Extracellular Vesicles and Infection: From Hijacked Machinery to Therapeutic Tools
Abstract
:1. Introduction
2. Biogenesis
3. Function and Composition of EVs
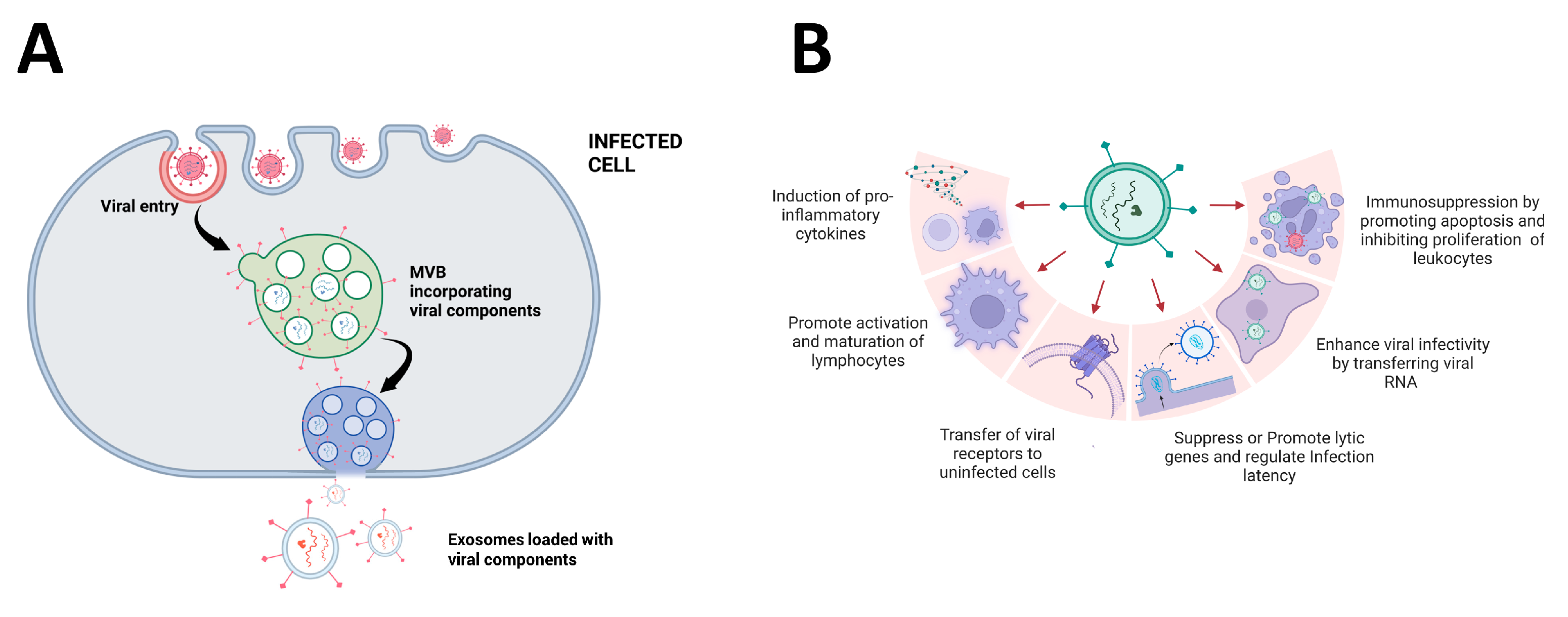
4. Promotion of Viral Entry and Replication
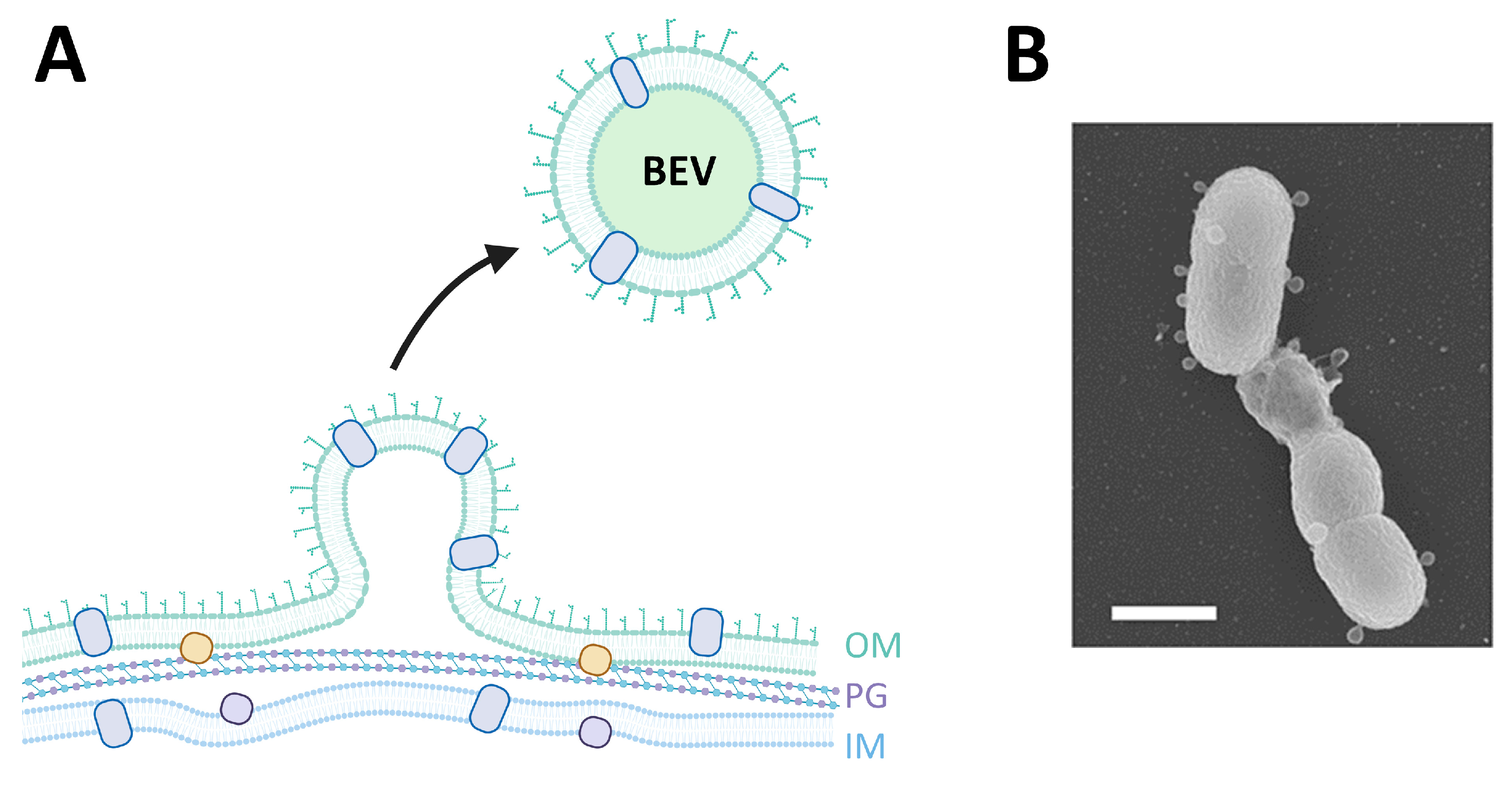
5. EVs in Bacterial Infection
6. Impact of EVs on the Immune System
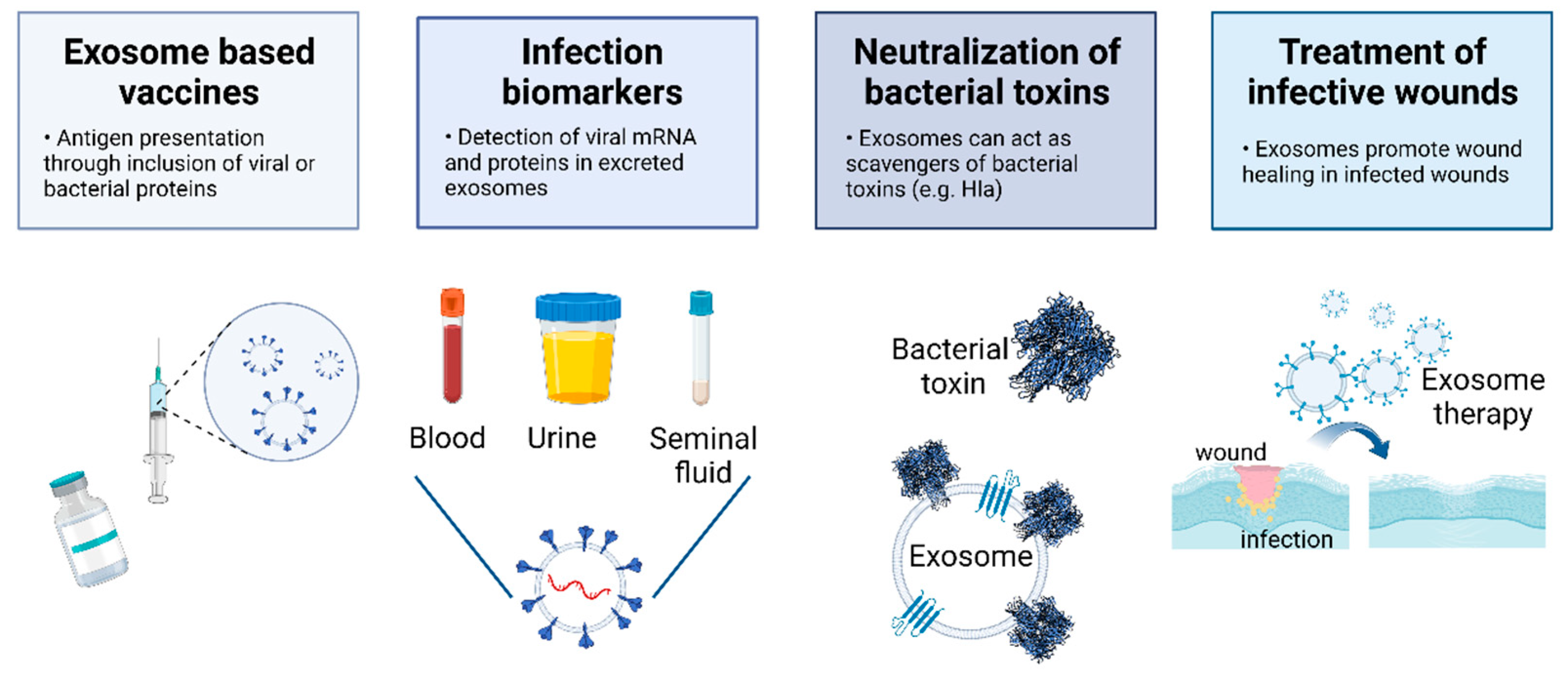
7. The Potential of EVs to Tackle Viral and Bacterial Infections
8. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Van Niel, G.; D’Angelo, G.; Raposo, G. Shedding Light on the Cell Biology of Extracellular Vesicles. Nat. Rev. Mol. Cell Biol. 2018, 19, 213–228. [Google Scholar] [CrossRef]
- Abels, E.R.; Breakefield, X.O. Introduction to Extracellular Vesicles: Biogenesis, RNA Cargo Selection, Content, Release, and Uptake. Cell. Mol. Neurobiol. 2016, 36, 301–312. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Richter, M.; Vader, P.; Fuhrmann, G. Approaches to Surface Engineering of Extracellular Vesicles. Adv. Drug Deliv. Rev. 2021, 173, 416–426. [Google Scholar] [CrossRef]
- Willms, E.; Cabañas, C.; Mäger, I.; Wood, M.J.A.; Vader, P. Extracellular Vesicle Heterogeneity: Subpopulations, Isolation Techniques, and Diverse Functions in Cancer Progression. Front. Immunol. 2018, 9, 738. [Google Scholar] [CrossRef] [Green Version]
- Doyle, L.; Wang, M. Overview of Extracellular Vesicles, Their Origin, Composition, Purpose, and Methods for Exosome Isolation and Analysis. Cells 2019, 8, 727. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fauré, J.; Lachenal, G.; Court, M.; Hirrlinger, J.; Chatellard-Causse, C.; Blot, B.; Grange, J.; Schoehn, G.; Goldberg, Y.; Boyer, V.; et al. Exosomes Are Released by Cultured Cortical Neurones. Mol. Cell. Neurosci. 2006, 31, 642–648. [Google Scholar] [CrossRef]
- Wollert, T.; Hurley, J.H. Molecular Mechanism of Multivesicular Body Biogenesis by ESCRT Complexes. Nature 2010, 464, 864–869. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alenquer, M.; Amorim, M.J. Exosome Biogenesis, Regulation, and Function in Viral Infection. Viruses 2015, 7, 5066–5083. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Spencer, N.; Yeruva, L. Role of Bacterial Infections in Extracellular Vesicles Release and Impact on Immune Response. Biomed. J. 2021, 44, 157–164. [Google Scholar] [CrossRef] [PubMed]
- Ramakrishnaiah, V.; Thumann, C.; Fofana, I.; Habersetzer, F.; Pan, Q.; De Ruiter, P.E.; Willemsen, R.; Demmers, J.A.A.; Raj, V.S.; Jenster, G.; et al. Exosome-Mediated Transmission of Hepatitis C Virus between Human Hepatoma Huh7.5 Cells. Proc. Natl. Acad. Sci. USA 2013, 110, 13109–13113. [Google Scholar] [CrossRef] [Green Version]
- Singh, P.P.; Smith, V.L.; Karakousis, P.C.; Schorey, J.S. Exosomes Isolated from Mycobacteria-Infected Mice or Cultured Macrophages Can Recruit and Activate Immune Cells In Vitro and In Vivo. J. Immunol. 2012, 189, 777–785. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bhatnagar, S.; Schorey, J.S. Exosomes Released from Infected Macrophages Contain Mycobacterium Avium Glycopeptidolipids and Are Proinflammatory. J. Biol. Chem. 2007, 282, 25779–25789. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Singh, P.P.; Li, L.; Schorey, J.S. Exosomal RNA from Mycobacterium Tuberculosis-Infected Cells Is Functional in Recipient Macrophages. Traffic 2015, 16, 555–571. [Google Scholar] [CrossRef] [Green Version]
- Anand, P.K.; Anand, E.; Bleck, C.K.E.; Anes, E.; Griffiths, G. Exosomal Hsp70 Induces a Pro-Inflammatory Response to Foreign Particles Including Mycobacteria. PLoS ONE 2010, 5, e10136. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.-J.; Chen, C.; Xie, P.-F.; Pan, Y.; Tan, Y.-H.; Tang, L.-J. Proteomic Analysis and Immune Properties of Exosomes Released by Macrophages Infected with Mycobacterium Avium. Microbes Infect. 2014, 16, 283–291. [Google Scholar] [CrossRef]
- Singh, P.P.; LeMaire, C.; Tan, J.C.; Zeng, E.; Schorey, J.S. Exosomes Released from m.Tuberculosis Infected Cells Can Suppress Ifn-γ Mediated Activation of Naïve Macrophages. PLoS ONE 2011, 6, e18564. [Google Scholar] [CrossRef] [Green Version]
- Rivera, J.; Cordero, R.J.B.; Nakouzi, A.S.; Frases, S.; Nicola, A.; Casadevall, A. Bacillus anthracis Produces Membrane-Derived Vesicles Containing Biologically Active Toxins. Proc. Natl. Acad. Sci. USA 2010, 107, 19002–19007. [Google Scholar] [CrossRef] [Green Version]
- Tsatsaronis, J.A.; Franch-Arroyo, S.; Resch, U.; Charpentier, E. Extracellular Vesicle RNA: A Universal Mediator of Microbial Communication? Trends Microbiol. 2018, 26, 401–410. [Google Scholar] [CrossRef]
- Shah, B.; Sullivan, C.J.; Lonergan, N.E.; Stanley, S.; Soult, M.C.; Britt, L.D. Circulating Bacterial Membrane Vesicles Cause Sepsis in Rats. Shock 2012, 37, 621–628. [Google Scholar] [CrossRef]
- Moulin, C.; Crupi, M.J.F.; Ilkow, C.S.; Bell, J.C.; Boulton, S. Extracellular Vesicles and Viruses: Two Intertwined Entities. Int. J. Mol. Sci. 2023, 24, 1036. [Google Scholar] [CrossRef]
- Martins, S.d.T.; Alves, L.R. Extracellular Vesicles in Viral Infections: Two Sides of the Same Coin? Front. Cell. Infect. Microbiol. 2020, 10, 593170. [Google Scholar] [CrossRef] [PubMed]
- Urbanelli, L.; Buratta, S.; Tancini, B.; Sagini, K.; Delo, F.; Porcellati, S.; Emiliani, C. The Role of Extracellular Vesicles in Viral Infection and Transmission. Vaccines 2019, 7, 102. [Google Scholar] [CrossRef] [Green Version]
- Sin, J.; McIntyre, L.; Stotland, A.; Feuer, R.; Gottlieb, R.A. Coxsackievirus B Escapes the Infected Cell in Ejected Mitophagosomes. J. Virol. 2017, 91, e01347-17. [Google Scholar] [CrossRef] [Green Version]
- Mao, L.; Wu, J.; Shen, L.; Yang, J.; Chen, J.; Xu, H. Enterovirus 71 Transmission by Exosomes Establishes a Productive Infection in Human Neuroblastoma Cells. Virus Genes 2016, 52, 189–194. [Google Scholar] [CrossRef] [PubMed]
- Dreux, M.; Garaigorta, U.; Boyd, B.; Décembre, E.; Chung, J.; Whitten-Bauer, C.; Wieland, S.; Chisari, F.V. Short-Range Exosomal Transfer of Viral RNA from Infected Cells to Plasmacytoid Dendritic Cells Triggers Innate Immunity. Cell Host Microbe 2012, 12, 558–570. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- De Jong, O.G.; van Balkom, B.W.M.; Schiffelers, R.M.; Bouten, C.V.C.; Verhaar, M.C. Extracellular Vesicles: Potential Roles in Regenerative Medicine. Front. Immunol. 2014, 5, 608. [Google Scholar] [CrossRef] [Green Version]
- Martin-Ventura, J.L.; Roncal, C.; Orbe, J.; Blanco-Colio, L.M. Role of Extracellular Vesicles as Potential Diagnostic and/or Therapeutic Biomarkers in Chronic Cardiovascular Diseases. Front. Cell Dev. Biol. 2022, 10, 813885. [Google Scholar] [CrossRef] [PubMed]
- Hwang, H.S.; Kim, H.; Han, G.; Lee, J.W.; Kim, K.; Kwon, I.C.; Yang, Y.; Kim, S.H. Extracellular Vesicles as Potential Therapeutics for Inflammatory Diseases. Int. J. Mol. Sci. 2021, 22, 5487. [Google Scholar] [CrossRef]
- Keller, M.D.; Ching, K.L.; Liang, F.X.; Dhabaria, A.; Tam, K.; Ueberheide, B.M.; Unutmaz, D.; Torres, V.J.; Cadwell, K. Decoy Exosomes Provide Protection against Bacterial Toxins. Nature 2020, 579, 260–264. [Google Scholar] [CrossRef]
- Kalluri, R.; LeBleu, V.S. The Biology, Function, and Biomedical Applications of Exosomes. Science 2020, 367, eaau6977. [Google Scholar] [CrossRef]
- Hessvik, N.P.; Llorente, A. Current Knowledge on Exosome Biogenesis and Release. Cell. Mol. Life Sci. 2018, 75, 193–208. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Teng, F.; Fussenegger, M. Shedding Light on Extracellular Vesicle Biogenesis and Bioengineering. Adv. Sci. 2021, 8, 202003505. [Google Scholar] [CrossRef] [PubMed]
- Williams, R.L.; Urbé, S. The Emerging Shape of the ESCRT Machinery. Nat. Rev. Mol. Cell Biol. 2007, 8, 355–368. [Google Scholar] [CrossRef]
- Henne, W.M.; Buchkovich, N.J.; Emr, S.D. The ESCRT Pathway. Dev. Cell 2011, 21, 77–91. [Google Scholar] [CrossRef] [Green Version]
- Vietri, M.; Radulovic, M.; Stenmark, H. The Many Functions of ESCRTs. Nat. Rev. Mol. Cell Biol. 2020, 21, 25–42. [Google Scholar] [CrossRef] [PubMed]
- Wurmser, A.E.; Emr, S.D. Phosphoinositide Signaling and Turnover: PtdIns(3)P, a Regulator of Membrane Traffic, Is Transported to the Vacuole and Degraded by a Process That Requires Lumenal Vacuolar Hydrolase Activities. EMBO J. 1998, 17, 4930–4942. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Odorizzi, G.; Babst, M.; Emr, S.D. Fab1p Ptdlns(3)P 5-Kinase Function Essential for Protein Sorting in the Multivesicular Body. Cell 1998, 95, 847–858. [Google Scholar] [CrossRef] [Green Version]
- Raiborg, C.; Grønvold Bache, K.; Mehlum, A.; Stang, E.; Stenmark, H. Hrs Recruits Clathrin to Early Endosomes. EMBO J. 2001, 20, 5008–5021. [Google Scholar] [CrossRef] [Green Version]
- Raiborg, C.; Wesche, J.; Malerød, L.; Stenmark, H. Flat Clathrin Coats on Endosomes Mediate Degradative Protein Sorting by Scaffolding Hrs in Dynamic Microdomains. J. Cell Sci. 2006, 119, 2414–2424. [Google Scholar] [CrossRef] [Green Version]
- Raiborg, C.; Bache, K.G.; Gillooly, D.J.; Madshus, I.H.; Stang, E.; Stenmark, H. Hrs Sorts Ubiquitinated Proteins into Clathrin-Coated Microdomains of Early Endosomes. Nat. Cell Biol. 2002, 4, 394–398. [Google Scholar] [CrossRef]
- Lu, Q.; Hope, L.W.; Brasch, M.; Reinhard, C.; Cohen, S.N. TSG101 Interaction with HRS Mediates Endosomal Trafficking and Receptor Down-Regulation. Proc. Natl. Acad. Sci. USA 2003, 100, 7626–7631. [Google Scholar] [CrossRef] [Green Version]
- Bache, K.G.; Brech, A.; Mehlum, A.; Stenmark, H. Hrs Regulates Multivesicular Body Formation via ESCRT Recruitment to Endosomes. J. Cell Biol. 2003, 162, 435–442. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Babst, M.; Katzmann, D.J.; Snyder, W.B.; Wendland, B.; Emr, S.D. Endosome-Associated Complex, ESCRT-II, Recruits Transport Machinery for Protein Sorting at the Multivesicular Body. Dev. Cell 2002, 3, 283–289. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Babst, M.; Katzmann, D.J.; Estepa-Sabal, E.J.; Meerloo, T.; Emr, S.D. ESCRT-III: An Endosome-Associated Heterooligomeric Protein Complex Required for MVB Sorting. Dev. Cell 2002, 3, 271–282. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Teo, H.; Perisic, O.; González, B.; Williams, R.L. ESCRT-II, an Endosome-Associated Complex Required for Protein Sorting: Crystal Structure and Interactions with ESCRT-III and Membranes. Dev. Cell 2004, 7, 559–569. [Google Scholar] [CrossRef] [Green Version]
- Akers, J.C.; Gonda, D.; Kim, R.; Carter, B.S.; Chen, C.C. Biogenesis of Extracellular Vesicles (EV): Exosomes, Microvesicles, Retrovirus-like Vesicles, and Apoptotic Bodies. J. Neurooncol. 2013, 113, 1–11. [Google Scholar] [CrossRef] [Green Version]
- Bonifacino, J.S.; Glick, B.S. The Mechanisms of Vesicle Budding and Fusion. Cell 2004, 116, 153–166. [Google Scholar] [CrossRef] [Green Version]
- Jahn, R.; Scheller, R.H. SNAREs-Engines for Membrane Fusion. Nat. Rev. Mol. Cell Biol. 2006, 7, 631–643. [Google Scholar] [CrossRef]
- Südhof, T.C.; Rothman, J.E. Membrane Fusion: Grappling with SNARE and SM Proteins. Science 2009, 323, 474–477. [Google Scholar] [CrossRef] [Green Version]
- Mathieu, M.; Martin-Jaular, L.; Lavieu, G.; Théry, C. Specificities of Secretion and Uptake of Exosomes and Other Extracellular Vesicles for Cell-to-Cell Communication. Nat. Cell Biol. 2019, 21, 9–17. [Google Scholar] [CrossRef]
- Théry, C.; Witwer, K.W.; Aikawa, E.; Alcaraz, M.J.; Anderson, J.D.; Andriantsitohaina, R.; Antoniou, A.; Arab, T.; Archer, F.; Atkin-Smith, G.K.; et al. Minimal Information for Studies of Extracellular Vesicles 2018 (MISEV2018): A Position Statement of the International Society for Extracellular Vesicles and Update of the MISEV2014 Guidelines. J. Extracell. Vesicles 2018, 7, 1535750. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nabhan, J.F.; Hu, R.; Oh, R.S.; Cohen, S.N.; Lu, Q. Formation and Release of Arrestin Domain-Containing Protein 1-Mediated Microvesicles (ARMMs) at Plasma Membrane by Recruitment of TSG101 Protein. Proc. Natl. Acad. Sci. USA 2012, 109, 4146–4151. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schlienger, S.; Campbell, S.; Claing, A. ARF1 Regulates the Rho/MLC Pathway to Control EGF-Dependent Breast Cancer Cell Invasion. Mol. Biol. Cell 2014, 25, 17–29. [Google Scholar] [CrossRef] [PubMed]
- Muralidharan-Chari, V.; Clancy, J.; Plou, C.; Romao, M.; Chavrier, P.; Raposo, G.; D’Souza-Schorey, C. ARF6-Regulated Shedding of Tumor-Cell Derived Plasma Membrane Microvesicles Vandhana. Curr. Biol. 2009, 19, 1875–1885. [Google Scholar] [CrossRef] [Green Version]
- Li, B.; Antonyak, M.A.; Zhang, J.; Cerione, R.A. RhoA Triggers a Specific Signaling Pathway That Generates Transforming Microvesicles in Cancer Cells Bo. Oncogene 2012, 35, 4740–4749. [Google Scholar] [CrossRef] [Green Version]
- Awojoodu, A.O.; Keegan, P.M.; Lane, A.R.; Zhang, Y.; Lynch, K.R.; Platt, M.O.; Botchwey, E.A. Acid Sphingomyelinase Is Activated in Sickle Cell Erythrocytes and Contributes to Inflammatory Microparticle Generation in SCD. Blood 2014, 124, 1941–1950. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bianco, F.; Perrotta, C.; Novellino, L.; Francolini, M.; Riganti, L.; Menna, E.; Saglietti, L.; Schuchman, E.H.; Furlan, R.; Clementi, E.; et al. Acid Sphingomyelinase Activity Triggers Microparticle Release from Glial Cells. EMBO J. 2009, 28, 1043–1054. [Google Scholar] [CrossRef] [Green Version]
- White, I.J.; Bailey, L.M.; Aghakhani, M.R.; Moss, S.E.; Futter, C.E. EGF Stimulates Annexin 1-Dependent Inward Vesiculation in a Multivesicular Endosome Subpopulation. EMBO J. 2006, 25, 1–12. [Google Scholar] [CrossRef] [Green Version]
- Wickman, G.; Julian, L.; Olson, M.F. How Apoptotic Cells Aid in the Removal of Their Own Cold Dead Bodies. Cell Death Differ. 2012, 19, 735–742. [Google Scholar] [CrossRef] [Green Version]
- Battistelli, M.; Falcieri, E. Apoptotic Bodies: Particular Extracellular Vesicles Involved in Intercellular Communication. Biology 2020, 9, 21. [Google Scholar] [CrossRef] [Green Version]
- Quek, C.; Hill, A.F. The Role of Extracellular Vesicles in Neurodegenerative Diseases. Biochem. Biophys. Res. Commun. 2017, 483, 1178–1186. [Google Scholar] [CrossRef]
- Pegtel, D.M.; Gould, S.J. Annual Review of Biochemistry Exosomes. Exosomes 2019, 88, 487–514. [Google Scholar] [CrossRef]
- Shen, B.; Fang, Y.; Wu, N.; Gould, S.J. Biogenesis of the Posterior Pole Is Mediated by the Exosome/Microvesicle Protein-Sorting Pathway. J. Biol. Chem. 2011, 286, 44162–44176. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Choi, Y.; Kim, S.M.; Heo, Y.; Lee, G.; Kang, J.Y.; Yoon, D.S. Nanoelectrical Characterization of Individual Exosomes Secreted by Aβ42-Ingested Cells Using Electrostatic Force Microscopy. Nanotechnology 2021, 32, 025705. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Balaji, V.; Kaniyappan, S.; Krüger, L.; Irsen, S.; Tepper, K.; Chandupatla, R.; Maetzler, W.; Schneider, A.; Mandelkow, E.; et al. The Release and Trans-Synaptic Transmission of Tau via Exosomes. Mol. Neurodegener. 2017, 12, 5. [Google Scholar] [CrossRef] [Green Version]
- Guo, M.; Wang, J.; Zhao, Y.; Feng, Y.; Han, S.; Dong, Q.; Cui, M.; Tieu, K. Microglial Exosomes Facilitate A-Synuclein Transmission in Parkinson’s Disease. Brain 2020, 143, 1476–1497. [Google Scholar] [CrossRef]
- Park, G.; Kim, B.S.; Kim, E. A Novel Function of FAF1, Which Induces Dopaminergic Neuronal Death through Cell-to-Cell Transmission. Cell Commun. Signal. 2020, 18, 133. [Google Scholar] [CrossRef]
- Street, J.M.; Koritzinsky, E.H.; Glispie, D.M.; Star, R.A.; Yuen, P.S.T. Urine Exosomes: An Emerging Trove of Biomarkers, 1st ed.; Elsevier Inc.: Bethesda, MD, USA, 2017; Volume 78. [Google Scholar]
- Machida, T.; Tomofuji, T.; Ekuni, D.; Maruyama, T.; Yoneda, T.; Kawabata, Y.; Mizuno, H.; Miyai, H.; Kunitomo, M.; Morita, M. MicroRNAs in Salivary Exosome as Potential Biomarkers of Aging. Int. J. Mol. Sci. 2015, 16, 21294–21309. [Google Scholar] [CrossRef] [Green Version]
- Qin, W.; Tsukasaki, Y.; Dasgupta, S.; Mukhopadhyay, N.; Ikebe, M.; Sauter, E.R. Exosomes in Human Breast Milk Promote EMT. Clin. Cancer Res. 2016, 22, 4517–4524. [Google Scholar] [CrossRef] [Green Version]
- Yagi, Y.; Ohkubo, T.; Kawaji, H.; Machida, A.; Miyata, H.; Goda, S.; Roy, S.; Hayashizaki, Y.; Suzuki, H.; Yokota, T. Next-Generation Sequencing-Based Small RNA Profiling of Cerebrospinal Fluid Exosomes. Neurosci. Lett. 2017, 636, 48–57. [Google Scholar] [CrossRef]
- Keller, S.; Rupp, C.; Stoeck, A.; Runz, S.; Fogel, M.; Lugert, S.; Hager, H.D.; Abdel-Bakky, M.S.; Gutwein, P.; Altevogt, P. CD24 Is a Marker of Exosomes Secreted into Urine and Amniotic Fluid. Kidney Int. 2007, 72, 1095–1102. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Peng, P.; Yan, Y.; Keng, S. Exosomes in the Ascites of Ovarian Cancer Patients: Origin and Effects on Anti-Tumor Immunity. Oncol. Rep. 2011, 25, 749–762. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Y.; Liu, Y.; Liu, H.; Tang, W.H. Exosomes: Biogenesis, Biologic Function and Clinical Potential. Cell Biosci. 2019, 9, 19. [Google Scholar] [CrossRef] [PubMed]
- Pascucci, L.; Coccè, V.; Bonomi, A.; Ami, D.; Ceccarelli, P.; Ciusani, E.; Viganò, L.; Locatelli, A.; Sisto, F.; Doglia, S.M.; et al. Paclitaxel Is Incorporated by Mesenchymal Stromal Cells and Released in Exosomes That Inhibit in Vitro Tumor Growth: A New Approach for Drug Delivery. J. Control. Release 2014, 192, 262–270. [Google Scholar] [CrossRef]
- Haney, M.J.; Klyachko, N.L.; Zhao, Y.; Gupta, R.; Plotnikova, E.G.; He, Z.; Patel, T.; Piroyan, A.; Sokolsky, M.; Kabanov, A.V.; et al. Exosomes as Drug Delivery Vehicles for Parkinson’s Disease Therapy. J. Control. Release 2015, 207, 18–30. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kalani, A.; Kamat, P.K.; Chaturvedi, P.; Tyagi, S.C.; Tyagi, N. Curcumin-Primed Exosomes Mitigate Endothelial Cell Dysfunction during Hyperhomocysteinemia. Life Sci. 2014, 107, 1–7. [Google Scholar] [CrossRef] [Green Version]
- Yang, T.; Martin, P.; Fogarty, B.; Brown, A.; Schurman, K.; Phipps, R.; Yin, V.P.; Lockman, P.; Bai, S. Exosome Delivered Anticancer Drugs across the Blood-Brain Barrier for Brain Cancer Therapy in Danio Rerio. Pharm. Res. 2015, 32, 2003–2014. [Google Scholar] [CrossRef] [Green Version]
- Clancy, J.W.; Schmidtmann, M.; D’Souza-Schorey, C. The Ins and Outs of Microvesicles. FASEB BioAdvances 2021, 3, 399–406. [Google Scholar] [CrossRef]
- Lv, Y.M.; Tan, J.; Miao, Y.; Zhang, Q. The Role of Microvesicles and Its Active Molecules in Regulating Cellular Biology. J. Cell. Mol. Med. 2019, 23, 7894–7904. [Google Scholar] [CrossRef] [Green Version]
- Zaborowski, M.P.; Balaj, L.; Breakefield, X.O.; Lai, C.P. Extracellular Vesicles: Composition, Biological Relevance, and Methods of Study. Bioscience 2015, 65, 783–797. [Google Scholar] [CrossRef] [Green Version]
- Li, X.; Corbett, A.L.; Little, J.P.; Garnis, C. Challenges and Opportunities in Exosome Research—Perspectives from Biology, Engineering, and Cancer Therapy. APL Bioeng. 2019, 3, 011503. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ekström, K.; Crescitelli, R.; Pétursson, H.I.; Johansson, J.; Lässer, C.; Bagge, R.O. Characterization of Surface Markers on Extracellular Vesicles Isolated from Lymphatic Exudate from Patients with Breast Cancer. BMC Cancer 2022, 22, 50. [Google Scholar] [CrossRef] [PubMed]
- Conde-Vancells, J.; Rodriguez-Suarez, E.; Embade, N.; Gil, D.; Matthiesen, R.; Valle, M.; Elortza, F.; Lu, S.C.; Mato, J.M.; Falcon-Perez, J.M. Characterization and Comprehensive Proteome Profiling of Exosomes Secreted by Hepatocytes. J. Proteome Res. 2008, 7, 5157–5166. [Google Scholar] [CrossRef] [Green Version]
- Wen, C.; Seeger, R.C.; Fabbri, M.; Wang, L.; Wayne, A.S.; Jong, A.Y. Biological Roles and Potential Applications of Immune Cell-Derived Extracellular Vesicles. J. Extracell. Vesicles 2017, 6, 1400370. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rontogianni, S.; Synadaki, E.; Li, B.; Liefaard, M.C.; Lips, E.H.; Wesseling, J.; Wu, W.; Altelaar, M. Proteomic Profiling of Extracellular Vesicles Allows for Human Breast Cancer Subtyping. Commun. Biol. 2019, 2, 1–13. [Google Scholar] [CrossRef] [Green Version]
- Meckes, D.G. Exosomal Communication Goes Viral. J. Virol. 2015, 89, 5200–5203. [Google Scholar] [CrossRef] [Green Version]
- Madison, M.N.; Jones, P.H.; Okeoma, C.M. Exosomes in Human Semen Restrict HIV-1 Transmission by Vaginal Cells and Block Intravaginal Replication of LP-BM5 Murine AIDS Virus Complex. Virology 2015, 482, 189–201. [Google Scholar] [CrossRef] [Green Version]
- van Dongen, H.M.; Masoumi, N.; Witwer, K.W.; Pegtel, D.M. Extracellular Vesicles Exploit Viral Entry Routes for Cargo Delivery. Microbiol. Mol. Biol. Rev. 2016, 80, 369–386. [Google Scholar] [CrossRef] [Green Version]
- Izquierdo-Useros, N.; Puertas, M.C.; Borràs, F.E.; Blanco, J.; Martinez-Picado, J. Exosomes and Retroviruses: The Chicken or the Egg? Cell. Microbiol. 2011, 13, 10–17. [Google Scholar] [CrossRef]
- Wubbolts, R.; Leckie, R.S.; Veenhuizen, P.T.M.; Schwarzmann, G.; Möbius, W.; Hoernschemeyer, J.; Slot, J.W.; Geuze, H.J.; Stoorvogel, W. Proteomic and Biochemical Analyses of Human B Cell-Derived Exosomes: Potential Implications for Their Function and Multivesicular Body Formation. J. Biol. Chem. 2003, 278, 10963–10972. [Google Scholar] [CrossRef] [Green Version]
- Théry, C.; Zitvogel, L.; Amigorena, S. Exosomes: Composition, Biogenesis and Function. Nat. Rev. Immunol. 2002, 2, 569–579. [Google Scholar] [CrossRef] [PubMed]
- Ott, D.E. Cellular Proteins Detected in HIV-1. Rev. Med. Virol. 2008, 18, 159–175. [Google Scholar] [CrossRef] [PubMed]
- Krishnamoorthy, L.; Bess, J.W.; Preston, A.B.; Nagashima, K.; Mahal, L.K. HIV-1 and Microvesicles from T Cells Share a Common Glycome, Arguing for a Common Origin. Nat. Chem. Biol. 2009, 5, 244–250. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Théry, C.; Boussac, M.; Véron, P.; Ricciardi-Castagnoli, P.; Raposo, G.; Garin, J.; Amigorena, S. Proteomic Analysis of Dendritic Cell-Derived Exosomes: A Secreted Subcellular Compartment Distinct from Apoptotic Vesicles. J. Immunol. 2001, 166, 7309–7318. [Google Scholar] [CrossRef] [Green Version]
- Wu, L.; KewalRamani, V.N. Dendritic-Cell Interactions with HIV: Infection and Viral Dissemination. Nat. Rev. Immunol. 2006, 6, 859–868. [Google Scholar] [CrossRef]
- Votteler, J.; Sundquist, W.I. Virus Budding and the ESCRT Pathway. Cell Host Microbe. 2013, 14, 232–241. [Google Scholar] [CrossRef] [Green Version]
- Meng, B.; Lever, A.M.L. The Interplay between ESCRT and Viral Factors in the Enveloped Virus Life Cycle. Viruses 2021, 13, 324. [Google Scholar] [CrossRef]
- Trajkovic, K.; Hsu, C.; Chiantia, S.; Rajendran, L.; Wenzel, D.; Wieland, F.; Schwille, P.; Brügger, B.; Simons, M. Ceramide Triggers Budding of Exosome Vesicles into Multivesicular Endosomes. Science 2008, 319, 1244–1247. [Google Scholar] [CrossRef]
- Perez-Hernandez, D.; Gutiérrez-Vázquez, C.; Jorge, I.; López-Martín, S.; Ursa, A.; Sánchez-Madrid, F.; Vázquez, J.; Yañez-Mó, M. The Intracellular Interactome of Tetraspanin-Enriched Microdomains Reveals Their Function as Sorting Machineries toward Exosomes. J. Biol. Chem. 2013, 288, 11649–11661. [Google Scholar] [CrossRef] [Green Version]
- Van Niel, G.; Charrin, S.; Simoes, S.; Romao, M.; Rochin, L.; Saftig, P.; Marks, M.S.; Rubinstein, E.; Raposo, G. The Tetraspanin CD63 Regulates ESCRT-Independent and -Dependent Endosomal Sorting during Melanogenesis. Dev. Cell 2011, 21, 708–721. [Google Scholar] [CrossRef] [Green Version]
- Savina, A.; Vidal, M.; Colombo, M.I. The Exosome Pathway in K562 Cells Is Regulated by Rab11. J. Cell Sci. 2002, 115, 2505–2515. [Google Scholar] [CrossRef] [PubMed]
- Bruce, E.A.; Digard, P.; Stuart, A.D. The Rab11 Pathway Is Required for Influenza A Virus Budding and Filament Formation. J. Virol. 2010, 84, 5848–5859. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rowe, R.K.; Suszko, J.W.; Pekosz, A. Roles for the Recycling Endosome, Rab8, and Rab11 in Hantavirus Release from Epithelial Cells. Virology 2008, 382, 239–249. [Google Scholar] [CrossRef] [Green Version]
- Utley, T.J.; Ducharme, N.A.; Varthakavi, V.; Shepherd, B.E.; Santangelo, P.J.; Lindquist, M.E.; Goldenring, J.R.; Crowe, J.E. Respiratory Syncytial Virus Uses a Vps4-Independent Budding Mechanism Controlled by Rab11-FIP2. Proc. Natl. Acad. Sci. USA 2008, 105, 10209–10214. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Abrami, L.; Brandi, L.; Moayeri, M.; Brown, M.J.; Krantz, B.A.; Leppla, S.H.; VanderGoot, F.G. Hijacking Multivesicular Bodies Enables Long-Term and Exosome-Mediated Long-Distance Action of Anthrax Toxin. Cell Rep. 2013, 5, 986–996. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bhuin, T.; Roy, J.K. Rab11 in Disease Progression. Int. J. Mol. Cell. Med. 2015, 4, 1–8. [Google Scholar]
- Ostrowski, M.; Carmo, N.B.; Krumeich, S.; Fanget, I.; Raposo, G.; Savina, A.; Moita, C.F.; Schauer, K.; Hume, A.N.; Freitas, R.P.; et al. Rab27a and Rab27b Control Different Steps of the Exosome Secretion Pathway. Nat. Cell Biol. 2010, 12, 19–30. [Google Scholar] [CrossRef] [Green Version]
- Bobrie, A.; Krumeich, S.; Reyal, F.; Recchi, C.; Moita, L.F.; Seabra, M.C.; Ostrowski, M.; Théry, C. Rab27a Supports Exosome-Dependent and -Independent Mechanisms That Modify the Tumor Microenvironment and Can Promote Tumor Progression. Cancer Res. 2012, 72, 4920–4930. [Google Scholar] [CrossRef] [Green Version]
- Fraile-Ramos, A.; Cepeda, V.; Elstak, E.; van der Sluijs, P. Rab27a Is Required for Human Cytomegalovirus Assembly. PLoS ONE 2010, 5, e15318. [Google Scholar] [CrossRef]
- Gerber, P.P.; Cabrini, M.; Jancic, C.; Paoletti, L.; Banchio, C.; Von Bilderling, C.; Sigaut, L.; Pietrasanta, L.I.; Duette, G.; Freed, E.O.; et al. Rab27a Controls HIV-1 Assembly by Regulating Plasma Membrane Levels of Phosphatidylinositol 4,5-Bisphosphate. J. Cell Biol. 2015, 209, 435–452. [Google Scholar] [CrossRef] [Green Version]
- Meng, B.; Ip, N.C.Y.; Prestwood, L.J.; Abbink, T.E.M.; Lever, A.M.L. Evidence That the Endosomal Sorting Complex Required for Transport-II (ESCRT-II) Is Required for Efficient Human Immunodeficiency Virus-1 (HIV-1) Production. Retrovirology 2015, 12, 72. [Google Scholar] [CrossRef] [Green Version]
- Cheruiyot, C.; Pataki, Z.; Ramratnam, B.; Li, M. Proteomic Analysis of Exosomes and Its Application in HIV-1 Infection. Proteom.-Clin. Appl. 2018, 12, e1700142. [Google Scholar] [CrossRef] [PubMed]
- Mori, Y.; Koike, M.; Moriishi, E.; Kawabata, A.; Tang, H.; Oyaizu, H.; Uchiyama, Y.; Yamanishi, K. Human Herpesvirus-6 Induces MVB Formation, and Virus Egress Occurs by an Exosomal Release Pathway. Traffic 2008, 9, 1728–1742. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aydin, Y.; Koksal, A.R.; Reddy, V.; Lin, D.; Osman, H.; Heidari, Z.; Rhadhi, S.M.; Wimley, W.C.; Parsi, M.A.; Dash, S. Extracellular Vesicle Release Promotes Viral Replication during Persistent Hcv Infection. Cells 2021, 10, 984. [Google Scholar] [CrossRef]
- Longatti, A.; Boyd, B.; Chisari, F.V. Virion-Independent Transfer of Replication-Competent Hepatitis C Virus RNA between Permissive Cells. J. Virol. 2015, 89, 2956–2961. [Google Scholar] [CrossRef] [Green Version]
- Chivero, E.T.; Bhattarai, N.; Rydze, R.T.; Winters, M.A.; Holodniy, M.; Stapleton, J.T. Human Pegivirus RNA Is Found in Multiple Blood Mononuclear Cells in Vivo and Serum-Derived Viral RNA-Containing Particles Are Infectious in Vitro. J. Gen. Virol. 2014, 95, 1307–1319. [Google Scholar] [CrossRef] [PubMed]
- Walls, A.C.; Park, Y.J.; Tortorici, M.A.; Wall, A.; McGuire, A.T.; Veesler, D. Structure, Function, and Antigenicity of the SARS-CoV-2 Spike Glycoprotein. Cell 2020, 181, 281–292.e6. [Google Scholar] [CrossRef]
- Cocozza, F.; Névo, N.; Piovesana, E.; Lahaye, X.; Buchrieser, J.; Schwartz, O.; Manel, N.; Tkach, M.; Théry, C.; Martin-Jaular, L. Extracellular Vesicles Containing ACE2 Efficiently Prevent Infection by SARS-CoV-2 Spike Protein-Containing Virus. J. Extracell. Vesicles 2020, 10, e12050. [Google Scholar] [CrossRef]
- Narayanan, A.; Iordanskiy, S.; Das, R.; Van Duyne, R.; Santos, S.; Jaworski, E.; Guendel, I.; Sampey, G.; Dalby, E.; Iglesias-Ussel, M.; et al. Exosomes Derived from HIV-1-Infected Cells Contain Trans-Activation Response Element RNA. J. Biol. Chem. 2013, 288, 20014–20033. [Google Scholar] [CrossRef] [Green Version]
- Anderson, M.R.; Kashanchi, F.; Jacobson, S. Exosomes in Viral Disease. Neurotherapeutics 2016, 13, 535–546. [Google Scholar] [CrossRef] [Green Version]
- Chahar, H.S.; Bao, X.; Casola, A. Exosomes and Their Role in the Life Cycle and Pathogenesis of RNA Viruses. Viruses 2015, 7, 3204–3225. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hoshina, S.; Sekizuka, T.; Kataoka, M.; Hasegawa, H.; Hamada, H.; Kuroda, M.; Katano, H. Profile of Exosomal and Intracellular MicroRNA in Gamma-Herpesvirus-Infected Lymphoma Cell Lines. PLoS ONE 2016, 11, e0162574. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chugh, P.E.; Sin, S.H.; Ozgur, S.; Henry, D.H.; Menezes, P.; Griffith, J.; Eron, J.J.; Damania, B.; Dittmer, D.P. Systemically Circulating Viral and Tumor-Derived MicroRNAs in KSHV-Associated Malignancies. PLoS Pathog. 2013, 9, e1003484. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Canitano, A.; Venturi, G.; Borghi, M.; Ammendolia, M.G.; Fais, S. Exosomes Released in Vitro from Epstein-Barr Virus (EBV)-Infected Cells Contain EBV-Encoded Latent Phase MRNAs. Cancer Lett. 2013, 337, 193–199. [Google Scholar] [CrossRef]
- Meckes, D.G.; Gunawardena, H.P.; Dekroon, R.M.; Heaton, P.R.; Edwards, R.H.; Ozgur, S.; Griffith, J.D.; Damania, B.; Raab-Traub, N. Modulation of B-Cell Exosome Proteins by Gamma Herpesvirus Infection. Proc. Natl. Acad. Sci. USA 2013, 110, E2925–E2933. [Google Scholar] [CrossRef] [Green Version]
- Edwards, R.H.; Marquitz, A.R.; Raab-Traub, N. Epstein-Barr Virus BART MicroRNAs Are Produced from a Large Intron Prior to Splicing. J. Virol. 2008, 82, 9094–9106. [Google Scholar] [CrossRef] [Green Version]
- Hooykaas, M.J.G.; Kruse, E.; Wiertz, E.J.H.J.; Lebbink, R.J. Comprehensive Profiling of Functional Epstein-Barr Virus MiRNA Expression in Human Cell Lines. BMC Genom. 2016, 17, 644. [Google Scholar] [CrossRef] [Green Version]
- Shinozaki-Ushiku, A.; Kunita, A.; Isogai, M.; Hibiya, T.; Ushiku, T.; Takada, K.; Fukayama, M. Profiling of Virus-Encoded MicroRNAs in Epstein-Barr Virus-Associated Gastric Carcinoma and Their Roles in Gastric Carcinogenesis. J. Virol. 2015, 89, 5581–5591. [Google Scholar] [CrossRef] [Green Version]
- Gallo, A.; Vella, S.; Miele, M.; Timoneri, F.; Di Bella, M.; Bosi, S.; Sciveres, M.; Conaldi, P.G. Global Profiling of Viral and Cellular Non-Coding RNAs in Epstein–Barr Virus-Induced Lymphoblastoid Cell Lines and Released Exosome Cargos. Cancer Lett. 2017, 388, 334–343. [Google Scholar] [CrossRef]
- Ranjit, S.; Kodidela, S.; Sinha, N.; Chauhan, S.; Kumar, S. Extracellular Vesicles from Human Papilloma Virus-Infected Cervical Cancer Cells Enhance HIV-1 Replication in Differentiated U1 Cell Line. Viruses 2020, 12, 239–259. [Google Scholar] [CrossRef] [Green Version]
- Martínez-Rojas, P.P.; Quiroz-García, E.; Monroy-Martínez, V.; Agredano-Moreno, L.T.; Jiménez-García, L.F.; Ruiz-Ordaz, B.H. Participation of Extracellular Vesicles from Zika-Virus-Infected Mosquito Cells in the Modification of Naïve Cells’ Behavior by Mediating Cell-to-Cell Transmission of Viral Elements. Cells 2020, 9, 123. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Birge, R.B.; Boeltz, S.; Kumar, S.; Carlson, J.; Wanderley, J.; Calianese, D.; Barcinski, M.; Brekken, R.A.; Huang, X.; Hutchins, J.T.; et al. Phosphatidylserine Is a Global Immunosuppressive Signal in Efferocytosis, Infectious Disease, and Cancer. Cell Death Differ. 2016, 23, 962–978. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bevers, E.M.; Williamson, P.L. Getting to the Outer Leaflet: Physiology of Phosphatidylserine Exposure at the Plasma Membrane. Physiol. Rev. 2016, 96, 605–645. [Google Scholar] [CrossRef] [PubMed]
- Brakhage, A.A.; Zimmermann, A.-K.; Rivieccio, F.; Visser, C.; Blango, M.G. Host-Derived Extracellular Vesicles for Antimicrobial Defense. Microlife 2021, 2, uqab003. [Google Scholar] [CrossRef] [PubMed]
- Timár, C.I.; Lorincz, Á.M.; Csépányi-Kömi, R.; Vályi-Nagy, A.; Nagy, G.; Buzás, E.I.; Iványi, Z.; Kittel, Á.; Powell, D.W.; McLeish, K.R.; et al. Antibacterial Effect of Microvesicles Released from Human Neutrophilic Granulocytes. Blood 2013, 121, 510–518. [Google Scholar] [CrossRef] [Green Version]
- Lőrincz, Á.M.; Bartos, B.; Szombath, D.; Szeifert, V.; Timár, C.I.; Turiák, L.; Drahos, L.; Kittel, Á.; Veres, D.S.; Kolonics, F.; et al. Role of Mac-1 Integrin in Generation of Extracellular Vesicles with Antibacterial Capacity from Neutrophilic Granulocytes. J. Extracell. Vesicles 2020, 9, 1698889. [Google Scholar] [CrossRef]
- Miao, Y.; Li, G.; Zhang, X.; Xu, H.; Abraham, S.N. A TRP Channel Senses Lysosome Neutralization by Pathogens to Trigger Their Expulsion. Cell 2015, 161, 1306–1319. [Google Scholar] [CrossRef] [Green Version]
- Briaud, P.; Carrolla, R.K. Extracellular Vesicle Biogenesis and Functions in Gram-Positive Bacteria. Infect. Immun. 2020, 88, e00433-20. [Google Scholar] [CrossRef]
- Chronopoulos, A.; Kalluri, R. Emerging Role of Bacterial Extracellular Vesicles in Cancer. Oncogene 2020, 39, 6951–6960. [Google Scholar] [CrossRef]
- Hosseini-Giv, N.; Basas, A.; Hicks, C.; El-Omar, E.; El-Assaad, F.; Hosseini-Beheshti, E. Bacterial Extracellular Vesicles and Their Novel Therapeutic Applications in Health and Cancer. Front. Cell. Infect. Microbiol. 2022, 12, 962216. [Google Scholar] [CrossRef]
- Toyofuku, M.; Nomura, N.; Eberl, L. Types and Origins of Bacterial Membrane Vesicles. Nat. Rev. Microbiol. 2019, 17, 13–24. [Google Scholar] [CrossRef] [PubMed]
- Tashiro, Y.; Uchiyama, H.; Nomura, N. Multifunctional Membrane Vesicles in Pseudomonas Aeruginosa. Environ. Microbiol. 2012, 14, 1349–1362. [Google Scholar] [CrossRef]
- Dakhlallah, D.A.; Wisler, J.; Gencheva, M.; Brown, C.M.; Leatherman, E.R.; Singh, K.; Brundage, K.; Karsies, T.; Dakhlallah, A.; Witwer, K.W.; et al. Circulating Extracellular Vesicle Content Reveals de Novo DNA Methyltransferase Expression as a Molecular Method to Predict Septic Shock. J. Extracell. Vesicles 2019, 8, 1669881. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tian, C.; Wang, K.; Zhao, M.; Cong, S.; Di, X.; Li, R. Extracellular Vesicles Participate in the Pathogenesis of Sepsis. Front. Cell. Infect. Microbiol. 2022, 12, 1018692. [Google Scholar] [CrossRef] [PubMed]
- Wang, E.; Liu, Y.; Qiu, X.; Tang, Y.; Wang, H.; Xiao, X.; Chen, F.; Lu, B. Bacteria-Released Outer Membrane Vesicles Promote Disseminated Intravascular Coagulation. Thromb. Res. 2019, 178, 26–33. [Google Scholar] [CrossRef]
- Kang, C.-S.; Ban, M.; Choi, E.-J.; Moon, H.-G.; Jeon, J.-S.; Kim, D.-K.; Park, S.-K.; Jeon, S.G.; Roh, T.-Y.; Myung, S.-J.; et al. Extracellular Vesicles Derived from Gut Microbiota, Especially Akkermansia muciniphila, Protect the Progression of Dextran Sulfate Sodium-Induced Colitis. PLoS ONE 2013, 8, e76520. [Google Scholar] [CrossRef] [Green Version]
- Lahiri, D.; Nag, M.; Dey, A.; Sarkar, T.; Pattnaik, S.; Ghosh, S.; Edinur, H.A.; Pati, S.; Kari, Z.A.; Ray, R.R. Exosome-Associated Host–Pathogen Interaction: A Potential Effect of Biofilm Formation. J. Anal. Sci. Technol. 2021, 12, 1–8. [Google Scholar] [CrossRef]
- Muhammad, M.H.; Idris, A.L.; Fan, X.; Guo, Y.; Yu, Y.; Jin, X.; Qiu, J.; Guan, X.; Huang, T. Beyond Risk: Bacterial Biofilms and Their Regulating Approaches. Front. Microbiol. 2020, 11, 928. [Google Scholar] [CrossRef]
- Davies, D. Understanding Biofilm Resistance to Antibacterial Agents. Nat. Rev. Drug Discov. 2003, 2, 114–122. [Google Scholar] [CrossRef]
- Flemming, H.C.; Neu, T.R.; Wozniak, D.J. The EPS Matrix: The “House of Biofilm Cells”. J. Bacteriol. 2007, 189, 7945–7947. [Google Scholar] [CrossRef] [Green Version]
- Toyofuku, M.; Roschitzki, B.; Riedel, K.; Eberl, L. Identification of Proteins Associated with the Pseudomonas Aeruginosa Biofilm Extracellular Matrix. J. Proteome Res. 2012, 11, 4906–4915. [Google Scholar] [CrossRef] [PubMed]
- Berlutti, F.; Morea, C.; Battistoni, A.; Sarli, S.; Cipriani, P.; Superti, F.; Ammendolia, M.G.; Valenti, P. Iron Availability Influences Aggregation, Biofilm, Adhesion and Invasion of Pseudomonas Aeruginosa and Burkholderia Cenocepacia. Int. J. Immunopathol. Pharmacol. 2005, 18, 661–670. [Google Scholar] [CrossRef] [PubMed]
- Schooling, S.R.; Beveridge, T.J. Membrane Vesicles: An Overlooked Component of the Matrices of Biofilms. J. Bacteriol. 2006, 188, 5945–5957. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Inagaki, S.; Onishi, S.; Kuramitsu, H.K.; Sharma, A. Porphyromonas Gingivalis Vesicles Enhance Attachment, and the Leucine-Rich Repeat BspA Protein Is Required for Invasion of Epithelial Cells by “Tannerella Forsythia”. Infect. Immun. 2006, 74, 5023–5028. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fleming, A.; Sampey, G.; Chung, M.C.; Bailey, C.; van Hoek, M.L.; Kashanchi, F.; Hakami, R.M. The Carrying Pigeons of the Cell: Exosomes and Their Role in Infectious Diseases Caused by Human Pathogens. Pathog. Dis. 2014, 71, 109–120. [Google Scholar] [CrossRef] [Green Version]
- Schorey, J.S.; Cheng, Y.; Singh, P.P.; Smith, V.L. Exosomes and Other Extracellular Vesicles in Host–Pathogen Interactions. EMBO Rep. 2015, 16, 24–43. [Google Scholar] [CrossRef] [Green Version]
- Van der Pol, E.; Böing, A.N.; Harrison, P.; Sturk, A.; Nieuwland, R. Classification, Functions, and Clinical Relevance of Extracellular Vesicles. Pharmacol. Rev. 2012, 64, 676–705. [Google Scholar] [CrossRef] [Green Version]
- Cvjetkovic, A.; Lötvall, J.; Lässer, C. The Influence of Rotor Type and Centrifugation Time on the Yield and Purity of Extracellular Vesicles. J. Extracell. Vesicles 2014, 3, 23111. [Google Scholar] [CrossRef] [Green Version]
- Böing, A.N.; van der Pol, E.; Grootemaat, A.E.; Coumans, F.A.W.; Sturk, A.; Nieuwland, R. Single-Step Isolation of Extracellular Vesicles by Size-Exclusion Chromatography. J. Extracell. Vesicles 2014, 3, 23430. [Google Scholar] [CrossRef] [PubMed]
- Oldstone, M.B.A. Viral Persistence: Parameters, Mechanisms and Future Predictions. Virology 2006, 344, 111–118. [Google Scholar] [CrossRef]
- Cox, J.E.; Sullivan, C.S. Balance and Stealth: The Role of Noncoding RNAs in the Regulation of Virus Gene Expression. Annu. Rev. Virol. 2014, 1, 89–109. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Han, Z.; Liu, X.; Chen, X.; Zhou, X.; Du, T.; Roizman, B.; Zhou, G. MiR-H28 and MiR-H29 Expressed Late in Productive Infection Are Exported and Restrict HSV-1 Replication and Spread in Recipient Cells. Proc. Natl. Acad. Sci. USA 2016, 113, E894–E901. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kalamvoki, M.; Du, T.; Roizman, B. Cells Infected with Herpes Simplex Virus 1 Export to Uninfected Cells Exosomes Containing STING, Viral MRNAs, and MicroRNAs. Proc. Natl. Acad. Sci. USA 2014, 111, E4991–E4996. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nandakumar, R.; Windross, S.J.; Paludan, S.R. Intercellular communication in the innate immune system through the cGAS-STING pathway. Methods Enzymol. 2019, 625, 1–11. [Google Scholar] [CrossRef]
- Nandakumar, R.; Tschismarov, R.; Meissner, F.; Prabakaran, T.; Krissanaprasit, A.; Farahani, E.; Zhang, B.-C.; Assil, S.; Martin, A.; Bertrams, W.; et al. Intracellular Bacteria Engage a STING–TBK1–MVB12b Pathway to Enable Paracrine CGAS–STING Signalling. Nat. Microbiol. 2019, 4, 701–713. [Google Scholar] [CrossRef]
- Khatua, A.K.; Taylor, H.E.; Hildreth, J.E.K.; Popik, W. Exosomes Packaging APOBEC3G Confer Human Immunodeficiency Virus Resistance to Recipient Cells. J. Virol. 2009, 83, 512–521. [Google Scholar] [CrossRef] [Green Version]
- Soros, V.B.; Yonemoto, W.; Greene, W.C. Newly Synthesized APOBEC3G Is Incorporated into HIV Virions, Inhibited by HIV RNA, and Subsequently Activated by RNase H. PLoS Pathog. 2007, 3, e15. [Google Scholar] [CrossRef] [Green Version]
- Chen, Y.; Wang, X.; Yu, Y.; Xiao, Y.; Huang, J.; Yao, Z.; Chen, X.; Zhou, T.; Li, P.; Xu, C. Serum Exosomes of Chronic Gastritis Patients Infected with Helicobacter pylori Mediate IL-1α Expression via IL-6 Trans-Signalling in Gastric Epithelial Cells. Clin. Exp. Immunol. 2018, 194, 339–349. [Google Scholar] [CrossRef] [Green Version]
- Zhang, W.; Jiang, X.; Bao, J.; Wang, Y.; Liu, H.; Tang, L. Exosomes in Pathogen Infections: A Bridge to Deliver Molecules and Link Functions. Front. Immunol. 2018, 9, 90. [Google Scholar] [CrossRef] [Green Version]
- De Carvalho, J.V.; De Castro, R.O.; Da Silva, E.Z.M.; Silveira, P.P.; Da Silva-Januário, M.E.; Arruda, E.; Jamur, M.C.; Oliver, C.; Aguiar, R.S.; DaSilva, L.L.P. Nef Neutralizes the Ability of Exosomes from CD4+ T Cells to Act as Decoys during HIV-1 Infection. PLoS ONE 2014, 9, e113691. [Google Scholar] [CrossRef] [Green Version]
- Cheng, Y.; Schorey, J.S. Extracellular Vesicles Deliver Mycobacterium RNA to Promote Host Immunity and Bacterial Killing. EMBO Rep. 2019, 20, e46613. [Google Scholar] [CrossRef] [PubMed]
- Bhatnagar, S.; Shinagawa, K.; Castellino, F.J.; Schorey, J.S. Exosomes Released from Macrophages Infected with Intracellular Pathogens Stimulate a Proinflammatory Response in Vitro and in Vivo. Blood 2007, 110, 3234–3244. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, C.; Chalasani, G.; Ng, Y.H.; Robbins, P.D. Exosomes Released from Mycoplasma Infected Tumor Cells Activate Inhibitory B Cells. PLoS ONE 2012, 7, e36138. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Radomskia, N.; Kargerhttps, A.; Franzkec, K.; Liebler-Tenoriod, E.; Jahnkea, R.; Matthiesena, S.; Knittlera, M.R. Chlamydia Psittaci-Infected Dendritic Cells Communicate with NK Cells via Exosomes To Activate Antibacterial Immunity. Infect. Immun. 2019, 88, e00541-19. [Google Scholar] [CrossRef] [PubMed]
- Vromman, F.; Perrinet, S.; Gehre, L.; Subtil, A. The DUF582 Proteins of Chlamydia trachomatis Bind to Components of the ESCRT Machinery, Which Is Dispensable for Bacterial Growth In Vitro. Front. Cell. Infect. Microbiol. 2016, 6, 123. [Google Scholar] [CrossRef] [Green Version]
- Herrmann, I.K.; Wood, M.J.A.; Fuhrmann, G. Extracellular Vesicles as a Next-Generation Drug Delivery Platform. Nat. Nanotechnol. 2021, 16, 748–759. [Google Scholar] [CrossRef]
- Murphy, D.E.; de Jong, O.G.; Brouwer, M.; Wood, M.J.; Lavieu, G.; Schiffelers, R.M.; Vader, P. Extracellular Vesicle-Based Therapeutics: Natural versus Engineered Targeting and Trafficking. Exp. Mol. Med. 2019, 51, 1–12. [Google Scholar] [CrossRef] [Green Version]
- Zhao, Y.; Li, X.; Zhang, W.; Yu, L.; Wang, Y.; Deng, Z.; Liu, M.; Mo, S.; Wang, R.; Zhao, J.; et al. Trends in the Biological Functions and Medical Applications of Extracellular Vesicles and Analogues. Acta Pharm. Sin. B 2021, 11, 2114–2135. [Google Scholar] [CrossRef]
- Wong, C.H.; Chen, Y.C. Clinical Significance of Exosomes as Potential Biomarkers in Cancer. World J. Clin. Cases 2019, 7, 171–190. [Google Scholar] [CrossRef]
- Yu, H.; Sun, T.; An, J.; Wen, L.; Liu, F.; Bu, Z.; Cui, Y.; Feng, J. Potential Roles of Exosomes in Parkinson’s Disease: From Pathogenesis, Diagnosis, and Treatment to Prognosis. Front. Cell Dev. Biol. 2020, 8, 86. [Google Scholar] [CrossRef] [Green Version]
- Li, K.L.; Huang, H.Y.; Ren, H.; Yang, X.L. Role of Exosomes in the Pathogenesis of Inflammation in Parkinson’s Disease. Neural Regen. Res. 2022, 17, 1898–1906. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Austin, R.H. Applications of Microfluidics in Stem Cell Biology. Bionanoscience 2012, 2, 277–286. [Google Scholar] [CrossRef]
- Kuate, S.; Cinatl, J.; Doerr, H.W.; Überla, K. Exosomal Vaccines Containing the S Protein of the SARS Coronavirus Induce High Levels of Neutralizing Antibodies. Virology 2007, 362, 26–37. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fu, Y.; Xiong, S. Tagged Extracellular Vesicles with the RBD of the Viral Spike Protein for Delivery of Antiviral Agents against SARS-CoV-2 Infection. J. Control. Release 2021, 335, 584–595. [Google Scholar] [CrossRef]
- Teng, Y.; Xu, F.; Zhang, X.; Mu, J.; Sayed, M.; Hu, X.; Lei, C.; Sriwastva, M.; Kumar, A.; Sundaram, K.; et al. Plant-Derived Exosomal MicroRNAs Inhibit Lung Inflammation Induced by Exosomes SARS-CoV-2 Nsp12. Mol. Ther. 2021, 29, 2424–2440. [Google Scholar] [CrossRef] [PubMed]
- Aljuhani, A.; Albalawi, O.; Albalawi, R.; Alsalama, R.; Alatawi, S.; Altemani, R.; Abdel-Maksoud, M.S.; Elsherbiny, N. Exosomes in COVID-19 Infection: Focus on Role in Diagnosis, Pathogenesis, Immunity, and Clinical Trials. Cell Biol. Int. 2023, 47, 1049–1067. [Google Scholar] [CrossRef] [PubMed]
- Jesus, S.; Soares, E.; Cruz, M.T.; Borges, O. Exosomes as Adjuvants for the Recombinant Hepatitis B Antigen: First Report. Eur. J. Pharm. Biopharm. 2018, 133, 1–11. [Google Scholar] [CrossRef]
- Foster, J.L.; Denial, S.J.; Temple, B.R.S.; Garcia, J.V. Mechanisms of HIV-1 Nef Function and Intracellular Signaling. J. Neuroimmune Pharmacol. 2011, 6, 230–246. [Google Scholar] [CrossRef] [Green Version]
- Ferrantelli, F.; Manfredi, F.; Chiozzini, C.; Anticoli, S.; Olivetta, E.; Arenaccio, C.; Federico, M. DNA Vectors Generating Engineered Exosomes Potential CTL Vaccine Candidates Against AIDS, Hepatitis B, and Tumors. Mol. Biotechnol. 2018, 60, 773–782. [Google Scholar] [CrossRef]
- Näslund, T.I.; Paquin-Proulx, D.; Paredes, P.T.; Vallhov, H.; Sandberg, J.K.; Gabrielsson, S. Exosomes from Breast Milk Inhibit HIV-1 Infection of Dendritic Cells and Subsequent Viral Transfer to CD4+ T Cells. Aids 2014, 28, 171–180. [Google Scholar] [CrossRef] [Green Version]
- Madison, M.N.; Roller, R.J.; Okeoma, C.M. Human Semen Contains Exosomes with Potent Anti-HIV-1 Activity. Retrovirology 2014, 11, 1–16. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Green, B.N.; Johnson, C.D.; Egan, J.T.; Rosenthal, M.; Griffith, E.A.; Evans, M.W. Methicillin-Resistant Staphylococcus aureus: An Overview for Manual Therapists. J. Chiropr. Med. 2012, 11, 64–76. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, A.S.; De Lencastre, H.; Garau, J.; Kluytmans, J.; Malhotra-Kumar, S.; Peschel, A.; Harbarth, S. Methicillin-Resistant Staphylococcus aureus. Nat. Rev. Dis. Prim. 2018, 4, 18033. [Google Scholar] [CrossRef] [Green Version]
- Qian, Z.; Bai, Y.; Zhou, J.; Li, L.; Na, J.; Fan, Y.; Guo, X.; Liu, H. A Moisturizing Chitosan-Silk Fibroin Dressing with Silver Nanoparticles-Adsorbed Exosomes for Repairing Infected Wounds. J. Mater. Chem. B 2020, 8, 7197–7212. [Google Scholar] [CrossRef] [PubMed]
- Parveen, S.; Subramanian, K. Emerging Roles of Extracellular Vesicles in Pneumococcal Infections: Immunomodulators to Potential Novel Vaccine Candidates. Front. Cell. Infect. Microbiol. 2022, 12, 836070. [Google Scholar] [CrossRef]
- Shirejini, S.Z.; Inci, F. The Yin and Yang of Exosome Isolation Methods: Conventional Practice, Microfluidics, and Commercial Kits. Biotechnol. Adv. 2022, 54, 107814. [Google Scholar] [CrossRef]
- Abreu, C.M.; Costa-Silva, B.; Reis, R.L.; Kundu, S.C.; Caballero, D. Microfluidic Platforms for Extracellular Vesicle Isolation, Analysis and Therapy in Cancer. Lab Chip 2022, 22, 1093–1125. [Google Scholar] [CrossRef]

| Formulation Name | EV Origin | Therapeutic Application | Clinical Trial Identifier |
|---|---|---|---|
| ExoFlo™ | BMMSC | Moderate-to-severe ARDS in patients with COVID-19 | NCT04493242 NCT04657458 * |
| ExoFlo™ | BMMSC | Post-Acute COVID-19 and chronic Post-COVID-19 syndrome. | NCT05116761 * |
| UCMSC-EV | UCMSC | COVID-19 pneumonia | NCT05787288 * |
| UCMSC-EV | UCMSC | Chronic cough after COVID-19 infection. | NCT05808400 * |
| EV-Pure™ and WJ-Pure™ | Placenta and UCMSC | Moderate or severe ARDS in patients with COVID-19 | NCT05387278 |
| Zofin™ | AFMSC | Treating COVID-19 long haulers | NCT04384445 * |
| Zofin™ | AFMSC | Patients with mild to moderate COVID-19 | NCT04657406 * |
| Zofin™ | AFMSC | NCT05228899 * | |
| CSTC-Exo | COVID-19-specific T cells of convalescent patients | Early-stage COVID-19 pneumonia | NCT04389385 |
| haMSC-Exos | ADMSC | Severe COVID-19-related pneumonia | NCT04276987 |
| EXO-CD24 | CD24 expressing 293-TREx™ derived from HEK-293 cells | Moderate or severe COVID-19 infection | NCT04747574 |
| EXO-CD24 | CD24 expressing 293-TREx™ derived from HEK-293 cells | Moderate or severe COVID-19 infection | NCT04969172 |
| EXO-CD24 | CD24 expressing 293-TREx™ derived from HEK-293 cells | Moderate or severe COVID-19 infection | NCT04902183 * |
| ARDOXSO | PMSC | ARDS or COVID-19 pneumonia | NCT04798716 * |
| MSC-Exosomes | MSC | Immune modulation in COVID-19 infection | NCT05191381 * |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gonçalves, D.; Pinto, S.N.; Fernandes, F. Extracellular Vesicles and Infection: From Hijacked Machinery to Therapeutic Tools. Pharmaceutics 2023, 15, 1738. https://doi.org/10.3390/pharmaceutics15061738
Gonçalves D, Pinto SN, Fernandes F. Extracellular Vesicles and Infection: From Hijacked Machinery to Therapeutic Tools. Pharmaceutics. 2023; 15(6):1738. https://doi.org/10.3390/pharmaceutics15061738
Chicago/Turabian StyleGonçalves, Diogo, Sandra N. Pinto, and Fábio Fernandes. 2023. "Extracellular Vesicles and Infection: From Hijacked Machinery to Therapeutic Tools" Pharmaceutics 15, no. 6: 1738. https://doi.org/10.3390/pharmaceutics15061738





