Pharmacogenetic Sex-Specific Effects of Methotrexate Response in Patients with Rheumatoid Arthritis
Abstract
:1. Introduction
2. Materials and Methods
2.1. Participant Recruitment
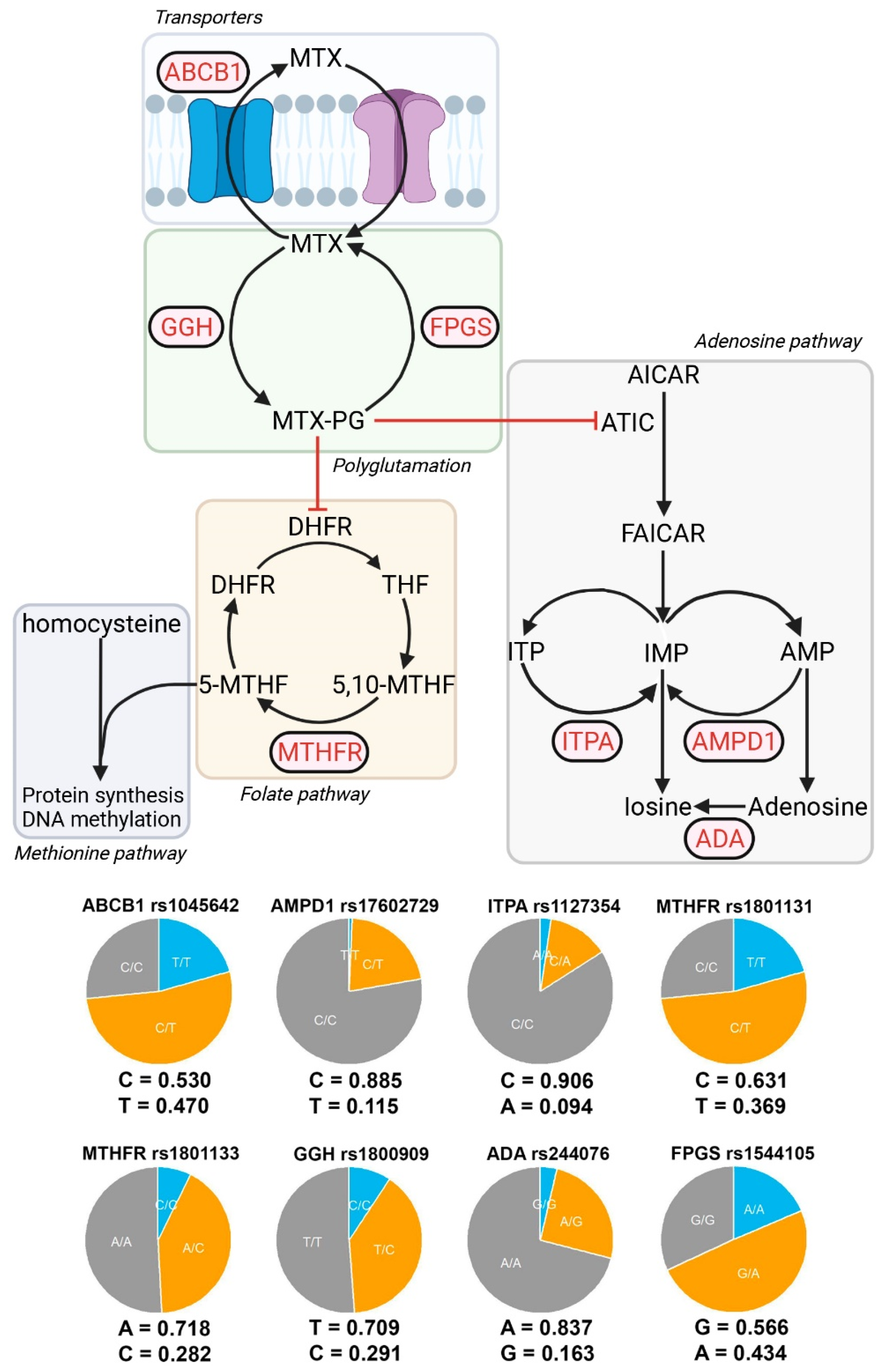
2.2. DNA Extraction and Genotyping
- -
- MTX extraction pumps: ABCB1 rs1045642 (3435C>T).
- -
- MTX polyglutamate formation: GGH rs1800909 (T16C) and FPGS rs1544105 (G2782A).
- -
- Folate cycle: MTHFR rs1801131 (C677T) and rs1801133 (A1298C).
- -
- Adenosine pathway: AMPD1 rs17602729 (34C>T), ITPA rs1127354 (94C>A), and ADA rs244076 (534A>G).
2.3. Variables of Interest
2.4. Statistical Analysis
3. Results
3.1. Clinical Characteristics
3.2. Allele Frequencies
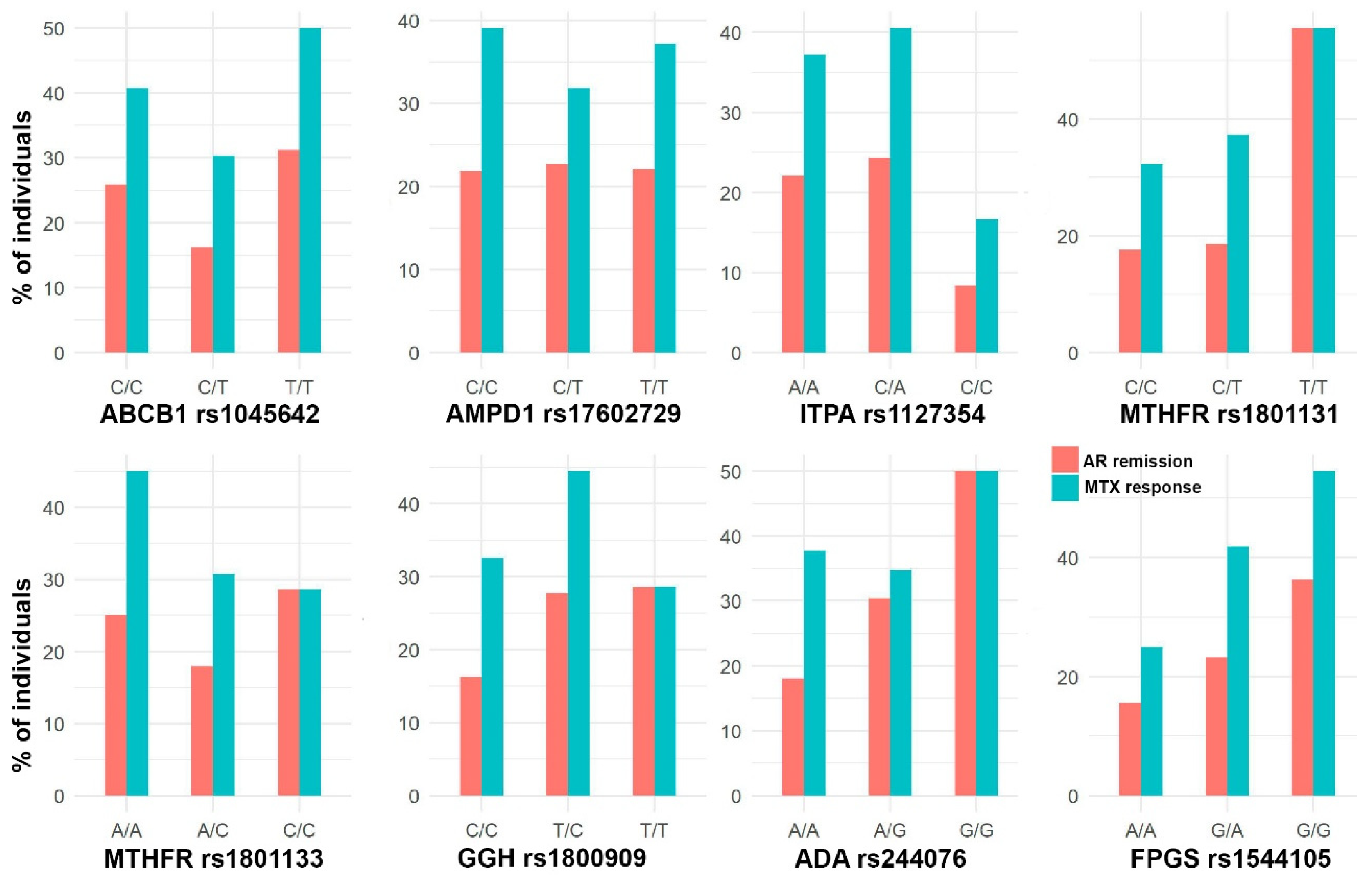
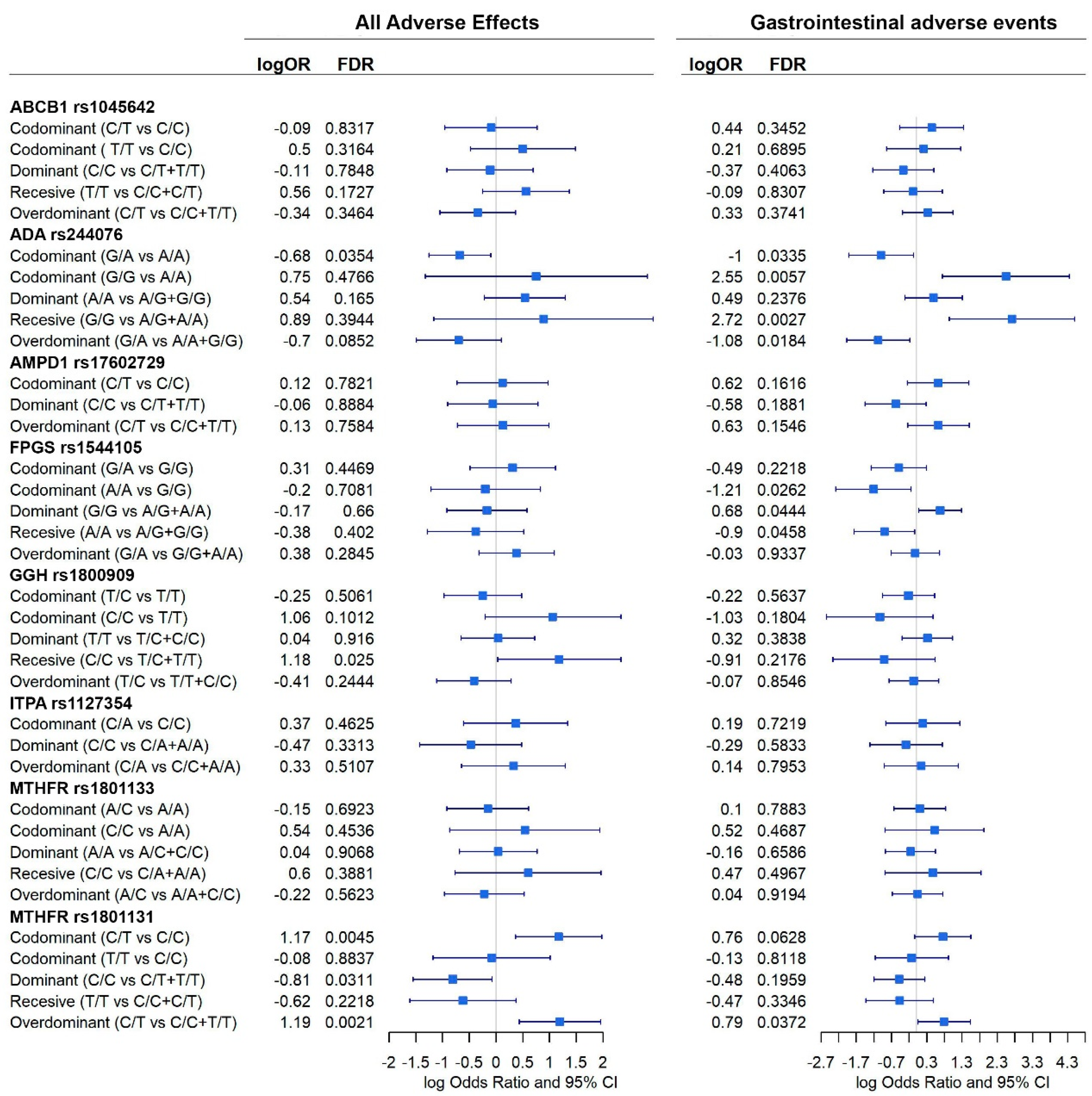
3.3. Association between Genetic Variants and MTX Efficacy
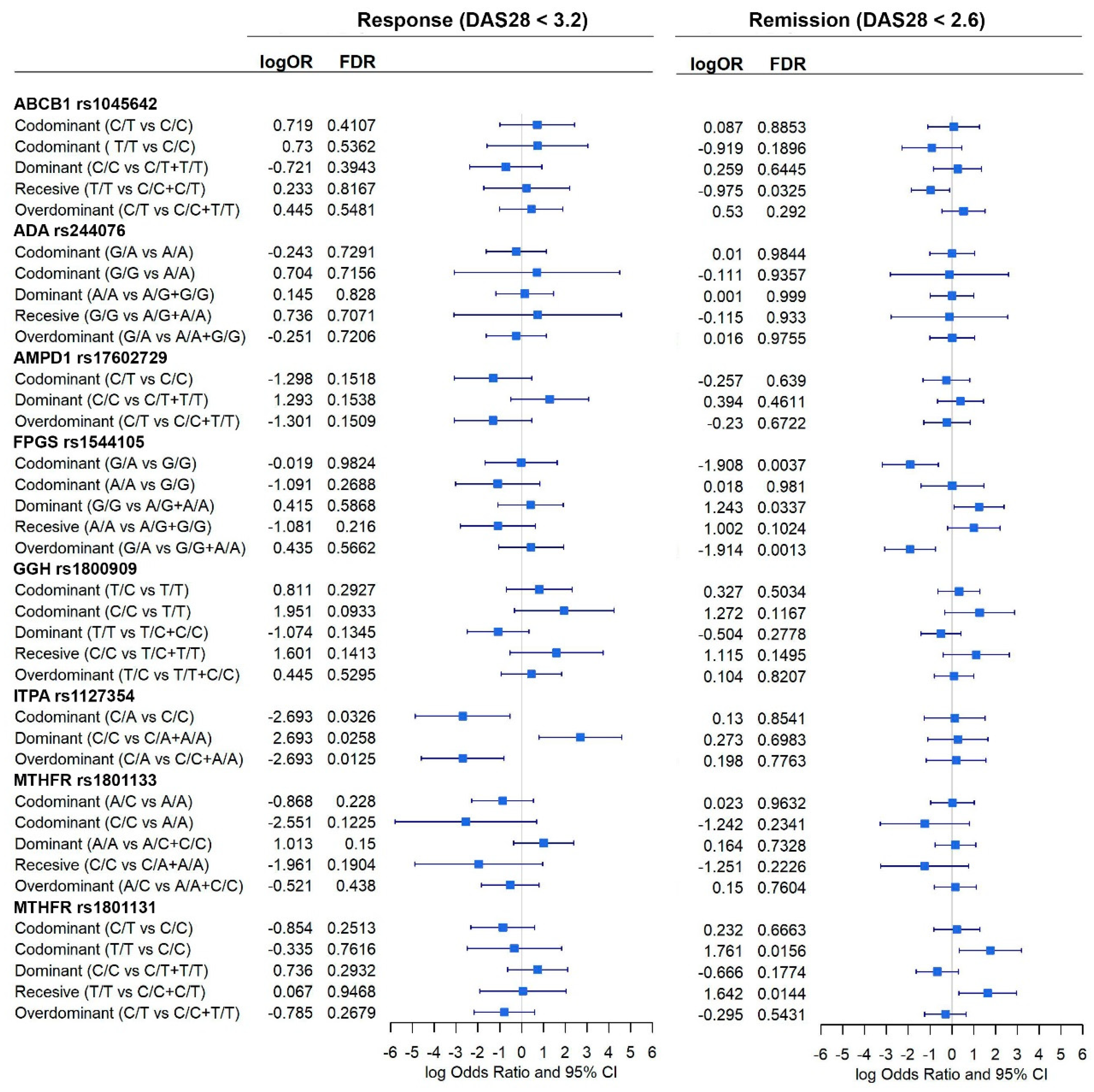
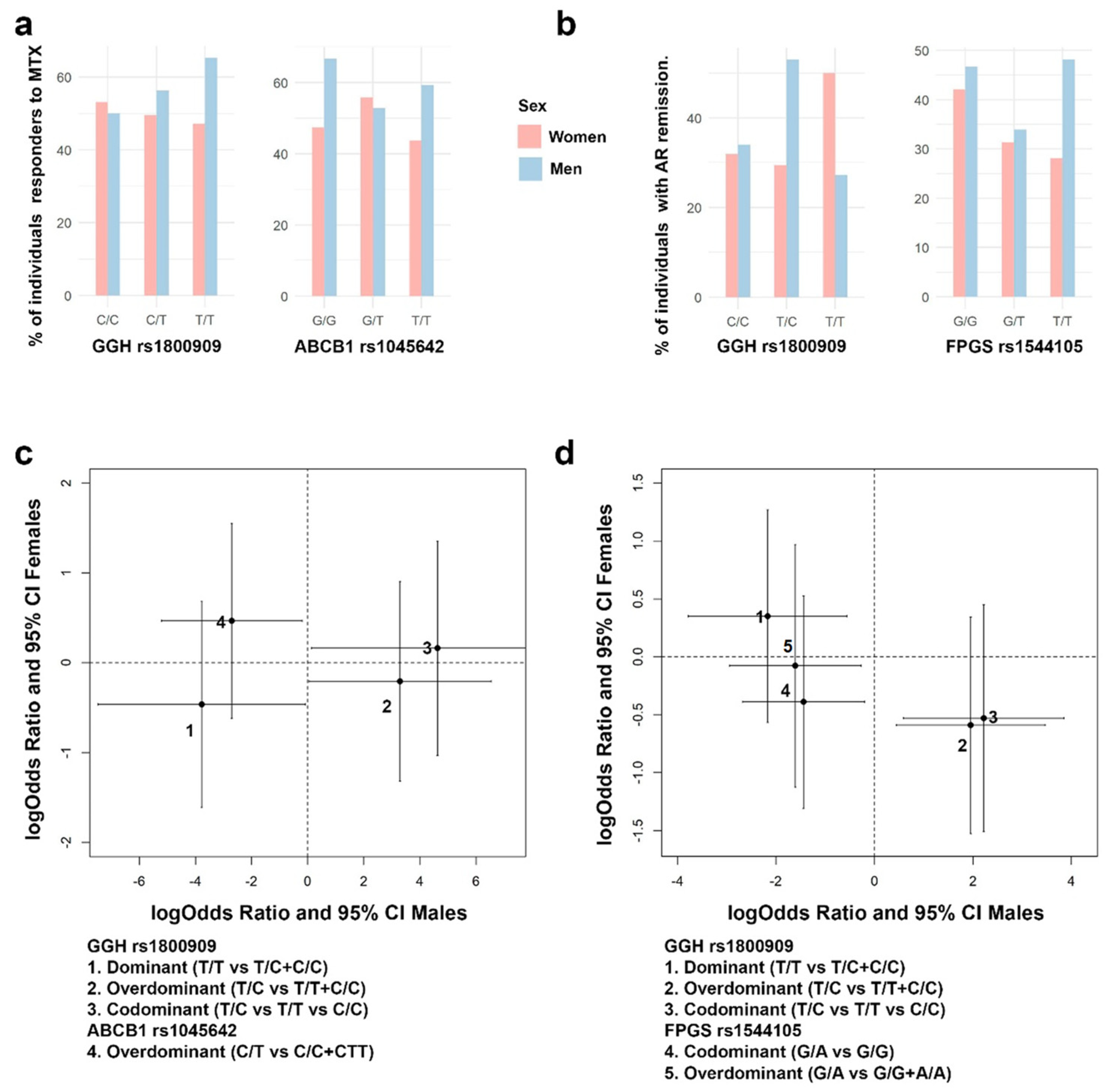
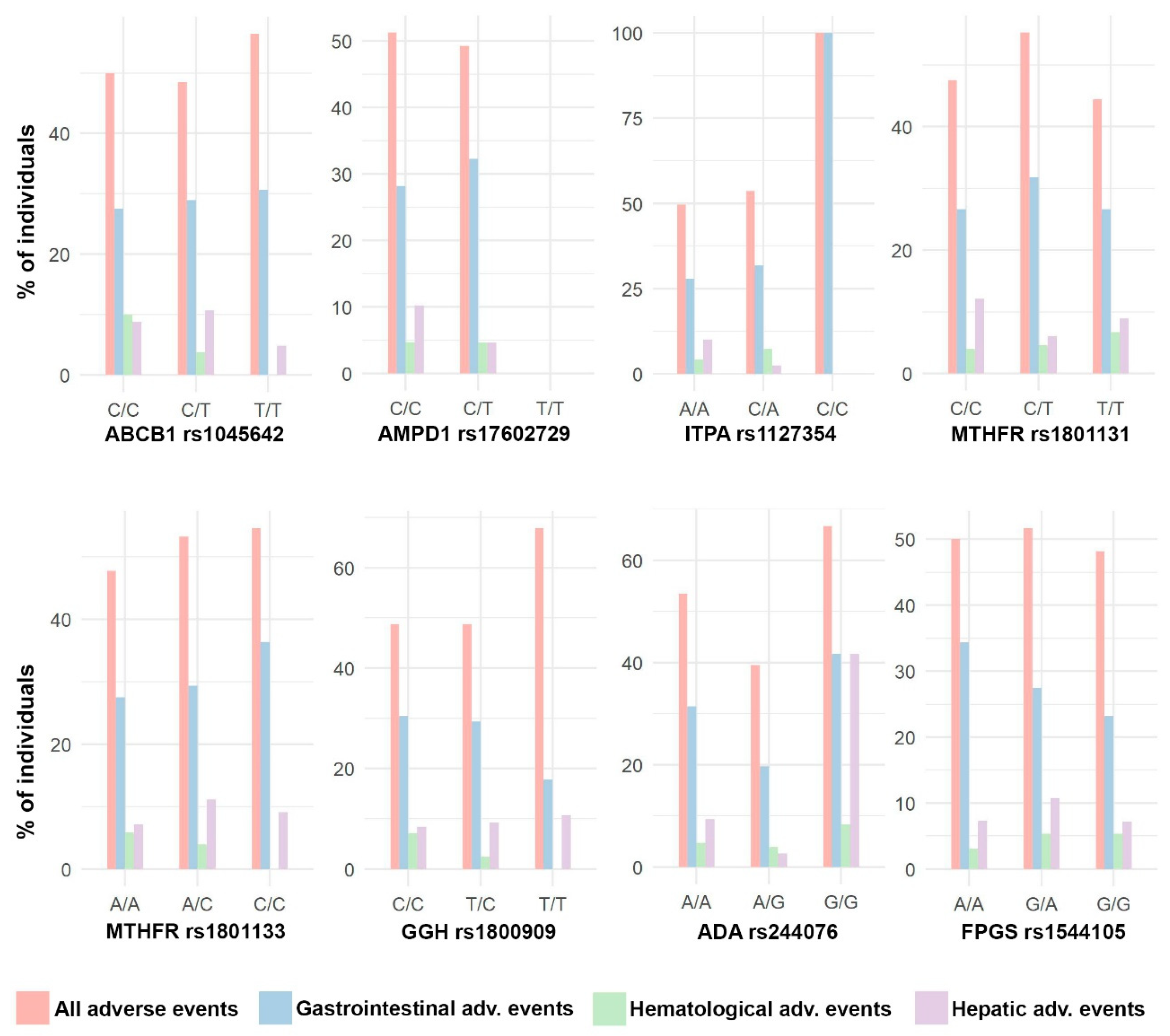
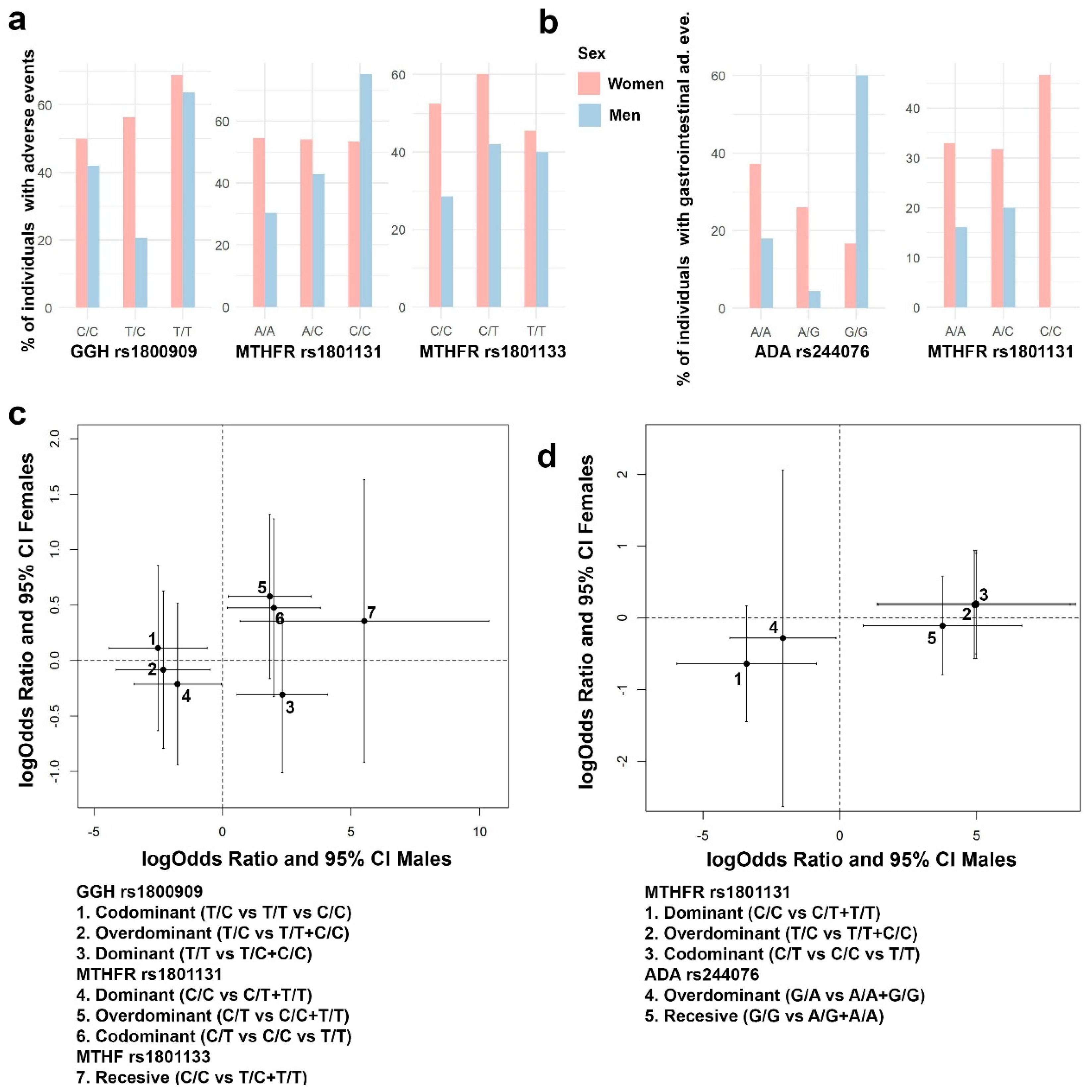
3.4. Association between Genetic Variants and MTX Adverse Events
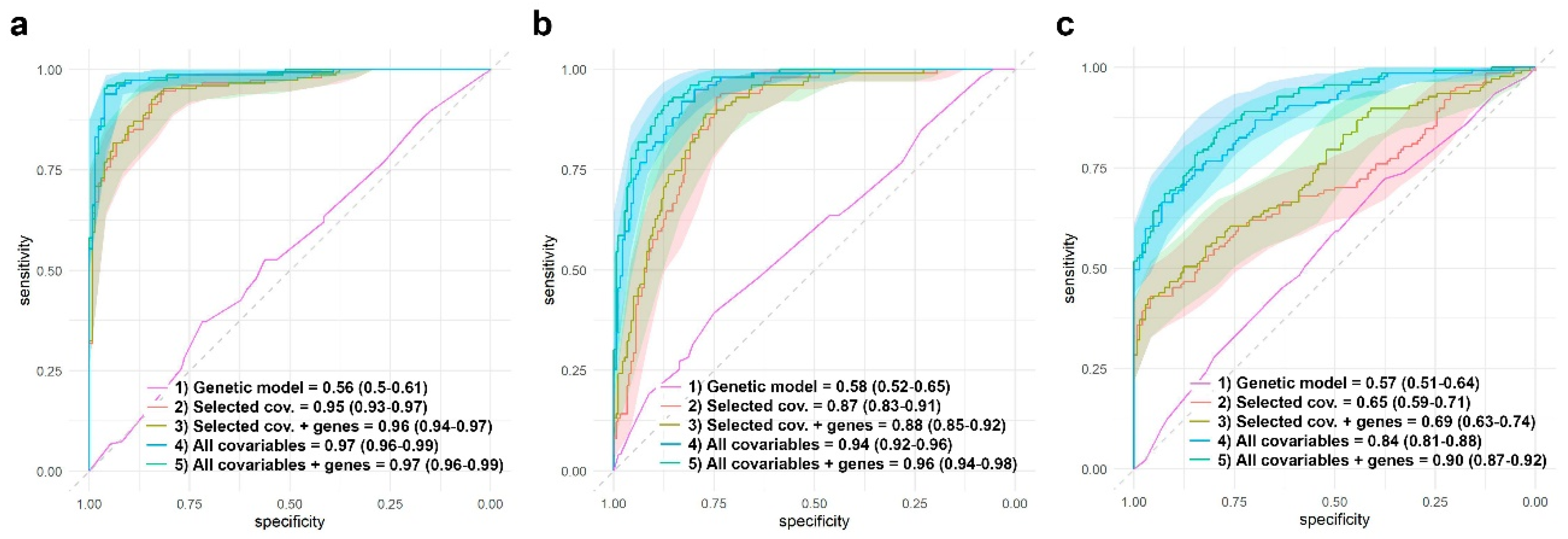
3.5. Assessing Genomic Effects to Predict RA Remission and Adverse Events
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Dinesh, P.; Rasool, M. Berberine mitigates IL-21/IL-21R mediated autophagic influx in fibroblast-like synoviocytes and regulates Th17/Treg imbalance in rheumatoid arthritis. Apoptosis 2019, 24, 644–661. [Google Scholar] [CrossRef] [PubMed]
- Smolen, J.S.; Aletaha, D.; Barton, A.; Burmester, G.R.; Emery, P.; Firestein, G.S.; Kavanaugh, A.; McInnes, I.B.; Solomon, D.H.; Strand, V.; et al. Rheumatoid arthritis. Nat. Rev. Dis. Prim. 2018, 4, 18001. [Google Scholar] [CrossRef] [PubMed]
- Smolen, J.S.; Aletaha, D. Rheumatoid arthritis therapy reappraisal: Strategies, opportunities and challenges. Nat. Rev. Rheumatol. 2015, 11, 276–289. [Google Scholar] [CrossRef] [PubMed]
- Ling, S.F.; Bluett, J. Pharmacogenetics of methotrexate response in rheumatoid arthritis: An update. Pharmacogenomics 2020, 21, 3–6. [Google Scholar] [CrossRef] [PubMed]
- Song, Z.; Hu, Y.; Liu, S.; Jiang, D.; Yi, Z.; Benjamin, M.M.; Zhao, R. The Role of Genetic Polymorphisms in High-Dose Methotrexate Toxicity and Response in Hematological Malignancies: A Systematic Review and Meta-Analysis. Front Pharm. 2021, 12, 757464. [Google Scholar] [CrossRef]
- Zhang, K.X.; Ip, C.K.; Chung, S.K.; Lei, K.K.; Zhang, Y.Q.; Liu, L.; Wong, V.K.W. Drug-resistance in rheumatoid arthritis: The role of p53 gene mutations, ABC family transporters and personal factors. Curr. Opin. Pharm. 2020, 54, 59–71. [Google Scholar] [CrossRef]
- Huang, J.; Fan, H.; Qiu, Q.; Liu, K.; Lv, S.; Li, J.; Yang, H.; Shu, X.; Xu, Y.; Lu, X.; et al. Are gene polymorphisms related to adverse events of methotrexate in patients with rheumatoid arthritis? A retrospective cohort study based on an updated meta-analysis. Ther. Adv. Chronic. Dis. 2020, 11, 2040622320916026. [Google Scholar] [CrossRef]
- He, X.; Sun, M.; Liang, S.; Li, M.; Li, L.; Yang, Y. Association between ABCB1 C3435T polymorphism and methotrexate treatment outcomes in rheumatoid arthritis patients: A meta-analysis. Pharmacogenomics 2019, 20, 381–392. [Google Scholar] [CrossRef]
- Taylor, J.C.; Bongartz, T.; Massey, J.; Mifsud, B.; Spiliopoulou, A.; Scott, I.C.; Wang, J.; Morgan, M.; Plant, D.; Colombo, M.; et al. Genome-wide association study of response to methotrexate in early rheumatoid arthritis patients. Pharm. J. 2018, 18, 528–538. [Google Scholar] [CrossRef]
- De Rotte, M.; Pluijm, S.M.F.; de Jong, P.H.P.; Bulatovic Calasan, M.; Wulffraat, N.M.; Weel, A.; Lindemans, J.; Hazes, J.M.W.; de Jonge, R. Development and validation of a prognostic multivariable model to predict insufficient clinical response to methotrexate in rheumatoid arthritis. PLoS ONE 2018, 13, e0208534. [Google Scholar] [CrossRef] [Green Version]
- Szostak, B.; Machaj, F.; Rosik, J.; Pawlik, A. Using pharmacogenetics to predict methotrexate response in rheumatoid arthritis patients. Expert. Opin. Drug. Metab. Toxicol. 2020, 16, 617–626. [Google Scholar] [CrossRef] [PubMed]
- Machaj, F.; Rosik, J.; Szostak, B.; Pawlik, A. The evolution in our understanding of the genetics of rheumatoid arthritis and the impact on novel drug discovery. Expert. Opin. Drug. Discov. 2020, 15, 85–99. [Google Scholar] [CrossRef] [PubMed]
- Sundbaum, J.K.; Baecklund, E.; Eriksson, N.; Hallberg, P.; Kohnke, H.; Wadelius, M. MTHFR, TYMS and SLCO1B1 polymorphisms and adverse liver effects of methotrexate in rheumatoid arthritis. Pharmacogenomics 2020, 21, 337–346. [Google Scholar] [CrossRef] [PubMed]
- Londono, J.; Saldarriaga, E.L.; Rueda, J.C.; Giraldo-Bustos, R.; Angarita, J.I.; Restrepo, L.; Ballesteros-Munoz, J.; Gonzalez, C.; Ospina, M.J.; Arias-Correal, S.; et al. Pharmacogenetic aspects of methotrexate in a cohort of Colombian patients with rheumatoid arthritis. Biomed. Rep. 2020, 13, 34. [Google Scholar] [CrossRef] [PubMed]
- Arida, A.; Nezos, A.; Papadaki, I.; Sfikakis, P.P.; Mavragani, C.P. Osteoprotegerin and MTHFR gene variations in rheumatoid arthritis: Association with disease susceptibility and markers of subclinical atherosclerosis. Sci. Rep. 2022, 12, 9534. [Google Scholar] [CrossRef]
- Melikoglu, M.A.; Balkan, E. Can we predict unresponsiveness to methotrexate in rheumatoid arthritis? A pharmacogenetic study. Inflammopharmacology 2022, 30, 193–197. [Google Scholar] [CrossRef]
- Dedmon, L.E. The genetics of rheumatoid arthritis. Rheumatology 2020, 59, 2661–2670. [Google Scholar] [CrossRef]
- Mezzalira, S.; Toffoli, G. The effects of sex on pharmacogenetically guided drug treatment. Pharmacogenomics 2021, 22, 959–962. [Google Scholar] [CrossRef]
- Sha, H.X.; Veerapen, K.; Chow, S.K.; Gun, S.C.; Lau, I.S.; Lim, R.L.H.; Zulkifli, Z.; Yow, Y.Y.; Peh, S.C.; Hwang, J.S. Genetic variations in methotrexate metabolic pathway genes influence methotrexate responses in rheumatoid arthritis patients in Malaysia. Sci. Rep. 2022, 12, 11844. [Google Scholar] [CrossRef]
- Escudero-Contreras, A.; Lopez-Medina, C.; Collantes-Estevez, E.; Ortega-Castro, R.; Calvo-Gutierrez, J.; Mena-Vazquez, N.; Panero-Lamothe, B.; Manzanares-Martin, B.; Caliz-Caliz, R.; Jimenez-Morales, A.; et al. Genetic Polymorphisms of GGH and ABCC2 Are Associated with Methotrexate Intolerance in Patients with Rheumatoid Arthritis. J. Clin. Med. 2021, 10, 4070. [Google Scholar] [CrossRef]
- Muralidharan, N.; Sundaram, R.; Kodidela, S.; Chengappa, K.G.; Mariaselvam, C.M.; Misra, D.P.; Negi, V.S. Folyl polyglutamate synthethase (FPGS) gene polymorphisms may influence methotrexate adverse events in South Indian Tamil Rheumatoid Arthritis patients. Pharm. J. 2020, 20, 342–349. [Google Scholar] [CrossRef] [PubMed]
- D’Cruz, L.G.; McEleney, K.G.; Tan, K.B.C.; Shukla, P.; Gardiner, P.V.; Connolly, P.; Conway, C.; Cobice, D.; Gibson, D.S. Clinical and Laboratory Associations with Methotrexate Metabolism Gene Polymorphisms in Rheumatoid Arthritis. J. Pers. Med. 2020, 10, 149. [Google Scholar] [CrossRef] [PubMed]
- Shao, W.; Yuan, Y.; Li, Y. Association Between MTHFR C677T Polymorphism and Methotrexate Treatment Outcome in Rheumatoid Arthritis Patients: A Systematic Review and Meta-Analysis. Genet. Test. Mol. Biomarkers. 2017, 21, 275–285. [Google Scholar] [CrossRef] [PubMed]
- Qiu, Q.; Huang, J.; Lin, Y.; Shu, X.; Fan, H.; Tu, Z.; Zhou, Y.; Xiao, C. Polymorphisms and pharmacogenomics for the toxicity of methotrexate monotherapy in patients with rheumatoid arthritis: A systematic review and meta-analysis. Med. Baltim. 2017, 96, e6337. [Google Scholar] [CrossRef] [PubMed]
- Qiu, Q.; Huang, J.; Shu, X.; Fan, H.; Zhou, Y.; Xiao, C. Polymorphisms and Pharmacogenomics for the Clinical Efficacy of Methotrexate in Patients with Rheumatoid Arthritis: A Systematic Review and Meta-analysis. Sci. Rep. 2017, 7, 44015. [Google Scholar] [CrossRef] [Green Version]
- Chen, Y.; Zou, K.; Sun, J.; Yang, Y.; Liu, G. Are gene polymorphisms related to treatment outcomes of methotrexate in patients with rheumatoid arthritis? A systematic review and meta-analysis. Pharmacogenomics 2017, 18, 175–195. [Google Scholar] [CrossRef]
- Li, X.; Hu, M.; Li, W.; Gu, L.; Chen, M.; Ding, H.; Vanarsa, K.; Du, Y. The association between reduced folate carrier-1 gene 80G/A polymorphism and methotrexate efficacy or methotrexate related-toxicity in rheumatoid arthritis: A meta-analysis. Int. Immunopharmacol. 2016, 38, 8–15. [Google Scholar] [CrossRef]
- Aletaha, D.; Neogi, T.; Silman, A.J.; Funovits, J.; Felson, D.T.; Bingham, C.O., 3rd; Birnbaum, N.S.; Burmester, G.R.; Bykerk, V.P.; Cohen, M.D.; et al. 2010 Rheumatoid arthritis classification criteria: An American College of Rheumatology/European League Against Rheumatism collaborative initiative. Arthritis Rheum. 2010, 62, 2569–2581. [Google Scholar] [CrossRef]
- Takanashi, S.; Kaneko, Y.; Takeuchi, T. CDAI and DAS28 in the management of rheumatoid arthritis in clinical practice. Ann. Rheum. Dis. 2020, 79, 671–674. [Google Scholar] [CrossRef]
- Van Riel, P.L.; Reekers, P.; van de Putte, L.B.; Gribnau, F.W. Association of HLA antigens, toxic reactions and therapeutic response to auranofin and aurothioglucose in patients with rheumatoid arthritis. Tissue Antigens 1983, 22, 194–199. [Google Scholar] [CrossRef]
- Van der Heijde, D.M.; van’t Hof, M.A.; van Riel, P.L.; Theunisse, L.A.; Lubberts, E.W.; van Leeuwen, M.A.; van Rijswijk, M.H.; van de Putte, L.B. Judging disease activity in clinical practice in rheumatoid arthritis: First step in the development of a disease activity score. Ann. Rheum. Dis. 1990, 49, 916–920. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Prevoo, M.L.; van’t Hof, M.A.; Kuper, H.H.; van Leeuwen, M.A.; van de Putte, L.B.; van Riel, P.L. Modified disease activity scores that include twenty-eight-joint counts. Development and validation in a prospective longitudinal study of patients with rheumatoid arthritis. Arthritis Rheum. 1995, 38, 44–48. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vandenbroucke, J.P.; von Elm, E.; Altman, D.G.; Gotzsche, P.C.; Mulrow, C.D.; Pocock, S.J.; Poole, C.; Schlesselman, J.J.; Egger, M.; Initiative, S. Strengthening the Reporting of Observational Studies in Epidemiology (STROBE): Explanation and elaboration. Int. J. Surg. 2014, 12, 1500–1524. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hakamata, J.; Hashiguchi, M.; Kaneko, Y.; Yamaoka, K.; Shimizu, M.; Maruyama, J.; Takeuchi, T.; Mochizuki, M. Risk factors for abnormal hepatic enzyme elevation by methotrexate treatment in patients with rheumatoid arthritis: A hospital based-cohort study. Mod. Rheumatol. 2018, 28, 611–620. [Google Scholar] [CrossRef]
| Type Study | Patients (n) | Country/ Region | Evaluation | Genes, SNP | Main Results | Ref. |
|---|---|---|---|---|---|---|
| Original article | 45 | Turkey | MTX efficacy through DAS28 | ABCB1 rs1045642 | 10 covariables and comorbidities included. ABCB1 heterozygous (C/T) is associated with non-response to MTX | [16] |
| Original article | 167 | Malaysia | MTX efficacy and toxicity | FPGS rs10106 | ATIC rs2372536 and rs4673993 associated with MTX adequate response in RA patients with Malay ancestry | [19] |
| GGH rs1145078, rs3758149 | ||||||
| ATIC rs2372536, rs4673993 | ||||||
| ITPA rs1127354 | ||||||
| Original article | 227 | Spain | Intolerance to MTX | RFC rs1051266 | Positive association with GGH AA + AG vs. GG. Inverse association with ABCC2 TT + TC vs. CC | [20] |
| GGH rs11545078 | ||||||
| MTHFR rs181133, rs1801131 | ||||||
| DHFR rs1105525 | ||||||
| SHMT1 rs1979277 | ||||||
| ITPA rs34743033 | ||||||
| ABCC2 rs717620 | ||||||
| ABCB1 rs1045642 | ||||||
| SLCOB1 rs11045879 | ||||||
| Original article | 381 | Sweden | Adverse liver effects | MTHFR rs1801131 | MTHFR nominally associated but did not pass correction for clinical factors | [13] |
| TYMS rs34743033 | ||||||
| SLCOB1 rs11045879 | ||||||
| Original article | 330 | India (Tamil) | Response to treatment (DAS28); Adverse events | FPGS rs1544105, rs10106 | FPGS heterozygous rs1544105 and rs10106 associated with adverse events | [21] |
| GGH rs11545078, rs3758149 | ||||||
| Original article | 400 | Colombia | Efficacy (DAS28 < 3.2); Toxicity | MTHFR rs1801133, rs1801131 | MTHFR rs1801133 and rs1801131 associated with efficacy of MTX. DHFR and ATIC associated with toxicity | [14] |
| ATIC rs2371536 | ||||||
| RFC1 rs1051266 | ||||||
| FPGS rs1554105 | ||||||
| DHFR rs10072026; | ||||||
| Original article | 100 | Northern Ireland | MTX-7-OH and MTX2PG–5PG metabolite levels | ABCC1 rs246240 | MTHFR rs1476413 TT CC alleles associated with plasma poly-glutamate metabolites | [22] |
| MTHFR rs1476413, rs4846051, rs17421511 | ||||||
| ABCG2 rs2231142 | ||||||
| ABCC2 rs717620, rs3740065 | ||||||
| SCLO1B1 rs4846051 | ||||||
| ABCB1 rs10280623 | ||||||
| ATIC rs16853826 | ||||||
| Original article | 114 | Japan | Hepatotoxicity; Hepatic enzyme elevation | ABCB1 rs1045642 | Multivariable analysis corrected for different confounding variables. ABCB1 and ATIC associated with MTX-induced abnormal hepatic enzyme elevation | [21] |
| ABCC2 rs717620 | ||||||
| RFC1 rs1051266 | ||||||
| FPGS rs1544105 | ||||||
| GGH rs3758149 | ||||||
| MTHFR rs1801131 | ||||||
| ATIC rs16853826 | ||||||
| Retrospective cohort study | 162 | China (Han Chinese) | MTX toxicity | MTHFR rs1801133, rs1801131 | MTHFR rs1801133 associated with MTX-related toxicity in East Asians. ATIC, ABCB1 and RFC1 associated in Europeans. RFC1 associated with MTX-related toxicity in the retrospective cohort study | [7] |
| ATIC rs2372536; | ||||||
| ABCB1 rs1045642 | ||||||
| MTR2756 rs1805087 | ||||||
| MTRR rs1801394 | ||||||
| FPGS rs10106 | ||||||
| AMPD1 rs17602729 | ||||||
| ITPA rs1127354 | ||||||
| GGH rs3758149, rs1800909, rs11545078 | ||||||
| ABCC2 rs4148396 | ||||||
| ABCG2 rs2231142 | ||||||
| FPGS rs1544105 | ||||||
| TYMS rs34743033 | ||||||
| ADORA2A rs2267076, rs2298383 | ||||||
| MTHFD1 rs2236225 | ||||||
| Meta-Analysis | 16 | Multiple | MTX efficacy (DAS28 < 3.2); MTX-related toxicity | MTHFR rs1801131, rs1801133 | Associated with toxicity but not efficacy in Asian and European populations. Model corrected for folic acid us | [23] |
| Meta-Analysis | 30 | Multiple | MTX efficacy (DAS28) | 34 genes 125 SNP included in the study | MTHFR rs1801131, ATIC rs2372536, SCL19A1 rs2838956, and SLC19A1 rs7499 associated with MTX efficacy | [24] |
| Meta-Analysis | 42 | Multiple | MTX toxicity | 28 genes with 88 SNP included in the study | RFC1 (rs1051266) was found to be associated with MTX toxicity | [25] |
| Meta-Analysis | 68 | Multiple | MTX efficacy (DAS28 < 3.2); MTX-related toxicity | 42 genes 152 SNP included in the study | 14 studies adjusted for sex and age. MTHFR rs1801131, TYMS, and AMPD1 associated with responsiveness. ATIC rs2372536 and ITPA rs1127354 associated with non-responsiveness. MTHFR rs1801131, ATIC rs2372536, and TYMS associated with adverse events. GGH, SLCO1B1 rs4149056, SLC19A1 rs2838956 rs1131596 rs2838958 associated with absenting overall adverse events | [26] |
| Meta-Analysis | 17 | European | MTX efficacy (DAS28 < 3.2); MTX-related toxicity | RFC1 rs1051266 | RFC1 associated with MTX efficacy in Asians | [27] |
| Asian | ||||||
| Meta-Analysis | 16 | Multiple | MTX treatment response (DAS28 > 3.2) | ABCB1 rs1045642 | ABCB1 associated with higher drug resistance. ABCG2 is associated with higher drug resistance | [6] |
| ABCC2 rs717620 | ||||||
| ABCC3; ABCC4; ABCC5; ABCG2 rs2231142 | ||||||
| Meta-Analysis | 12 | Multiple | MTX efficacy (DAS28 < 3.2); MTX-related toxicity | ABCB1 rs1045642 | ABCB1 Associated with MTX efficacy under a recessive model. Association found mainly in Asian not in European populations | [8] |
| Meta-Analysis | 39 | European | MTX toxicity | MTHFR rs1801133, rs1801131 | MTHFR rs1801133 associated with MTX-related toxicity in East Asians. ATIC, ABCB1, and RFC1 associated in Europeans. RFC1 associated with MTX-related toxicity in the retrospective cohort study | [7] |
| ATIC rs2372536; | ||||||
| ABCB1 rs1045642 | ||||||
| MTR2756 rs1805087 | ||||||
| MTRR rs1801394 | ||||||
| FPGS rs10106 | ||||||
| AMPD1 rs17602729 | ||||||
| ITPA rs1127354 | ||||||
| GGH rs3758149, rs1800909, rs11545078 | ||||||
| ABCC2 rs4148396 | ||||||
| ABCG2 rs2231142 | ||||||
| FPGS rs1544105 | ||||||
| TYMS rs34743033 | ||||||
| ADORA2A rs2267076, rs2298383 | ||||||
| MTHFD1 rs2236225 |
| Response to MTX | RA Remission | Adverse Events | |||||
|---|---|---|---|---|---|---|---|
| Individuals | DAS28 < 3.2 | DAS28 > 3.2 | DAS28 < 2.6 | DAS28 > 2.6 | YES | NO | |
| Number | 301 | 148 | 135 | 99 | 202 | 152 | 149 |
| Demographic | |||||||
| Age | 49 (41–58) | 53 (45–63.3) | 47 (38–56) | 52 (44.5–64) | 48 (38.5–56) | 48 (39.5–56) | 52 (41–62) |
| Sex (Women) | 202 (67.1%) | 94 (63.5%) | 94 (69.6) | 61 (61.6%) | 141 (69.8%) | 113 (74.3%) | 89 (59.7%) |
| Educational attainment | |||||||
| Before high school | 98 (32.5%) | 44 (29.7%) | 44 (32.6%) | 29 (29.2%) | 69 (34.1%) | 46 (30.2%) | 52 (34.8%) |
| High school | 105 (34.9%) | 54 (36.5%) | 49 (36.3%) | 33 (33.1%) | 72 (35.6%) | 54 (35.5%) | 51 (34.3%) |
| University studies | 98 (32.5%) | 50 (33.8%) | 42 (32.6%) | 37 (37.4%) | 61 (30.2%) | 52 (34.2%) | 46 (30.9%) |
| Activity | |||||||
| Sedentary | 57 (18.9%) | 28 (18.9) | 23 (17.1%) | 19 (19.2%) | 38 (18.8%) | 32 (21.1%) | 25 (16.8%) |
| Mixed Active | 153 (50.8%) | 71 (47.9%) | 72 (53.3%) | 46 (46.4%) | 107 (52.9%) | 79 (51.9%) | 74 (49.7%) |
| Active | 91 (30.2%) | 49 (33.1%) | 40 (29.6%) | 34 (34.3%) | 57 (28.2) | 41 (26.9%) | 50 (33.6%) |
| Lifestyle | |||||||
| Smokers | |||||||
| Never | 118 (39.2%) | 57 (38.5%) | 49 (36.3%) | 41 (41.4%) | 77 (38.1%) | 62 (40.7%) | 56 (37.6%) |
| Ex-smoker | 97 (32.2%) | 63 (42.6%) | 33 (24.4%) | 41 (41.4%) | 56 (27.8%) | 46 (30.3%) | 51 (34.2%) |
| Active smoker | 86 (28.6%) | 28 (18.9%) | 53 (39.3%) | 17 (17.2%) | 69 (34.2%) | 44 (28.9%) | 42 (28.2%) |
| Alcohol | |||||||
| Never | 181 (60.1%) | 83 (56.1%) | 86 (63.7%) | 55 (55.5%) | 126 (62.4%) | 97 (63.8%) | 84 (56.4%) |
| Ex-alcohol drinker | 33 (10.9%) | 18 (12.2%) | 14 (10.4%) | 8 (8.1%) | 25 (12.4%) | 15 (7.4%) | 18 (12.1%) |
| Active drinker | 87 (28.9%) | 47 (31.8%) | 35 (25.6%) | 36 (36.3%) | 51 (25.2%) | 40 (19.8%) | 47 (31.5%) |
| Tea consumption | 29 (9.6%) | 11 (7.4%) | 16 (11.9%) | 9 (9.1%) | 20 (9.9%) | 17 (11.2%) | 12 (8.1%) |
| Coffee consumption | 185 (61.7%) | 80 (54.1%) | 76 (56.3%) | 57 (57.6%) | 128 (63.4%) | 93 (61.2%) | 92 (61.7%) |
| Comorbidities | |||||||
| Any comorb. | 234 (77.7%) | 113 (76.4%) | 105 (77.8%) | 74 (74.7%) | 160 (79.2%) | 121 (79.6%) | 113 (75.8%) |
| Cardiovascular comorb. | 237 (78.7%) | 32 (21.6%) | 28 (20.7%) | 73 (73.7%) | 164 (81.2%) | 123 (80.9%) | 114 (76.5%) |
| Number of risk factors | |||||||
| 0 | 64 (21.3%) | 40 (27.0%) | 20 (14.8) | 26 (26.3%) | 38 (18.8%) | 29 (19.1%) | 35 (23.5%) |
| 1 | 94 (31.2%) | 55 (37.2%) | 39 (28.9%) | 41 (41.4%) | 53 (26.2%) | 39 (25.7%) | 55 (36.9%) |
| 2 | 79 (26.2%) | 37 (25%) | 42 (31.1%) | 21 (21.2%) | 58 (28.7%) | 45 (29.6%) | 34 (22.8%) |
| 3 | 52 (17.3%) | 23 (15.5%) | 32 (23.7%) | 10 (10.1%) | 42 (20.8%) | 30 (19.7%) | 22 (14.8%) |
| 4 | 10 (3.3%) | 3 (2%) | 7 (5.2%) | 1 (1%) | 9 (4.5%) | 7 (4.6%) | 3 (2.0%) |
| 5 | 2 (0.6%) | 0 | 2 (1.5%) | 0 | 2 (0.9%) | 2 (1.3%) | 0 |
| High blood pressure | 114 (37.9%) | 46 (31.1%) | 58 (42.9%) | 30 (30.3%) | 84 (41.6%) | 61 (40.1%) | 53 (35.6%) |
| Diabetes Mellitus | 49 (16.3%) | 20 (13.5%) | 29 (31.5%) | 13 (13.1%) | 36 (17.8%) | 32 (21.1%) | 17 (11.4%) |
| Hypercholesterolemia | 137 (45.5%) | 59 (39.9%) | 78 (57.8%) | 35 (35.4%) | 102 (50.5%) | 74 (48.7%) | 63 (42.3%) |
| Obesity | 72 (23.9%) | 31 (20.9%) | 41 (30.4%) | 22 (22.2%) | 50 (24.8%) | 46 (30.3%) | 26 (17.4%) |
| Ischemic heart disease | 20 (6.7%) | 12 (8.1%) | 8 (5.9%) | 8 (8.1%) | 12 (5.9%) | 12 (7.9%) | 8 (5.4%) |
| Any cardiovascular pathology | 63 (20.9%) | 34 (22.9%) | 39 (38.9%) | 24 (24.2%) | 39 (19.3%) | 30 (19.7%) | 33 (22.1%) |
| Vascular incidents | 46 (15.3%) | 19 (12.8%) | 27 (20%) | 17 (17.2%) | 29 (14.4%) | 26 (17.1%) | 20 (13.4%) |
| Thyroid diseases | 35 (11.6%) | 17 (11.5%) | 18 (13.3%) | 11 (11.1%) | 24 (11.9%) | 21 (13.8%) | 14 (9.4%) |
| Renal impairment | 12 (4.0%) | 5 (3.4%) | 7 (5.2%) | 3 (3.1%) | 9 (4.5%) | 9 (5.9%) | 3 (2.0%) |
| Osteoporosis | 73 (24.3%) | 33 (22.3%) | 31 (22.9%) | 22 (22.2%) | 51 (25.2%) | 38 (25.0%) | 35 (23.5%) |
| Tuberculosis | 42 (14.0%) | 8 (5.4%) | 33 (22.9%) | 4 (4.1%) | 38 (18.8%) | 26 (17.1%) | 16 (10.7%) |
| Baldness | 26 (8.6%) | 8 (5.4%) | 13 (9.6%) | 3 (3.0%) | 23 (11.4%) | 25 (16.4%) | 1 (0.7%) |
| Hypertransaminasemia | 27 (9.0%) | 12 (8.2%) | 15 (11.1%) | 5 (5.1%) | 22 (10.9%) | 27 (17.8%) | 0 |
| Depression | 63 (20.9%) | 25 (16.9%) | 28 (20.7%) | 13 (13.1%) | 50 (24.8%) | 39 (25.6%) | 24 (16.1%) |
| Diseases Manifestations | |||||||
| DAS28- | 4.3 (3.8–5.1) | 3.8 (3.6–4.1) | 4.9 (4.5–5.4) | 3.8 (3.5–4.2) | 4.7 (4.1–5.3) | 4.4 (3.9–5.1) | 4.1 (3.7–4.8) |
| Extraarticular manifestations | 71 (23.6%) | 27 (18.2%) | 44 (32.65) | 12 (12.1%) | 59 (29.2) | 44 (28.9) | 27 (18.1%) |
| Radiological erosions in hands | 163 (54.1%) | 65 (43.9%) | 98 (72.6%) | 28 (28.3%) | 135 (66.8%) | 88 (57.9%) | 75 (50.3%) |
| Sjogren syndrome | 33 (11.0%) | 15 (10.1%) | 18 (13.3) | 9 (9.1%) | 24 (11.9%) | 18 (11.8%) | 15 (10.1%) |
| Carpal tunnel syndrome | 20 (9.9%) | 7 (4.7%) | 13 (9.6$) | 4 (4.1%) | 16 (7.9%) | 11 (7.2%) | 9 (6.0%) |
| Pulmonary affection | 14 (4.7%) | 5 (3.45) | 9 (6.7%) | 3 (3.0%) | 11 (5.4%) | 9 (5.9%) | 5 (3.4%) |
| Pharmacological variables | |||||||
| Use DMARD previously | 81 (26.9%) | 34 (22.9%) | 47 (34.8%) | 16 (16.2%) | 65 (32.2%) | 47 (30.9%) | 34 (22.9%) |
| Number previous DMARD | |||||||
| 0 | 223 (74.1%) | 124 (83.8%) | 99 (73.3%) | 83 (83.8%) | 140 (69.3%) | 108 (71.1%) | 115 (77.2%) |
| 1 | 54 (17.9%) | 21 (14.2%) | 33 (24.4%) | 13 (13.1%) | 41 (20.3%) | 29 (19.1%) | 25 (16.8%) |
| 2 | 16 (5.3%) | 8 (5.4%) | 8 (5.9%) | 3 (3.0%) | 13 (6.4%) | 9 (5.9%) | 7 (4.7%) |
| 3 | 6 (2%) | 0 | 6 (4.4%) | 0 | 6 (3.0%) | 4 (2.6%) | 2 (1.3%) |
| 4 | 2 (0.7%) | 0 | 2 (1.5%) | 0 | 2 (1%) | 2 (1.3%) | 0 |
| Time between diagnosis and treatment with DMARD (days) | 0 (0–3) | 0 (0–2) | 1 (0–3) | 0 (0–2.1) | 1 (0–9) | 0 (0–4) | 0 (0–3) |
| Initial dose of prednisona (mg/day) | 10 (5–10) | 5 (5–10) | 10 (5–10) | 5 (5–10) | 10 (5–10) | 10 (5–10 | 10 (5–10 |
| Dose of folic acid (mg/week) | 5 (5–5) | 5 (5–5) | 5 (5–10) | 5 (5–5) | 5 (5–10) | 5 (5–5) | 5 (5–10) |
| Maximum dose of MTX (mg/week) | 15 (15–20) | 15 (15–20) | 20 (15–20) | 15 (12.5–15) | 17.5 (15–20) | 15 (15–20) | 15 (15–20) |
| Maximum tolerated dose of MTX (mg/week) | 15 (15–20) | 15 (12.5–17.5) | 15 (15–20) | 15 (12.5–15) | 17.5 (15–20) | 15 (15–20) | 15 (15–20) |
| Time between diagnosis and treatment with MTX (days) | 1 (0–15) | 0 (0–7.2) | 2 (0–14) | 0 (0–4) | 2 (0–28.7) | 1 (0–22.5) | 1 (0–9) |
| MTX monotherapy duration (months) | 51 (20–101) | 86 (43–125) | 24 (10–60) | 84 (38–118.5) | 42 (15–88) | 47 (14.7–96.2) | 59 (27–103) |
| Antibodies | |||||||
| Rheumatoid Factor | |||||||
| Positive | 242 (80.4%) | 111 (75%) | 131 (97%) | 78 (78.9%) | 164 (81.2%) | 120 (78.9%) | 122 (81.9%) |
| Mean Value (DE) | 61.6 (21–176) | 51.2 (20.2–125) | 75 (30.1–198) | 44.9 (16.1–105.3) | 85.3 (25.6–202) | 63.8 (19.2–207.6) | 60 (25–159) |
| Antipeptide citrullinate (ACPA) | 193 (64.1%) | 91 (61.5%) | 102 (75.6%) | 59 (59.6%) | 134 (66.3%) | 97 (63.8%) | 96 (64.4%) |
| Antinuclear antibodies | 27 (9%) | 9 (6.1%) | 18 (13.3%) | 4 (4.0%) | 23 (11.2%) | 14 (9.2%) | 13 (8.7%) |
| Description of Adverse Effect | N | % |
|---|---|---|
| Adverse effects (total) | 152 | 50.50 |
| Haematologics | 14 | 4.65 |
| Leukopenia (alone or as part of pancytopenia) | 9 | 2.99 |
| Thrombocytopenia (alone or as part of pancytopenia) | 12 | 3.99 |
| Pancytopenia | 7 | 2.33 |
| Hepatic (elevated transaminases) | 27 | 8.97 |
| 2 × ULN | 13 | 4.32 |
| 3 × ULN | 8 | 2.66 |
| >3 × ULN | 6 | 1.99 |
| Gastrointestinal | 87 | 28.90 |
| Nausea, vomiting, fullness | 80 | 26.58 |
| Diarrhea | 10 | 3.32 |
| Neurological/general | 55 | 18.27 |
| Alopecia | 26 | 8.64 |
| Cutaneous | 13 | 4.32 |
| Rash | 8 | 2.66 |
| Others | 5 | 1.66 |
| Oral ulcers | 20 | 6.64 |
| Accelerated nodulosis | 6 | 1.99 |
| Interstitial involvement | 2 | 0.66 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ceballos, F.C.; Chamizo-Carmona, E.; Mata-Martín, C.; Carrasco-Cubero, C.; Aznar-Sánchez, J.J.; Veroz-González, R.; Rojas-Herrera, S.; Dorado, P.; LLerena, A. Pharmacogenetic Sex-Specific Effects of Methotrexate Response in Patients with Rheumatoid Arthritis. Pharmaceutics 2023, 15, 1661. https://doi.org/10.3390/pharmaceutics15061661
Ceballos FC, Chamizo-Carmona E, Mata-Martín C, Carrasco-Cubero C, Aznar-Sánchez JJ, Veroz-González R, Rojas-Herrera S, Dorado P, LLerena A. Pharmacogenetic Sex-Specific Effects of Methotrexate Response in Patients with Rheumatoid Arthritis. Pharmaceutics. 2023; 15(6):1661. https://doi.org/10.3390/pharmaceutics15061661
Chicago/Turabian StyleCeballos, Francisco C., Eugenio Chamizo-Carmona, Carmen Mata-Martín, Carmen Carrasco-Cubero, Juan J. Aznar-Sánchez, Raúl Veroz-González, Sara Rojas-Herrera, Pedro Dorado, and Adrián LLerena. 2023. "Pharmacogenetic Sex-Specific Effects of Methotrexate Response in Patients with Rheumatoid Arthritis" Pharmaceutics 15, no. 6: 1661. https://doi.org/10.3390/pharmaceutics15061661
APA StyleCeballos, F. C., Chamizo-Carmona, E., Mata-Martín, C., Carrasco-Cubero, C., Aznar-Sánchez, J. J., Veroz-González, R., Rojas-Herrera, S., Dorado, P., & LLerena, A. (2023). Pharmacogenetic Sex-Specific Effects of Methotrexate Response in Patients with Rheumatoid Arthritis. Pharmaceutics, 15(6), 1661. https://doi.org/10.3390/pharmaceutics15061661








