A.U.; Johnson, J.; Spindler, R.; Tomalia, D.A.; Baker, J.R. Efficient Transfer of Genetic Material into Mammalian Cells Using Starburst Polyamidoamine Dendrimers. Proc. Natl. Acad. Sci. USA 1996, 93, 4897–4902. [Google Scholar] [CrossRef]
- Zhou, J.; Wu, J.; Hafdi, N.; Behr, J.-P.; Erbacher, P.; Peng, L. PAMAM Dendrimers for Efficient SiRNA Delivery and Potent Gene Silencing. Chem. Commun. 2006, 22, 2362–2364. [Google Scholar] [CrossRef]
- Loup, C.; Zanta, M.-A.; Caminade, A.-M.; Majoral, J.-P.; Meunier, B. Preparation of Water-Soluble Cationic Phosphorus-Containing Dendrimers as DNA Transfecting Agents. Chem. Eur. J. 1999, 5, 3644–3650. [Google Scholar] [CrossRef]
- Pedziwiatr, E.; Chonco, L.; Shcharbin, D.; de Jesus, E.; Bermejo, J.F.; Ortega, P.; de la Mata, F.J.; Flores, J.C.; Gomez, R.; Klajnert, B.; et al. Carbosilane Dendrimers as Drug Carriers. Eur. J. Clin. Investig. 2007, 37, 79. [Google Scholar] [CrossRef]
- Zinselmeyer, B.H.; Mackay, S.P.; Schatzlein, A.G.; Uchegbu, I.F. The Lower-Generation Polypropylenimine Dendrimers Are Effective Gene-Transfer Agents. Pharm. Res. 2002, 19, 960–967. [Google Scholar] [CrossRef]
- Sarbolouki, M.N.; Sadeghizadeh, M.; Yaghoobi, M.M.; Karami, A.; Lohrasbi, T. Dendrosomes: A Novel Family of Vehicles for Transfection and Therapy. J. Chem. Technol. Biotechnol. 2000, 75, 919–922. [Google Scholar] [CrossRef]
- Dutta, T.; Burgess, M.; McMillan, N.A.J.; Parekh, H.S. Dendrosome-Based Delivery of SiRNA against E6 and E7 Oncogenes in Cervical Cancer. Nanomed. Nanotechnol. Biol. Med. 2010, 6, 463–470. [Google Scholar] [CrossRef]
- Joester, D.; Losson, M.; Pugin, R.; Heinzelmann, H.; Walter, E.; Merkle, H.P.; Diederich, F. Amphiphilic Dendrimers: Novel Self-Assembling Vectors for Efficient Gene Delivery. Angew. Chem. Int. Ed. 2003, 42, 1486–1490. [Google Scholar] [CrossRef] [PubMed]
- Wood, K.C.; Little, S.R.; Langer, R.; Hammond, P.T. A Family of Hierarchically Self-Assembling Linear-Dendritic Hybrid Polymers for Highly Efficient Targeted Gene Delivery. Angew. Chem. Int. Ed. 2005, 44, 6704–6708. [Google Scholar] [CrossRef] [PubMed]
- Wu, G.Y.; Wu, C.H. Receptor-Mediated in Vitro Gene Transformation by a Soluble DNA Carrier System. J. Biol. Chem. 1987, 262, 4429–4432. [Google Scholar] [CrossRef] [PubMed]
- Boussif, O.; Lezoualc’h, F.; Zanta, M.A.; Mergny, M.D.; Scherman, D.; Demeneix, B.; Behr, J.P. A Versatile Vector for Gene and Oligonucleotide Transfer into Cells in Culture and in Vivo: Polyethylenimine. Proc. Natl. Acad. Sci. USA 1995, 92, 7297–7301. [Google Scholar] [CrossRef]
- McKinlay, C.J.; Vargas, J.R.; Blake, T.R.; Hardy, J.W.; Kanada, M.; Contag, C.H.; Wender, P.A.; Waymouth, R.M. Charge-Altering Releasable Transporters (CARTs) for the Delivery and Release of MRNA in Living Animals. Proc. Natl. Acad. Sci. USA 2017, 114, E448–E456. [Google Scholar] [CrossRef]
- Benner, N.L.; Near, K.E.; Bachmann, M.H.; Contag, C.H.; Waymouth, R.M.; Wender, P.A. Functional DNA Delivery Enabled by Lipid-Modified Charge-Altering Releasable Transporters (CARTs). Biomacromolecules 2018, 19, 2812–2824. [Google Scholar] [CrossRef]
- Bangham, A.D.; Horne, R.W. Negative Staining of Phospholipids and Their Structural Modification by Surface-Active Agents as Observed in the Electron Microscope. J. Mol. Biol. 1964, 8, 660–668. [Google Scholar] [CrossRef]
- Buhleier, E.; Wehner, W.; Voegtle, F. “Cascade”- and “Nonskid-Chain-Like” Syntheses of Molecular Cavity Topologies. Synthesis 1978, 155, 155–158. [Google Scholar] [CrossRef]
- Denkewalter, R.G.; Kole, J.; Lukasavage, W.J. Macromolecular Highly Branched Homogeneous Compound Based on Lysine Units. U.S. Patent 4 289 872, 15 September 1981. [Google Scholar]
- Tomalia, D.A.; Baker, H.; Dewald, J.; Hall, M.; Kallos, G.; Martin, S.; Roeck, J.; Ryder, J.; Smith, P. A New Class of Polymers: Starburst-Dendritic Macromolecules. Polym. J. 1985, 17, 117–132. [Google Scholar] [CrossRef]
- Newkome, G.R.; Yao, Z.; Baker, G.R.; Gupta, V.K. Cascade Molecules: A New Approach to Micelles. A [27]-Arborol. J. Org. Chem. 1985, 50, 2003–2004. [Google Scholar] [CrossRef]
- Percec, V.; Heck, J. Side Chain Liquid Crystal Polymers Containing Mesogenic Units Based on Half Disc and Rod-Like Moieties. Polym. Prepr. Am. Chem. Soc. Div. Polym. Chem. 1989, 30, 450–451. [Google Scholar]
- Ortega, P.; Bermejo, J.F.; Chonco, L.; de Jesus, E.; de la Mata, F.J.; Fernández, G.; Flores, J.C.; Gómez, R.; Serramía, M.J.; Muñoz-Fernandez, M.A. Novel Water-Soluble Carbosilane Dendrimers: Synthesis and Biocompatibility. Eur. J. Inorg. Chem. 2006, 2006, 1388–1396. [Google Scholar] [CrossRef]
- Bermejo, J.F.; Ortega, P.; Chonco, L.; Eritja, R.; Samaniego, R.; Müllner, M.; de Jesus, E.; de la Mata, F.J.; Flores, J.C.; Gomez, R.; et al. Water-Soluble Carbosilane Dendrimers: Synthesis Biocompatibility and Complexation with Oligonucleotides; Evaluation for Medical Applications. Chem. Eur. J. 2007, 13, 483–495. [Google Scholar] [CrossRef]
- Chonco, L.; Bermejo-Martín, J.F.; Ortega, P.; Shcharbin, D.; Pedziwiatr, E.; Klajnert, B.; de la Mata, F.J.; Eritja, R.; Gómez, R.; Bryszewska, M.; et al. Water-Soluble Carbosilane Dendrimers Protect Phosphorothioate Oligonucleotides from Binding to Serum Proteins. Org. Biomol. Chem. 2007, 5, 1886–1893. [Google Scholar] [CrossRef]
- Mata, F.D.L.M.D.L.; Ramírez, R.G.; Serrano, J.C.F.; Alcañiz, E.D.J.; López, P.O.; Fernández, M.Á.M.; Martín, J.F.B.; Lobera, M.J.S.; Gómez-Chacón, G.F.; Jiménez, L.C.; et al. Nuevos Dendrímeros Carbosilanos, su Preparación y Sus Usos. WO2007010080A2, 25 January 2007. Available online: https://patents.google.com/patent/WO2007010080A2/en?oq=WO2007010080 (accessed on 18 April 2023).
- Weber, N.; Ortega, P.; Clemente, M.I.; Shcharbin, D.; Bryszewska, M.; de la Mata, F.J.; Gómez, R.; Muñoz-Fernández, M.A. Characterization of Carbosilane Dendrimers as Effective Carriers of SiRNA to HIV-Infected Lymphocytes. J. Control. Release 2008, 132, 55–64. [Google Scholar] [CrossRef]
- Zinselmeyer, B.H.; Mackay, S.P.; Uchegbu, I.F.; Schatzlein, A.G. Computer Aided Optimisation of Synthetic Gene Delivery Agents Using PPI Dendrimers. Br. J. Cancer 2002, 86, S105. [Google Scholar] [CrossRef]
- Khan, O.F.; Zaia, E.W.; Yin, H.; Bogorad, R.L.; Pelet, J.M.; Webber, M.J.; Zhuang, I.; Dahlman, J.E.; Langer, R.; Anderson, D.G. Ionizable Amphiphilic Dendrimer-Based Nanomaterials with Alkyl-Chain-Substituted Amines for Tunable SiRNA Delivery to the Liver Endothelium In Vivo. Angew. Chem. Int. Ed. 2014, 53, 14397–14401. [Google Scholar] [CrossRef]
- Khan, O.F.; Zaia, E.W.; Jhunjhunwala, S.; Xue, W.; Cai, W.; Yun, D.S.; Barnes, C.M.; Dahlman, J.E.; Dong, Y.; Pelet, J.M.; et al. Dendrimer-Inspired Nanomaterials for the in Vivo Delivery of SiRNA to Lung Vasculature. Nano Lett. 2015, 15, 3008–3016. [Google Scholar] [CrossRef]
- Posadas, I.; Lopez-Hernandez, B.; Clemente, M.I.; Jimenez, J.L.; Ortega, P.; de la Mata, J.; Gomez, R.; Munoz-Fernandez, M.A.; Cena, V. Highly Efficient Transfection of Rat Cortical Neurons Using Carbosilane Dendrimers Unveils a Neuroprotective Role for HIF-1 Alpha in Early Chemical Hypoxia-Mediated Neurotoxicity. Pharm. Res. 2009, 26, 1181–1191. [Google Scholar] [CrossRef] [PubMed]
- Posadas, I.; Guerra, F.; Cena, V. Nonviral Vectors for the Delivery of Small Interfering RNAs to the CNS. Nanomedicine 2010, 5, 1219–1236. [Google Scholar] [CrossRef]
- Rodrigo, A.C.; Rivilla, I.; Perez-Martinez, F.C.; Monteagudo, S.; Ocana, V.; Guerra, J.; Garcia-Martinez, J.C.; Merino, S.; Sanchez-Verdu, P.; Cena, V.; et al. Efficient, Non-Toxic Hybrid PPV-PAMAM Dendrimer as a Gene Carrier for Neuronal Cells. Biomacromolecules 2011, 12, 1205–1213. [Google Scholar] [CrossRef] [PubMed]
- Perez-Martinez, F.; Guerra, J.; Posadas, I.; Cena, V. Barriers to Non-Viral Vector-Mediated Gene Delivery in the Nervous System. Pharm. Res. 2011, 28, 1843–1858. [Google Scholar] [CrossRef]
- Perez-Martinez, F.; Carrion, B.; Cena, V. The Use of Nanoparticles for Gene Therapy in the Nervous System. J. Alzheimer’s Dis. 2012, 31, 697–710. [Google Scholar] [CrossRef] [PubMed]
- Perez-Carrion, M.D.; Perez-Martinez, F.C.; Merino, S.; Sanchez-Verdu, P.; Martinez-Hernandez, J.; Lujan, R.; Cena, V. Dendrimer-Mediated SiRNA Delivery Knocks down Beclin 1 and Potentiates NMDA-Mediated Toxicity in Rat Cortical Neurons. J. Neurochem. 2012, 120, 259–268. [Google Scholar] [CrossRef]
- Perez-Martinez, F.; Ocana, A.; Perez-Carrion, M.; Cena, V. Dendrimers As Vectors for Genetic Material Delivery to the Nervous System. Curr. Med. Chem. 2012, 19, 5101–5108. [Google Scholar] [CrossRef]
- Perez-Carrion, M.D.; Cena, V. Knocking Down HMGB1 Using Dendrimer-Delivered SiRNA Unveils Its Key Role in NMDA-Induced Autophagy in Rat Cortical Neurons. Pharm. Res. 2013, 30, 2584–2595. [Google Scholar] [CrossRef]
- Posadas, I.; Monteagudo, S.; Cena, V. Nanoparticles for Brain-Specific Drug and Genetic Material Delivery, Imaging and Diagnosis. Nanomedicine 2016, 11, 833–849. [Google Scholar] [CrossRef]
- Janiszewska, J.; Posadas, I.; Jativa, P.; Bugaj-Zarebska, M.; Urbanczyk-Lipkowska, Z.; Cena, V. Second Generation Amphiphilic Poly-Lysine Dendrons Inhibit Glioblastoma Cell Proliferation without Toxicity for Neurons or Astrocytes. PLoS ONE 2016, 11, e0165704. [Google Scholar] [CrossRef]
- Posadas, I.; Romero-Castillo, L.; El Brahmi, N.; Manzanares, D.; Mignani, S.; Majoral, J.-P.; Cena, V. Neutral High-Generation Phosphorus Dendrimers Inhibit Macrophage-Mediated Inflammatory Response In Vitro and In Vivo. Proc. Natl. Acad. Sci. USA 2017, 114, E7660–E7669. [Google Scholar] [CrossRef] [PubMed]
- Cena, V.; Jativa, P. Nanoparticle Crossing of Blood-Brain Barrier: A Road to New Therapeutic Approaches to Central Nervous System Diseases. Nanomedicine 2018, 13, 1513–1516. [Google Scholar] [CrossRef]
- Stenstrom, P.; Manzanares, D.; Zhang, Y.; Cena, V.; Malkoch, M. Evaluation of Amino-Functional Polyester Dendrimers Based on Bis-MPA as Nonviral Vectors for SiRNA Delivery. Molecules 2018, 23, 2028. [Google Scholar] [CrossRef] [PubMed]
- de la Torre, C.; Cena, V. The Delivery Challenge in Neurodegenerative Disorders: The Nanoparticles Role in Alzheimer’s Disease Therapeutics and Diagnostics. Pharmaceutics 2018, 10, 190. [Google Scholar] [CrossRef] [PubMed]
- Mignani, S.; Shi, X.; Cena, V.; Shcharbin, D.; Bryszewska, M.; Majoral, J.-P. In Vivo Therapeutic Applications of Phosphorus Dendrimers: State of the Art. Drug Discov. Today 2020, 26, 677–689. [Google Scholar] [CrossRef]
- Posadas, I.; Romero-Castillo, L.; Ronca, R.-A.; Karpus, A.; Mignani, S.; Majoral, J.-P.; Munoz-Fernandez, M.; Cena, V. Engineered Neutral Phosphorous Dendrimers Protect Mouse Cortical Neurons and Brain Organoids from Excitotoxic Death. Int. J. Mol. Sci. 2022, 23, 4391. [Google Scholar] [CrossRef]
- Discher, B.M.; Won, Y.-Y.; Ege, D.S.; Lee, J.C.-M.; Bates, F.S.; Discher, D.E.; Hammer, D.A. Polymersomes: Tough Vesicles Made from Diblock Copolymers. Science 1999, 284, 1143–1146. [Google Scholar] [CrossRef]
- Leung, A.K.K.; Hafez, I.M.; Baoukina, S.; Belliveau, N.M.; Zhigaltsev, I.V.; Afshinmanesh, E.; Tieleman, D.P.; Hansen, C.L.; Hope, M.J.; Cullis, P.R. Lipid Nanoparticles Containing SiRNA Synthesized by Microfluidic Mixing Exhibit an Electron-Dense Nanostructured Core. J. Phys. Chem. C 2012, 116, 18440–18450. [Google Scholar] [CrossRef]
- Leung, A.K.K.; Tam, Y.Y.C.; Chen, S.; Hafez, I.M.; Cullis, P.R. Microfluidic Mixing: A General Method for Encapsulating Macromolecules in Lipid Nanoparticle Systems. J. Phys. Chem. B 2015, 119, 8698–8706. [Google Scholar] [CrossRef]
- Evers, M.J.W.; Kulkarni, J.A.; van der Meel, R.; Cullis, P.R.; Vader, P.; Schiffelers, R.M. State-of-the-Art Design and Rapid-Mixing Production Techniques of Lipid Nanoparticles for Nucleic Acid Delivery. Small Methods 2018, 2, 1700375. [Google Scholar] [CrossRef]
- Kulkarni, J.A.; Witzigmann, D.; Chen, S.; Cullis, P.R.; van der Meel, R. Lipid Nanoparticle Technology for Clinical Translation of SiRNA Therapeutics. Acc. Chem. Res. 2019, 52, 2435–2444. [Google Scholar] [CrossRef]
- Kulkarni, J.A.; Witzigmann, D.; Leung, J.; Tam, Y.Y.C.; Cullis, P.R. On the Role of Helper Lipids in Lipid Nanoparticle Formulations of SiRNA. Nanoscale 2019, 11, 21733–21739. [Google Scholar] [CrossRef]
- Aharoni, S.M.; Crosby, C.R., III; Walsh, E.K. Size and Solution Properties of Globular tert-Butyloxycarbonyl-poly (α,ω-L-lysine). Macromolecules 1982, 15, 1093–1098. [Google Scholar] [CrossRef]
- Tomalia, D.A.; Naylor, A.M.; Goddard, W.A., III. Starburst Dendrimers: Molecular-Level Control of Size, Shape, Surface Chemistry, Topology, and Flexibility from Atom to Macroscopic Matter. Angew. Chem. Int. Ed. Engl. 1990, 29, 138–175. [Google Scholar] [CrossRef]
- Fischer, M.; Voegtle, F. Dendrimers: From Design to Application-A Progress Report. Angew. Chem. Int. Ed. 1999, 38, 884–905. [Google Scholar] [CrossRef]
- Newkome, G.R.; He, E.; Moorefield, C.N. Suprasupermolecules with Novel Properties: Metallodendrimers. Chem. Rev. 1999, 99, 1689–1746. [Google Scholar] [CrossRef] [PubMed]
- Bosman, A.W.; Jamssen, H.M.; Meijer, E.W. About Dendrimers: Structure, Physical Properties, and Applications. Chem. Rev. 1999, 99, 1665–1688. [Google Scholar] [CrossRef]
- Grayson, S.M.; Frechet, J.M.J. Convergent Dendrons and Dendrimers: From Synthesis to Applications. Chem. Rev. 2001, 101, 3819–3867. [Google Scholar] [CrossRef]
- Majoral, J.-P.; Caminade, A.M. Dendrimers Containing Heteroatoms (Si, P, B, Ge, or Bi). Chem. Rev. 1999, 99, 845–880. [Google Scholar] [CrossRef]
- Watson, M.D.; Fechtenkotter, A.; Muellen, K. Big is Beautiful-“Aromaticity” Revisited from Viewpoint of Macromoilecular and Supramolecular Benzene Chemistry. Chem. Rev. 2001, 101, 1267–1300. [Google Scholar] [CrossRef]
- Astruc, D.; Chardac, F. Dendritic Catalysts and Dendrimers in Catalysis. Chem. Rev. 2001, 101, 2991–3023. [Google Scholar] [CrossRef] [PubMed]
- Astruc, D.; Boisselier, E.; Omelas, C. Dendrimers Designed for Functions: From Physical, Photophysical, and Supramolecular Properties to Applications in Sensis, Catalysis, Molecular Electronics, Photonics, and Nanomedicine. Chem. Rev. 2010, 110, 1857–1959. [Google Scholar] [CrossRef] [PubMed]
- Li, W.-S.; Aida, T. Dendrimer Porphyrins and Phthalocyanines. Chem. Rev. 2009, 109, 6047–6076. [Google Scholar] [CrossRef] [PubMed]
- Carlmark, A.; Hawker, C.; Hult, A.; Malkoch, M. New methodologies in the Constructions of Dendritic Materials. Chem. Soc. Rev. 2009, 38, 352–362. [Google Scholar] [CrossRef]
- Schluter, A.D.; Rabe, J.P. Dendronized Polymers: Synthesis, Characterization, Assembly at Interfaces and Manipulation. Angew. Chem. Int. Ed. 2000, 39, 864–883. [Google Scholar] [CrossRef]
- Voegtle, F.; Richard, G.; Werner, N. Dendrimer Chemistry; Wiley-VCH: Weinheim, Germany, 2009. [Google Scholar]
- Frechet, J.M.J.; Tomalia, D.A. Dendrimers and Other Dendritic Polymers; John Wiley & Sons, Ltd.: Chichester, UK, 2001. [Google Scholar]
- Tomalia, D.A.; Christensen, J.B.; Boas, U. Dendrimers, Dendrons and Dendritic Polymers; Cambridge University Press: Cambridge, UK, 2012. [Google Scholar]
- Klajnert, B.; Peng, L.; Cena, V. (Eds.) Dendrimers in Biomedical Applications; RSC Publishing: Cambridge, UK, 2013. [Google Scholar]
- Newkome, G.R.; Moorefield, C.N.; Voegtle, F. Dendritic Molecules; VCH: Weinheim, Germany, 1996. [Google Scholar]
- Newkome, G.R.; Moorefield, C.N.; Voegtle, F. Dendrimers and Dendrons; Wiley-VCH: Weinheim, Germany, 2001. [Google Scholar]
- Rosen, B.M.; Wilson, C.J.; Wilson, D.A.; Peterca, M.; Imam, M.R.; Percec, V. Dendron-Mediated Self-Assembly, Disassembly, and Self-Organization of Complex Systems. Chem. Rev. 2009, 109, 6275–6540. [Google Scholar] [CrossRef]
- Percec, V.; Xiao, Q. The Legacy of Rosalind E. Franklin: Landmark Contributions to Two Nobel Prizes. Chem 2021, 7, 529–536. [Google Scholar] [CrossRef]
- Percec, V.; Dulcey, A.E.; Balagurusamy, V.S.K.; Miura, Y.; Smidrkal, J.; Peterca, M.; Nummelin, A.; Edlund, U.; Hudson, S.D.; Heiney, P.A.; et al. Self-Assembly of Amphiphilic Dendritic Dipeptides into Helical Pores. Nature 2004, 430, 764–768. [Google Scholar] [CrossRef]
- Percec, V.; Glodde, M.; Bera, T.K.; Miura, Y.; Shiyanowskaya, I.; Singer, K.D.; Balagurusamy, V.S.K.; Heiney, P.A.; Schnell, I.; Rapp, A.; et al. Self-Organization of Supramolecular Helical Dendrimers into Complex Electronic Materials. Nature 2002, 419, 384–387. [Google Scholar] [CrossRef]
- Percec, V. Merging Macromolecular and Supramolecular Chemistry into Bioinspired Synthesis of Complex Systems. Isr. J. Chem. 2020, 60, 48–66. [Google Scholar] [CrossRef]
- Percec, V.; Xiao, Q. Helical Chirality of Supramolecular Columns and Spheres Self-Organizes Complex Liquid Crystals, Crystals and Quasicrystals. Isr. J. Chem. 2021, 61, 530–556. [Google Scholar] [CrossRef]
- Percec, V.; Heck, J.; Johansson, G.; Tomazos, D.; Kawasumi, M.; Ungar, G. Molecular-Recognition-Directed Self-Assembly of Supramolecular Polymers. J. Macromol. Sci. Part A Pure Appl. Chem. 1994, 31, 1031–1070. [Google Scholar] [CrossRef]
- Percec, V.; Xiao, Q. Helical Self-Organizations and Emerging Functions in Architectures, Biological and Synthetic Macromolecules. Bull. Chem. Soc. Jpn. 2021, 94, 900–928. [Google Scholar] [CrossRef]
- Hudson, S.D.; Jung, H.-T.; Percec, V.; Cho, W.-D.; Johansson, G.; Ungar, G.; Balagurusamy, V.S.K. Direct Visualization of Individual Cylindrical and Spherical Supramolecular Dendrimers. Science 1997, 278, 449–452. [Google Scholar] [CrossRef]
- Yeardley, D.J.P.; Ungar, G.; Percec, V.; Holerca, M.N.; Johansson, G. Spherical Supramolecular Minidendrimers Self-Organized in an “Inverse Micellar”-like Thermotropic Body-Centered Cubic Liquid Crystalline Phase. J. Am. Chem. Soc. 2000, 122, 1684–1689. [Google Scholar] [CrossRef]
- Dukeson, D.R.; Ungar, G.; Balagurusamy, V.S.K.; Percec, V.; Johansson, G.A.; Glodde, M. Application of Isomorphous Replacement in the Structure Determination of a Cubic Liquid Crystal Phase and Location of Counterions. J. Am. Chem. Soc. 2003, 125, 15974–15980. [Google Scholar] [CrossRef]
- Balagurusamy, V.S.K.; Ungar, G.; Percec, V.; Johansson, G. Rational Design of the First Spherical Supramolecular Dendrimers Self-Organized in a Novel Thermotropic Cubic Liquid-Crystalline Phase and the Determination of Their Shape by X-ray Analysis. J. Am. Chem. Soc. 1997, 119, 1539–1555. [Google Scholar] [CrossRef]
- Peterca, M.; Imam, M.R.; Hudson, S.D.; Partridge, B.E.; Sahoo, D.; Heiney, P.A.; Klein, M.L.; Percec, V. Complex Arrangement of Orthogonal Nanoscale Columns via a Supramolecular Orientational Memory Effect. ACS Nano 2016, 10, 10480–10488. [Google Scholar] [CrossRef]
- Sahoo, D.; Peterca, M.; Aqad, E.; Partridge, B.E.; Heiney, P.A.; Graf, R.; Spiess, H.W.; Zeng, X.; Percec, V. Tetrahedral Arrangements of Perylene Bisimide Columns via Supramolecular Orientational Memory. ACS Nano 2017, 11, 983–991. [Google Scholar] [CrossRef]
- Sahoo, D.; Peterca, M.; Imam, M.R.; Partridge, B.E.; Xiao, Q.; Percec, V. Conformationally Flexible Dendronized Cyclotetraveratrylenes (CTTV)s Self-Organize a Large Diversity of Chiral Columnar, Frank-Kasper and Quasicrystal Phases. Giant 2022, 10, 100096. [Google Scholar] [CrossRef]
- Sahoo, D.; Peterca, M.; Aqad, E.; Partridge, B.E.; Klein, M.L.; Percec, V. Losing Supramolecular Orientational Memory via Self-Organization of a Misfolded Secondary Structure. Polym. Chem. 2018, 9, 2370–2381. [Google Scholar] [CrossRef]
- Wilson, D.A.; Andreopoulou, A.K.; Peterca, M.; Leowanawat, P.; Sahoo, D.; Partridge, B.E.; Huang, N.; Heiney, P.A.; Percec, V. Supramolecular Spheres Self-Assembled from Conical Dendrons are Chiral. J. Am. Chem. Soc. 2019, 141, 6162–6166. [Google Scholar] [CrossRef] [PubMed]
- Sahoo, D.; Peterca, M.; Aqad, E.; Partridge, B.E.; Heiney, P.A.; Graf, R.; Spiess, H.; Zeng, X.; Percec, V. Hierarchical Self-Organization of Perylene Bisimides into Supramolecular Spheres and Periodic Arrays Thereof. J. Am. Chem. Soc. 2016, 138, 14798–14807. [Google Scholar] [CrossRef] [PubMed]
- Percec, V.; Rudick, J.G.; Peterca, M.; Yurchenko, M.E.; Smidrkal, J.; Heiney, P.A. Supramolecular Structural Diversity among First-Generation Hybrid Dendrimers and Twin Dendrons. Chem. Eur. J. 2008, 14, 3355–3362. [Google Scholar] [CrossRef] [PubMed]
- Percec, V.; Cho, W.-D.; Möller, M.; Prokhorova, S.A.; Ungar, G.; Yeardley, D.J.P. Design and Structural Analysis of the First Spherical Monodendron Self-Organizable in a Cubic Lattice. J. Am. Chem. Soc. 2000, 122, 4249–4250. [Google Scholar] [CrossRef]
- Percec, V.; Ahn, C.-H.; Barboiu, B. Self-Encapsulation, Acceleration and Control in the Radical Polymerization of Monodendritic Monomers via Self-Assembly. J. Am. Chem. Soc. 1997, 119, 12978–12979. [Google Scholar] [CrossRef]
- Percec, V.; Ahn, C.-H.; Ungar, G.; Yeardley, D.J.P.; Möller, M.; Sheiko, S.S. Controlling Polymer Shape through the Self-Assembly of Dendritic Side-Groups. Nature 1998, 391, 161–164. [Google Scholar] [CrossRef]
- Percec, V.; Ahn, C.-H.; Cho, W.-D.; Jamieson, A.M.; Kim, J.; Leman, T.; Schmidt, M.; Gerle, M.; Möller, M.; Prokhorova, S.A.; et al. Visualizable Cylindrical Macromolecules with Controlled Stiffness from Backbones Containing Libraries of Self-Assembling Dendritic Side Groups. J. Am. Chem. Soc. 1998, 120, 8619–8631. [Google Scholar] [CrossRef]
- Holerca, M.N.; Peterca, M.; Partridge, B.E.; Xiao, Q.; Lligadas, G.; Monteiro, M.J.; Percec, V. Monodisperse Macromolecules by Self-Interrupted Living Polymerization. J. Am. Chem. Soc. 2020, 142, 15265–15270. [Google Scholar] [CrossRef]
- Percec, V.; Cho, W.-D.; Mosier, P.E.; Ungar, G.; Yeardley, D.J.P. Structural Analysis of Cylindrical and Spherical Supramolecular Dendrimers Quantifies the Concept of Monodendron Shape Control by Generation Number. J. Am. Chem. Soc. 1998, 120, 11061–11070. [Google Scholar] [CrossRef]
- Percec, V.; Cho, W.-D.; Ungar, G. Increasing the Diameter of Cylindrical and Spherical Supramolecular Dendrimers by Decreasing the Solid Angle of Their Monodendrons via Periphery Functionalization. J. Am. Chem. Soc. 2000, 122, 10273–10281. [Google Scholar] [CrossRef]
- Percec, V.; Cho, W.-D.; Ungar, G.; Yeardley, D.J.P. Synthesis and Structural Analysis of Two Constitutional Isomeric Libraries of AB2-Based Monodendrons and Supramolecular Dendrimers. J. Am. Chem. Soc. 2001, 123, 1302–1315. [Google Scholar] [CrossRef]
- Percec, V.; Mitchell, C.M.; Cho, W.-D.; Uchida, S.; Glodde, M.; Ungar, G.; Zeng, X.; Liu, Y.; Balagurusamy, V.S.K.; Heiney, P.A. Designing Libraries of First Generation AB3 and AB2 Self-Assembling Dendrons via the Primary Structure Generated from Combinations of (AB)y–AB3 and (AB)y–AB2 Building Blocks. J. Am. Chem. Soc. 2004, 126, 6078–6094. [Google Scholar] [CrossRef] [PubMed]
- Percec, V.; Holerca, M.N.; Nummelin, S.; Morrison, J.J.; Glodde, M.; Smidrkal, J.; Peterca, M.; Rosen, B.M.; Uchida, S.; Balagurusamy, V.S.K.; et al. Exploring and Expanding the Structural Diversity of Self-Assembling Dendrons through Combinations of AB, Constitutional Isomeric AB2, and AB3 Biphenyl-4-Methyl Ether Building Blocks. Chem. Eur. J. 2006, 12, 6216–6241. [Google Scholar] [CrossRef]
- Percec, V.; Peterca, M.; Sienkowska, M.J.; Ilies, M.A.; Aqad, E.; Smidrkal, J.; Heiney, P.A. Synthesis and Retrostructural Analysis of Libraries of AB3 and Constitutional Isomeric AB2 Phenylpropyl Ether-Based Supramolecular Dendrimers. J. Am. Chem. Soc. 2006, 128, 3324–3334. [Google Scholar] [CrossRef]
- Percec, V.; Won, B.C.; Peterca, M.; Heiney, P.A. Expanding the Structural Diversity of Self-Assembling Dendrons and Supramolecular Dendrimers via Complex Building Blocks. J. Am. Chem. Soc. 2007, 129, 11265–11278. [Google Scholar] [CrossRef]
- Rosen, B.M.; Wilson, D.A.; Wilson, C.J.; Peterca, M.; Won, B.C.; Huang, C.; Lipski, L.R.; Zeng, X.; Ungar, G.; Heiney, P.A.; et al. Predicting the Structure of Supramolecular Dendrimers via the Analysis of Libraries of AB3 and Constitutional Isomeric AB2 Biphenylpropyl Ether Self-Assembling Dendrons. J. Am. Chem. Soc. 2009, 131, 17500–17521. [Google Scholar] [CrossRef]
- Ungar, G.; Liu, Y.; Zeng, X.; Percec, V.; Cho, W.-D. Giant Supramolecular Liquid Crystal Lattice. Science 2003, 299, 1208–1211. [Google Scholar] [CrossRef]
- Sahoo, D.; Imam, M.R.; Peterca, M.; Partridge, B.E.; Wilson, D.A.; Zeng, X.; Ungar, G.; Heiney, P.A.; Percec, V. Hierarchical Self-Organization of Chiral Columns from Chiral Supramolecular Spheres. J. Am. Chem. Soc. 2018, 140, 13478–13487. [Google Scholar] [CrossRef]
- Percec, V.; Huang, N.; Xiao, Q.; Partridge, B.E.; Sahoo, D.; Imam, M.R.; Peterca, M.; Graf, R.; Spiess, H.-W.; Zeng, X.; et al. Self-Organization of Rectangular Bipyramidal Helical Columns by Supramolecular Orientational Memory Epitaxially Nucleated from a Frank-Kasper σ Phase. Giant 2022, 9, 100084. [Google Scholar] [CrossRef]
- Duan, H.; Hudson, S.D.; Ungar, G.; Holerca, M.N.; Percec, V. Definitive Support by Transmission Electron Microscopy, Electron Diffraction and Electron Density Calculations for the Formation of a BCC Lattice from Poly{N-[3,4,5-tris(n-dodecan-1-yloxy)benzoyl]ethyleneimine}. Chem. Eur. J. 2001, 7, 4134–4141. [Google Scholar] [CrossRef] [PubMed]
- Percec, V.; Heck, J.; Johansson, G.; Tomazos, D.; Ungar, G. Towards Tobacco Mosaic Virus-like Self-Assembled Supramolecular Architectures. Macromol. Symp. 1994, 77, 237–265. [Google Scholar] [CrossRef]
- Zeng, X.; Ungar, G.; Liu, Y.; Percec, V.; Dulcey, A.E.; Hobbs, J.K. Supramolecular dendritic liquid quasicrystals. Nature 2004, 428, 157–160. [Google Scholar] [CrossRef]
- Holerca, M.N.; Sahoo, D.; Partridge, B.E.; Peterca, M.; Zeng, X.; Ungar, G.; Percec, V. Dendronized Poly(2-Oxazoline) Displays within Only Five Monomer Repeat Units Liquid Quasicrystal, A15 and σ Frank–Kasper Phases. J. Am. Chem. Soc. 2018, 140, 16941–16947. [Google Scholar] [CrossRef] [PubMed]
- Ungar, G.; Percec, V.; Zeng, X.; Leowanawat, P. Liquid Quasicrystals. Isr. J. Chem. 2011, 51, 1206–1215. [Google Scholar] [CrossRef]
- Imam, M.R.; Peterca, M.; Xiao, Q.; Percec, V. Enhancing Conformational Flexibility of Dendronized Triphenylene via Diethylene Glycol Linkers Lowers Transitions of Helical Columnar, Frank-Kasper, and Quasicrystal Phases. Giant 2022, 10, 100098. [Google Scholar] [CrossRef]
- Yue, K.; Huang, M.; Marson, R.L.; He, J.; Huang, J.; Zhou, Z.; Wang, J.; Liu, C.; Yan, X.; Wu, K.; et al. Geometry Induced Sequence of Nanoscale Frank–Kasper and Quasicrystal Mesophases in Giant Surfactants. Proc. Natl. Acad. Sci. USA 2016, 113, 14195–14200. [Google Scholar] [CrossRef]
- Percec, V.; Sahoo, D. Discotic liquid crystals 45 years later. Dendronized discs and crowns increase liquid crystal complexity to columnar from spheres, cubic Frank-Kasper, liquid quasicrystals and memory-effect induced columnar-bundles. Giant 2022, 12, 100127. [Google Scholar] [CrossRef]
- Percec, V.; Chu, P.; Kawasumi, M. Toward ‘Willowlike’ Thermotropic Dendrimers. Macromolecules 1994, 27, 4441–4453. [Google Scholar] [CrossRef]
- Percec, V. Molecular Design of Novel Liquid Crystalline Polymers with Complex Architecture: Macrocyclics and Dendrimers. Pure Appl. Chem. 1995, 67, 2031–2038. [Google Scholar] [CrossRef]
- Percec, V.; Kawasumi, M. Synthesis and characterization of a thermotropic nematic liquid crystalline dendrimeric polymer. Macromolecules 1992, 25, 3843–3850. [Google Scholar] [CrossRef]
- Percec, V.; Sahoo, D.; Adamson, J. Stimuli-Responsive Principles of Supramolecular Organizations Emerging from Self-Assembling and Self-Organizable Dendrons, Dendrimers, and Dendronized Polymers. Polymers 2023, 15, 1832. [Google Scholar] [CrossRef] [PubMed]
- Percec, V.; Lee, M.; Heck, J.; Blackwell, H.E.; Ungar, G.; Alvarez-Castillo, A. Re-entrant Isotropic Phase in a Supramolecular Disc-like Oligomer of 4-[3,4,5-Tris(N-Dodecanyloxy)Benzoyloxy]-4′-[(2-Vinyloxy)Ethoxy]Biphenyl. J. Mater. Chem. 1992, 2, 931–938. [Google Scholar] [CrossRef]
- Percec, V.; Heck, J.; Tomazos, D.; Falkenberg, F.; Blackwell, H.; Ungar, G. Self-assembly of Taper-Shaped Monoesters of Oligo(ethylene oxide) with 3,4,5-Tris(p-dodecyloxybenzyloxy)benzoic Acid and of Their Polymethacrylates into Tubular Supramolecular Architectures Displaying a Columnar Mesophase. J. Chem. Soc. Perkin Trans. 1993, 1, 2799–2811. [Google Scholar] [CrossRef]
- Percec, V.; Tomazos, D.; Heck, J.; Blackwell, H.; Ungar, G. Self-assembly of Taper-Shaped Monoesters of Oligo(ethylene oxide) with 3,4,5-Tris(n-dodecan-1-yloxy)benzoic Acid and of Their Polymethacrylates into Tubular Supramolecular Architectures Displaying a Columnar Hexagonal Mesophase. J. Chem. Soc. Perkin Trans. 1994, 2, 31–44. [Google Scholar] [CrossRef]
- Kwon, Y.K.; Chvalun, S.N.; Schneider, A.-I.; Blackwell, J.; Percec, V.; Heck, J.A. Supramolecular Tubular Structures of a Polymethacrylate with Tapered Side Groups in Aligned Hexagonal Phases. Macromolecules 1994, 27, 6129–6132. [Google Scholar] [CrossRef]
- Kwon, Y.K.; Chvalun, S.N.; Blackwell, J.; Percec, V.; Heck, J.A. Effect of Temperature on the Supramolecular Tubular Structure in Oriented Fibers of a Poly(methacrylate) with Tapered Side Groups. Macromolecules 1995, 28, 1552–1558. [Google Scholar] [CrossRef]
- Percec, V.; Schlueter, D.; Kwon, Y.K.; Blackwell, J.; Moeller, M.; Slangen, P.J. Dramatic Stabilization of a Hexagonal Columnar Mesophase Generated from Supramolecular and Macromolecular Columns by the Semifluorination of the Alkyl Groups of Their Tapered Building Blocks. Macromolecules 1995, 28, 8807–8818. [Google Scholar] [CrossRef]
- Kwon, Y.K.; Danko, C.; Chvalun, S.; Blackwell, J.; Heck, J.A.; Percec, V. Comparison of the Supramolecular Structures Formed by a Polymethacrylate with a Highly Tapered Side-Chain and its Monomeric Precursor. Macromol. Symp. 1994, 87, 103–114. [Google Scholar] [CrossRef]
- Chvalun, S.N.; Scherbina, M.A.; Yakunin, A.N.; Blackwell, J.; Percec, V. Structure of gyroid mesophase formed by monodendrons with fluorinated alkyl tails. Polym. Sci. Ser. A 2007, 49, 158–167. [Google Scholar] [CrossRef]
- Blackwell, J.; Chvalun, S.N.; Cho, J.D.; Kwon, Y.K.; Percec, V.; Heck, J.A. X-ray Analyses of the Supramolecular Structures Formed by Polymers with Highly Tapered Side Groups; Case Western Reserve University: Cleveland, OH, USA, 1997; Volume 214, p. 32-Poly. [Google Scholar]
- Chvalun, S.N.; Blackwell, J.; Cho, J.D.; Kwon, Y.K.; Percec, V.; Heck, J.A. X-ray Analysis of the Internal Rearrangement of the Self-Assembling Columnar Structure Formed by a Highly Tapered Molecule. Polymers 1998, 39, 4515–4522. [Google Scholar] [CrossRef]
- Chvalun, S.N.; Blackwell, J.; Kwon, Y.K.; Percec, V. Small Angle X-ray Analysis of the Effect of Temperature on the Self-Assembling Columnar Structures Formed by a Polymethacrylate with Highly Tapered Side Groups and by One of its Low Molar Mass Precursors. Macromol. Symp. 1997, 118, 663–675. [Google Scholar] [CrossRef]
- Percec, V.; Schlueter, D.; Ronda, J.C.; Johansson, G.; Ungar, G.; Zhou, J.P. Tubular Architectures from Polymers with Tapered Side Groups. Assembly of Side Groups via a Rigid Helical Chain Conformation and Flexible Helical Chain Conformation Induced via Assembly of Side Groups. Macromolecules 1996, 29, 1464–1472. [Google Scholar] [CrossRef]
- Percec, V.; Schlueter, D. Mechanistic Investigations on the Formation of Supramolecular Cylindrical Shaped Oligomers and Polymers by Living Ring Opening Metathesis Polymerization of a 7-Oxanorbornene Monomer Substituted with Two Tapered Monodendrons. Macromolecules 1997, 30, 5783–5790. [Google Scholar] [CrossRef]
- Percec, V.; Johansson, G.; Schlueter, D.; Ronda, J.C.; Ungar, G. Molecular Recognition Directed Self-Assembly of Tubular Supramolecular Architectures from Building Blocks Containing Monodendrons as Exo-Receptors and Crown- or Pseudo-Crown-Ethers as Endo-Receptors. Macromol. Symp. 1996, 101, 43–60. [Google Scholar] [CrossRef]
- Percec, V.; Ahn, C.-H.; Cho, W.-D.; Johansson, G.; Schlueter, D. Design of New Macromolecular Architectures by Using Quasi-Equivalent Monodendrons as Building Blocks. Macromol. Symp. 1997, 118, 33–43. [Google Scholar] [CrossRef]
- Percec, V.; Schlueter, D.; Ungar, G.; Cheng, S.Z.D.; Zhang, A. Hierarchical Control of Internal Superstructure, Diameter, and Stability of Supramolecular and Macromolecular Columns Generated from Tapered Monodendritic Building Blocks. Macromolecules 1998, 31, 1745–1762. [Google Scholar] [CrossRef]
- Percec, V.; Johansson, G.; Ungar, G.; Zhou, J. Fluorophobic Effect Induces the Self-Assembly of Semifluorinated Tapered Monodendrons Containing Crown Ethers into Supramolecular Columnar Dendrimers Which Exhibit a Homeotropic Hexagonal Columnar Liquid Crystalline Phase. J. Am. Chem. Soc. 1996, 118, 9855–9866. [Google Scholar] [CrossRef]
- Johansson, G.; Percec, V.; Ungar, G.; Zhou, J.P. Fluorophobic Effect in the Self-Assembly of Polymers and Model Compounds Containing Tapered Groups into Supramolecular Columns. Macromolecules 1996, 29, 646–660. [Google Scholar] [CrossRef]
- Johansson, G.; Percec, V.; Ungar, G.; Smith, K. Fluorophobic Effect Generates a Systematic Approach to the Synthesis of the Simplest Class of Rodlike Liquid Crystals Containing a Single Benzene Unit. Chem. Mater. 1997, 9, 164–175. [Google Scholar] [CrossRef]
- Percec, V.; Heck, J. Side-Chain Liquid Crystal Polymers Containing Hemiphasmidic Mesogens. Polym. Prepr. Am. Chem. Soc. Div. Polym. Chem. 1991, 32, 263–264. [Google Scholar]
- Percec, V.; Heck, J. Molecular Recognition Directed Supramolecular Polymer Architectures. Polym. Prepr. Am. Chem. Soc. Div. Polym. Chem. 1991, 32, 698–699. [Google Scholar]
- Percec, V.; Heck, J.; Ungar, G. Towards Tobacco Mosaic Virus-Like Supramolecular Synthetic Polymers. Polym. Prepr. Am. Chem. Soc. Div. Polym. Chem. 1992, 33, 152–153. [Google Scholar]
- Percec, V.; Heck, J. Molecular Design of Externally Regulated Self-Assembled Supramolecular Ionic Channels. Polym. Prepr. Am. Chem. Soc. Div. Polym. Chem. 1992, 33, 217–218. [Google Scholar]
- Percec, V.; Heck, J.; Johansson, G.; Ungar, G. Towards Tobacco Mosai Virus-Like Self-Assembled Supramolecular Architectures. Polym. Prepr. Am. Chem. Soc. Div. Polym. Chem. 1993, 34, 116–117. [Google Scholar]
- Percec, V.; Heck, J. Liquid Crystalline Polymers Containing Mesogenic Units Based on Half-Disk and Rod-Like Moieties. 1. Synthesis and Characterization of 4-(11-Undecan-1yloxy)-4′-[3,4,5-Tri(p-n-Dodecan-1-yloxybenzyloxy)benzoate]Biphenyl Side Groups. J. Polym. Sci. Part A Polym. Chem. 1991, 29, 591–597. [Google Scholar] [CrossRef]
- Percec, V.; Heck, J. Liquid Crystalline Polymers Containing Mesogenic Units Based on Half-Disk and Rod-Like Moieties. 2. Synthesis and Characterization of Poly(2-[3,4,5-Tri[p(N-Dodecan-1yloxy)-Benzyloxy)Benzoate]-7-[p-11-Undecan-yloxy)Benzoare]Naphthalene]Methyl Siloxane. Polym. Bull. 1990, 24, 255–262. [Google Scholar] [CrossRef]
- Percec, V.; Heck, J. Liquid Crystalline Polymers Containing Mesogenic Units Based on Half-Disk and Rod-Like Moieties. 3. Synthesis and Characterization of Polymethylsiloxanes and Copolymethysiloxanes Based on 4-[3,4,5-Tri(p-N-Dodecan-1-yloxybenzyloxy)benzoate]-4′-[p-Allyloxybenzoate)biphenyl and 4-(3,4,5-Tri(p-N-dodecan-1-yloxy-benzyloxy)benzoate]-4′-(p-allyloxybenzoate)thiodiphenyl Side Groups. Polym. Bull. 1991, 25, 55–62. [Google Scholar] [CrossRef]
- Percec, V.; Heck, J. Liquid Crystalline Polymers Containing Mesogenic Units Based on Half-Disk and Rod-Like Moieties. 4. Side-Chain Liquid-Crystalline Polymethylsiloxanes Containing Hemiphasmidic Mesogens Based on 4-[3,4,5-Tri(Alkan-1-yloxy)benzoate]Biphenyl Groups. Polym. Bull. 1991, 25, 431–438. [Google Scholar] [CrossRef]
- Percec, V.; Heck, J.; Ungar, G. Liquid Crystalline Polymers Containing Mesogenic Units Based on Half-Disk and Rod-Like Moieties. 5. Side-Chain Liquid-Crystalline Poly(methylsiloxane)s Containing Hemiphasmidic Mesogens Based on 4-[[3,4,5-Tris(Alkan-1-yloxy)benzoyl]oxy]-4′-[[p-(Propan-1-yloxy)benzoyl]oxy]Biphenyl Groups. Macromolecules 1991, 24, 4957–4962. [Google Scholar] [CrossRef]
- Percec, V.; Heck, J. Side-chain liquid-crystal polymers containing mesogenic units based on 1/2 disk and rod-like moieties. In Abstracts of Papers of the American Chemical Society; ACS: Washington, DC, USA, 1989; Volume 30, p. 450. [Google Scholar]
- Ho, M.-S.; Partridge, B.E.; Sun, H.-J.; Sahoo, D.; Leowanawat, P.; Peterca, M.; Graf, R.; Spiess, H.W.; Zeng, X.; Ungar, G.; et al. Screening Libraries of Semifluorinated Arylene Bisimides to Discover and Predict Thermodynamically Controlled Helical Crystallization. ACS Comb. Sci. 2016, 18, 723–739. [Google Scholar] [CrossRef]
- Peterca, M.; Sahoo, D.; Imam, M.R.; Xiao, Q.; Percec, V. Searching for the simplest self-assembling dendron to study helical self-organization and supramolecular polymerization. Giant 2022, 12, 100118. [Google Scholar] [CrossRef]
- Peterca, M.; Sahoo, D.; Emad, A.; Peterca, M.; Percec, V. Molecular design principles of helical pyramidal chirality self-organized from achiral hexakis(alkyloxy)triphenylene. Giant 2023, 13, 100138. [Google Scholar] [CrossRef]
- Sahoo, D.; Emad, A.; Peterca, M.; Percec, V. A highly ordered 8/1 helical pyramidal column self-organized from the crown conformation of achiral hexa(butyloxy)triphenylene. Giant 2023, 13, 100135. [Google Scholar] [CrossRef]
- Rudick, J.G.; Percec, V. Helical chirality in dendronized in dendronized polyarylacetylenes. New J. Chem. 2007, 31, 1083–1096. [Google Scholar] [CrossRef]
- Rudick, J.G.; Percec, V. Induced Helical Backbone conformations of self-organizable dendronized polymers. Acc. Chem. Res. 2008, 41, 1641–1652. [Google Scholar] [CrossRef]
- Percec, V. Bioinspired supramolecular liquid crystals. Philos. Trans. R. Soc. A Math. Phys. Eng. Sci. 2006, 364, 2709–2719. [Google Scholar] [CrossRef] [PubMed]
- Tomalia, D.A.; Sheetz, D.P. Homopolymerization of 2-Alkyl- and 2-Aryl-2-Oxazolines. J. Polym. Sci. Part A-1 Polym. Chem. 1966, 4, 2253–2265. [Google Scholar] [CrossRef]
- Kagiya, T.; Narisawa, S.; Maeda, T.; Fukui, K. Ring-Opening Polymerization of 2-Substituted 2-Oxazolines. J. Polym. Sci. Part B Polym. Lett. 1966, 4, 441–445. [Google Scholar] [CrossRef]
- Seeliger, W.; Aufderhaar, E.; Diepers, W.; Feinauer, R.; Nehring, R.; Thier, W.; Hellmann, H. Recent Syntheses and Reactions of Cyclic Imidic Esters. Angew. Chem. Int. Ed. Engl. 1966, 5, 875–888. [Google Scholar] [CrossRef]
- Bassiri, T.G.; Levy, A.; Litt, M. Polymerization of Cyclic Imino Ethers. I. Oxazolines. J. Polym. Sci. Part B Polym. Lett. 1967, 5, 871–879. [Google Scholar] [CrossRef]
- Holerca, M.N.; Sahoo, D.; Peterca, M.; Partridge, B.E.; Heiney, P.A.; Percec, V. A Tetragonal Phase Self-Organized from Unimolecular Spheres Assembled from a Substituted Poly(2-Oxazoline). Macromolecules 2017, 50, 375–385. [Google Scholar] [CrossRef]
- Percec, V.; Holerca, M.N.; Magonov, S.N.; Yeardley, D.J.P.; Ungar, G.; Duan, H.; Hudson, S.D. Poly(Oxazolines)s with Tapered Minidendritic Side Groups. The Simplest Cylindrical Models to Investigate the Formation of Two-Dimensional and Three-Dimensional Order by Direct Visualization. Biomacromolecules 2001, 2, 706–728. [Google Scholar] [CrossRef] [PubMed]
- Percec, V.; Holerca, M.N.; Uchida, S.; Yeardley, D.J.P.; Ungar, G. Poly(Oxazoline)s with Tapered Minidendritic Side Groups as Models for the Design of Synthetic Macromolecules with Tertiary Structure. A Demonstration of the Limitations of Living Polymerization in the Design of 3-D Structures Based on Single Polymer Chains. Biomacromolecules 2001, 2, 729–740. [Google Scholar] [CrossRef]
- Lehn, J.-M. Toward Self-Organization and Complex Matter. Science 2002, 295, 2400–2403. [Google Scholar] [CrossRef] [PubMed]
- Lehn, J.-M. Supramolecular Chemistry: Where from? Where to? Chem. Soc. Rev. 2017, 46, 2378–2379. [Google Scholar] [CrossRef] [PubMed]
- Lehn, J.-M. Beyond Chemical Synthesis: Self-Organization? Isr. J. Chem. 2018, 58, 136–141. [Google Scholar] [CrossRef]
- Taylor, W.R. A “Periodic Table” for Protein Structures. Nature 2002, 416, 657–660. [Google Scholar] [CrossRef]
- Moutevelis, E.; Woolfson, D.N. A Periodic Table of Coiled-Coil Protein Structures. J. Mol. Biol. 2009, 385, 726–732. [Google Scholar] [CrossRef]
- Ahnert, S.E.; Marsh, J.A.; Hernández, H.; Robinson, C.V.; Teichmann, S.A. Principles of Assembly Reveal a Periodic Table of Protein Complexes. Science 2015, 350, aaa2245. [Google Scholar] [CrossRef]
- Tomalia, D.A. In Quest of a Systematic Framework for Unifying and Defining Nanoscience. J. Nanopart. Res. 2009, 11, 1251–1310. [Google Scholar] [CrossRef] [PubMed]
- Tomalia, D.A.; Khanna, S.N. A Systematic Framework and Nanoperiodic Concept for Unifying Nanoscience: Hard/Soft Nanoelements, Superatoms, Meta-Atoms, New Emerging Properties, Periodic Property Patterns, and Predictive Mendeleev-like Nanoperiodic Tables. Chem. Rev. 2016, 116, 2705–2774. [Google Scholar] [CrossRef] [PubMed]
- Le Gall, T.; Berchel, M.; Davies, L.; Mottais, A.; Ghanem, R.; Fautrel, A.; Gill, D.; Hyde, S.; Lehn, P.; Lehn, J.-M.; et al. Aerosol-Mediated Non-Viral Lung Gene Therapy: The Potential of Aminoglycoside-Based Cationic Liposomes. Pharmaceutics 2022, 14, 25. [Google Scholar] [CrossRef]
- Esfand, R.; Tomalia, D.A. Poly(Amidoamine) (PAMAM) Dendrimers: From Biomimicry to Drug Delivery and Biomedical Applications. Drug Discov. Today 2001, 6, 427–436. [Google Scholar] [CrossRef] [PubMed]
- Svenson, S.; Tomalia, D.A. Dendrimers in Biomedical Applications—Reflections on the Field. Adv. Drug Deliv. Rev. 2005, 57, 2106–2129. [Google Scholar] [CrossRef] [PubMed]
- Elsabahy, M.; Wooley, K.L. Design of Polymeric Nanoparticles for Biomedical Delivery Applications. Chem. Soc. Rev. 2012, 41, 2545–2561. [Google Scholar] [CrossRef]
- Bergmann, G.H.F.; Finkelmann, H.; Percec, V.; Zhao, M. Liquid-Crystalline Main-Chain Elastomers. Macromol. Rapid Commun. 1997, 18, 353–360. [Google Scholar] [CrossRef]
- Percec, V.; Hahn, B. Liquid Crystalline Polymers Containing Heterocycloalkanediyl Groups as Mesogens. 7. Molecular Weight and Composition Effects on the Phase Transitions of Poly(Methylsiloxane)s and Poly(Methylsiloxane-Co-Dimethylsiloxane)s Containing 2-[4-(2(S)-Methyl-1-Butoxy)Phenyl]-5-(11-Undecanyl)-1,3,2-Dioxaborinane Side Groups. Macromolecules 1989, 22, 1588–1599. [Google Scholar] [CrossRef]
- Johansson, G.; Percec, V.; Ungar, G.; Abramic, D. Molecular Recognition Directed Self-Assembly of Tubular Liquid Crystalline and Crystalline Supramolecular Architectures from Taper Shaped (15-Crown-5)Methyl 3,4,5-Tris(p-Alkyloxybenzyloxy)Benzoates and (15-Crown-5)Methyl 3,4,5-Tris(p-Dodecyloxy)Benzoate. J. Chem. Soc. Perkin Trans. 1994, 1, 447–459. [Google Scholar] [CrossRef]
- Percec, V.; Cho, W.-D.; Ungar, G.; Yeardley, D.J.P. From Molecular Flat Tapers, Discs, and Cones to Supramolecular Cylinders and Spheres Using Fréchet-Type Monodendrons Modified on Their Periphery. Angew. Chem. Int. Ed. 2000, 39, 1597–1602. [Google Scholar] [CrossRef]
- Rosen, B.M.; Percec, V. A Reaction to Stress. Nature 2007, 446, 381–382. [Google Scholar] [CrossRef]
- Ungar, G.; Abramic, D.; Percec, V.; Heck, J.A. Self-Assembly of Twin Tapered Bisamides into Supramolecular Columns Exhibiting Hexagonal Columnar Mesophases. Structural Evidence for a Microsegregated Model of the Supramolecular Column. Liq. Cryst. 1996, 21, 73–86. [Google Scholar] [CrossRef]
- Percec, V.; Grigoras, C.; Kim, H.-J. Toward Self-Assembling Dendritic Macromolecules from Conventional Monomers by a Combination of Living Radical Polymerization and Irreversible Terminator Multifunctional Initiator. J. Polym. Sci. A Polym. Chem. 2004, 42, 505–513. [Google Scholar] [CrossRef]
- Percec, V.; Peterca, M.; Tadjiev, T.; Zeng, X.; Ungar, G.; Leowanawat, P.; Aqad, E.; Imam, M.R.; Rosen, B.M.; Akbey, U.; et al. Self-Assembly of Dendronized Perylene Bisimides into Complex Helical Columns. J. Am. Chem. Soc. 2011, 133, 12197–12219. [Google Scholar] [CrossRef] [PubMed]
- Percec, V.; Asgarzadeh, F. Metal-Catalyzed Living Radical Graft Copolymerization of Olefins Initiated from the Structural Defects of Poly(Vinyl Chloride). J. Polym. Sci. A Polym. Chem. 2001, 39, 1120–1135. [Google Scholar] [CrossRef]
- Percec, V.; Glodde, M.; Peterca, M.; Rapp, A.; Schnell, I.; Spiess, H.W.; Bera, T.K.; Miura, Y.; Balagurusamy, V.S.K.; Aqad, E.; et al. Self-Assembly of Semifluorinated Dendrons Attached to Electron-Donor Groups Mediates Their π-Stacking via a Helical Pyramidal Column. Chem. Eur. J. 2006, 12, 6298–6314. [Google Scholar] [CrossRef]
- Wilson, D.A.; Wilson, C.J.; Rosen, B.M.; Percec, V. Two-Step, One-Pot Ni-Catalyzed Neopentylglycolborylation and Complementary Pd/Ni-Catalyzed Cross-Coupling with Aryl Halides, Mesylates, and Tosylates. Org. Lett. 2008, 10, 4879–4882. [Google Scholar] [CrossRef]
- Rosen, B.M.; Huang, C.; Percec, V. Sequential Ni-Catalyzed Borylation and Cross-Coupling of Aryl Halides via in Situ Prepared Neopentylglycolborane. Org. Lett. 2008, 10, 2597–2600. [Google Scholar] [CrossRef]
- Nguyen, N.H.; Levere, M.E.; Percec, V. SET-LRP of Methyl Acrylate to Complete Conversion with Zero Termination. J. Polym. Sci. A Polym. Chem. 2012, 50, 860–873. [Google Scholar] [CrossRef]
- Moreno, A.; Lligadas, G.; Adamson, J.; Maurya, D.S.; Percec, V. Assembling Complex Macromolecules and Self-Organizations of Biological Relevance with Cu(I)-Catalyzed Azide-Alkyne, Thio-Bromo, and TERMINI Double “Click” Reactions. Polymers 2023, 15, 1075. [Google Scholar] [CrossRef]
- Rosen, B.M.; Percec, V. Single Electron-Transfer and Single-Electron Transfer Degenerative Chain Transfer Living Radical Polymerization. Chem. Rev. 2009, 109, 5069–5119. [Google Scholar] [CrossRef] [PubMed]
- Rosen, B.M.; Quasdorf, K.W.; Wilson, D.A.; Zhang, N.; Resmerita, A.-M.; Garg, N.K.; Percec, V. Nickel-Catalyzed Cross-Couplings Involving Carbon–Oxygen Bonds. Chem. Rev. 2011, 111, 1346–1416. [Google Scholar] [CrossRef] [PubMed]
- Zhang, N.; Samantha, S.R.; Rosen, B.M.; Percec, V. Single-Electron Transfer in Radical and Radical-Mediated Organic, Materials and Polymer Synthesis. Chem. Rev. 2014, 114, 5848–5958. [Google Scholar] [CrossRef] [PubMed]
- Jiang, X.; Rosen, B.M.; Percec, V. Mimicking “Nascent” Cu(0) Mediated SET-LRP of Methyl Acrylate in DMSO Leads to Complete Conversion in Several Minutes: MIMICS of “Nascent” Cu(0) in SET-LRP of MA. J. Polym. Sci. A Polym. Chem. 2010, 48, 403–409. [Google Scholar] [CrossRef]
- Nguyen, N.H.; Kulis, J.; Sun, H.-J.; Jia, Z.; Van Beusekom, B.; Levere, M.E.; Wilson, D.A.; Monteiro, M.J.; Percec, V. A Comparative Study of the SET-LRP of Oligo(Ethylene Oxide) Methyl Ether Acrylate in DMSO and in H2O. Polym. Chem. 2013, 4, 144–155. [Google Scholar] [CrossRef]
- Percec, V.; Ungar, G.; Peterca, M. Self-Assembly in Action. Science 2006, 313, 55–56. [Google Scholar] [CrossRef]
- Peterca, M.; Percec, V. Recasting Metal Alloy Phases with Block Copolymers. Science 2010, 330, 333–334. [Google Scholar] [CrossRef]
- Peterca, M.; Imam, M.R.; Leowanawat, P.; Rosen, B.M.; Wilson, D.A.; Wilson, C.J.; Zeng, X.; Ungar, G.; Heiney, P.A.; Percec, V. Self-Assembly of Hybrid Dendrons into Doubly Segregated Supramolecular Polyhedral Columns and Vesicles. J. Am. Chem. Soc. 2010, 132, 11288–11305. [Google Scholar] [CrossRef]










Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lu, J.; Atochina-Vasserman, E.N.; Maurya, D.S.; Shalihin, M.I.; Zhang, D.; Chenna, S.S.; Adamson, J.; Liu, M.; Shah, H.U.R.; Shah, H.; et al. Screening Libraries to Discover Molecular Design Principles for the Targeted Delivery of mRNA with One-Component Ionizable Amphiphilic Janus Dendrimers Derived from Plant Phenolic Acids. Pharmaceutics 2023, 15, 1572. https://doi.org/10.3390/pharmaceutics15061572
Lu J, Atochina-Vasserman EN, Maurya DS, Shalihin MI, Zhang D, Chenna SS, Adamson J, Liu M, Shah HUR, Shah H, et al. Screening Libraries to Discover Molecular Design Principles for the Targeted Delivery of mRNA with One-Component Ionizable Amphiphilic Janus Dendrimers Derived from Plant Phenolic Acids. Pharmaceutics. 2023; 15(6):1572. https://doi.org/10.3390/pharmaceutics15061572
Chicago/Turabian StyleLu, Juncheng, Elena N. Atochina-Vasserman, Devendra S. Maurya, Muhammad Irhash Shalihin, Dapeng Zhang, Srijay S. Chenna, Jasper Adamson, Matthew Liu, Habib Ur Rehman Shah, Honey Shah, and et al. 2023. "Screening Libraries to Discover Molecular Design Principles for the Targeted Delivery of mRNA with One-Component Ionizable Amphiphilic Janus Dendrimers Derived from Plant Phenolic Acids" Pharmaceutics 15, no. 6: 1572. https://doi.org/10.3390/pharmaceutics15061572
APA StyleLu, J., Atochina-Vasserman, E. N., Maurya, D. S., Shalihin, M. I., Zhang, D., Chenna, S. S., Adamson, J., Liu, M., Shah, H. U. R., Shah, H., Xiao, Q., Queeley, B., Ona, N. A., Reagan, E. K., Ni, H., Sahoo, D., Peterca, M., Weissman, D., & Percec, V. (2023). Screening Libraries to Discover Molecular Design Principles for the Targeted Delivery of mRNA with One-Component Ionizable Amphiphilic Janus Dendrimers Derived from Plant Phenolic Acids. Pharmaceutics, 15(6), 1572. https://doi.org/10.3390/pharmaceutics15061572





