Fabrication of Co-Assembly from Berberine and Tannic Acid for Multidrug-Resistant Bacteria Infection Treatment
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials
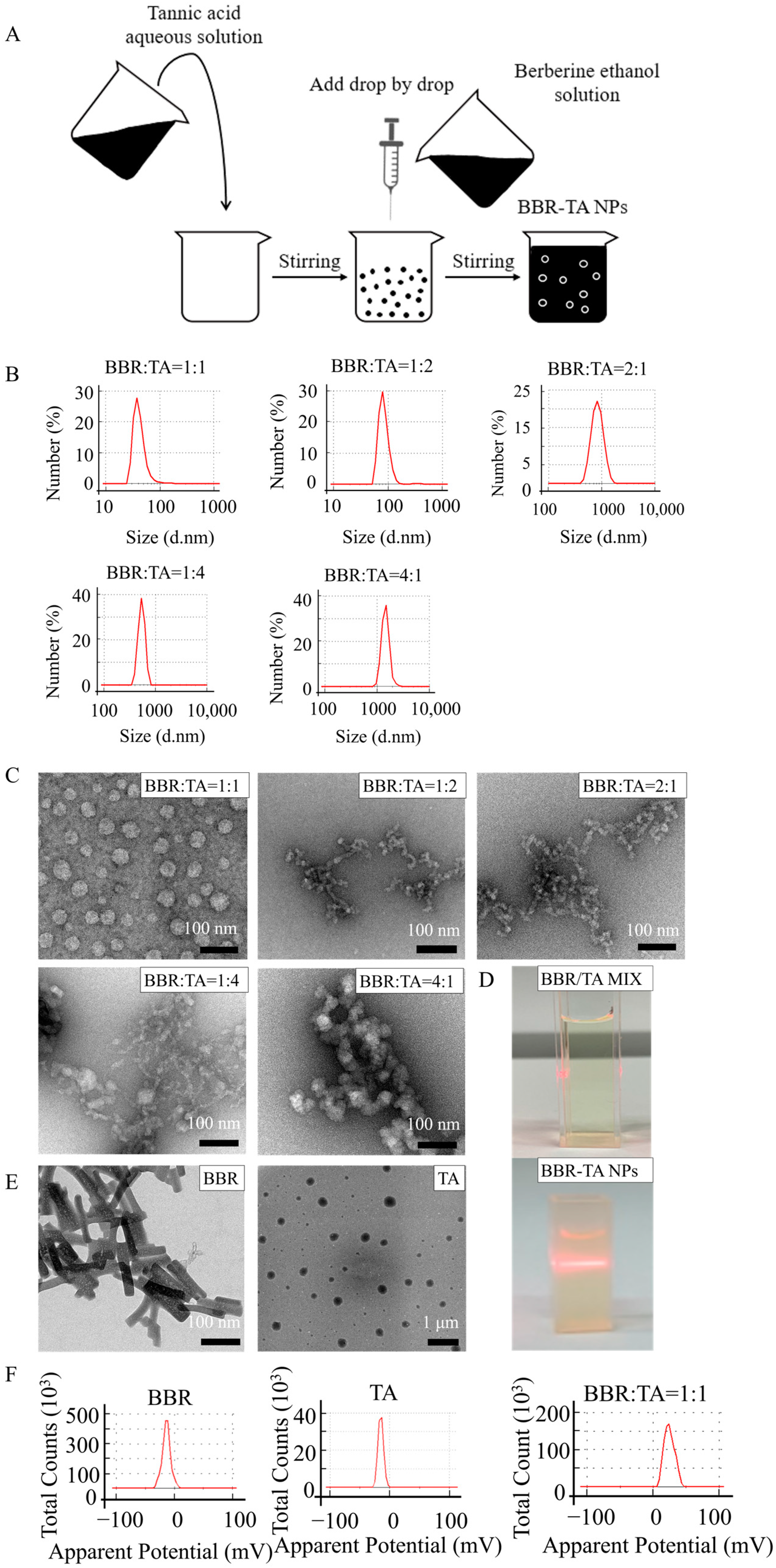
2.2. Preparation of BBR-TA NPs
2.3. Characterization of BBR-TA NPs
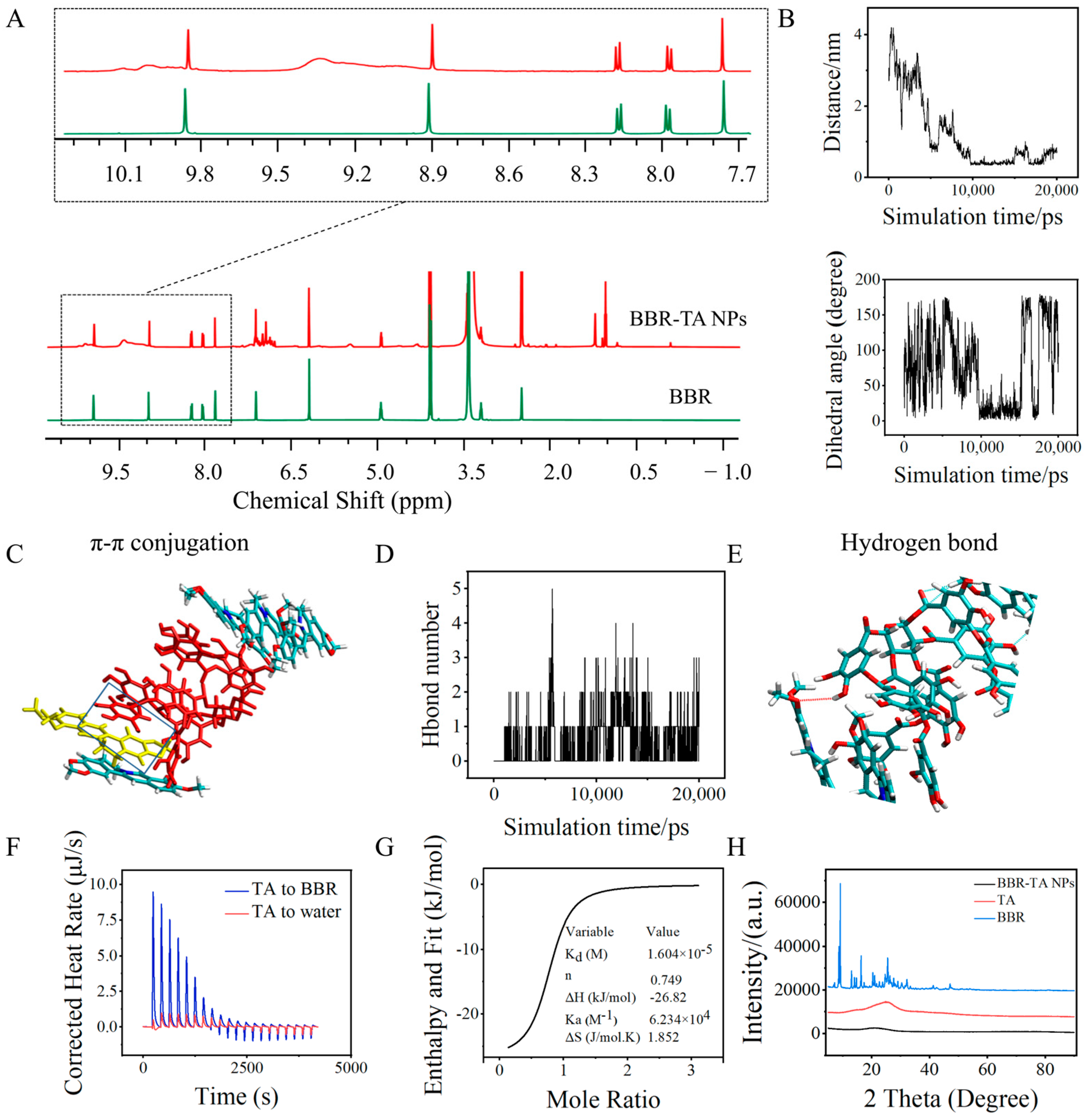
2.4. Molecular Dynamics Simulations
2.5. Bacteria and Culture Conditions
2.6. Determination of Inhibition Ratio
2.7. Bacterial Cell Membrane Integrity
2.8. Visual Assay of the Scavenging Effect of Biofilm
2.9. Determination of Intracellular ATP in Bacteria
2.10. Cell Cycle
2.11. Visual Assay of the Interaction Force between Bacteria and BBR-TA NPs
2.12. Cytotoxicity Evaluation
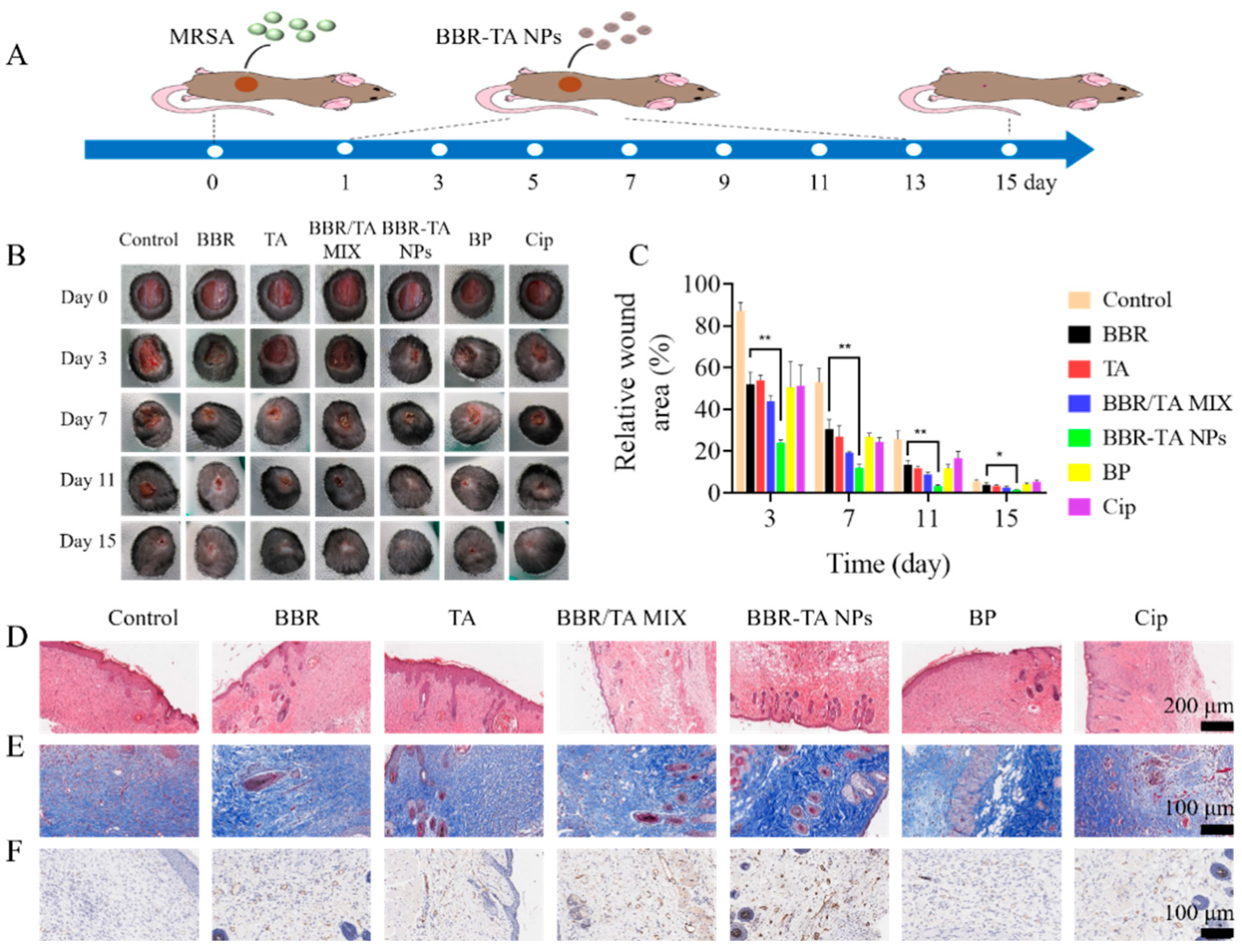
2.13. In Vivo Therapeutic Efficacy in Bacteria-Infected Wound Mice Model
2.14. Statistical Analysis
3. Results and Discussion
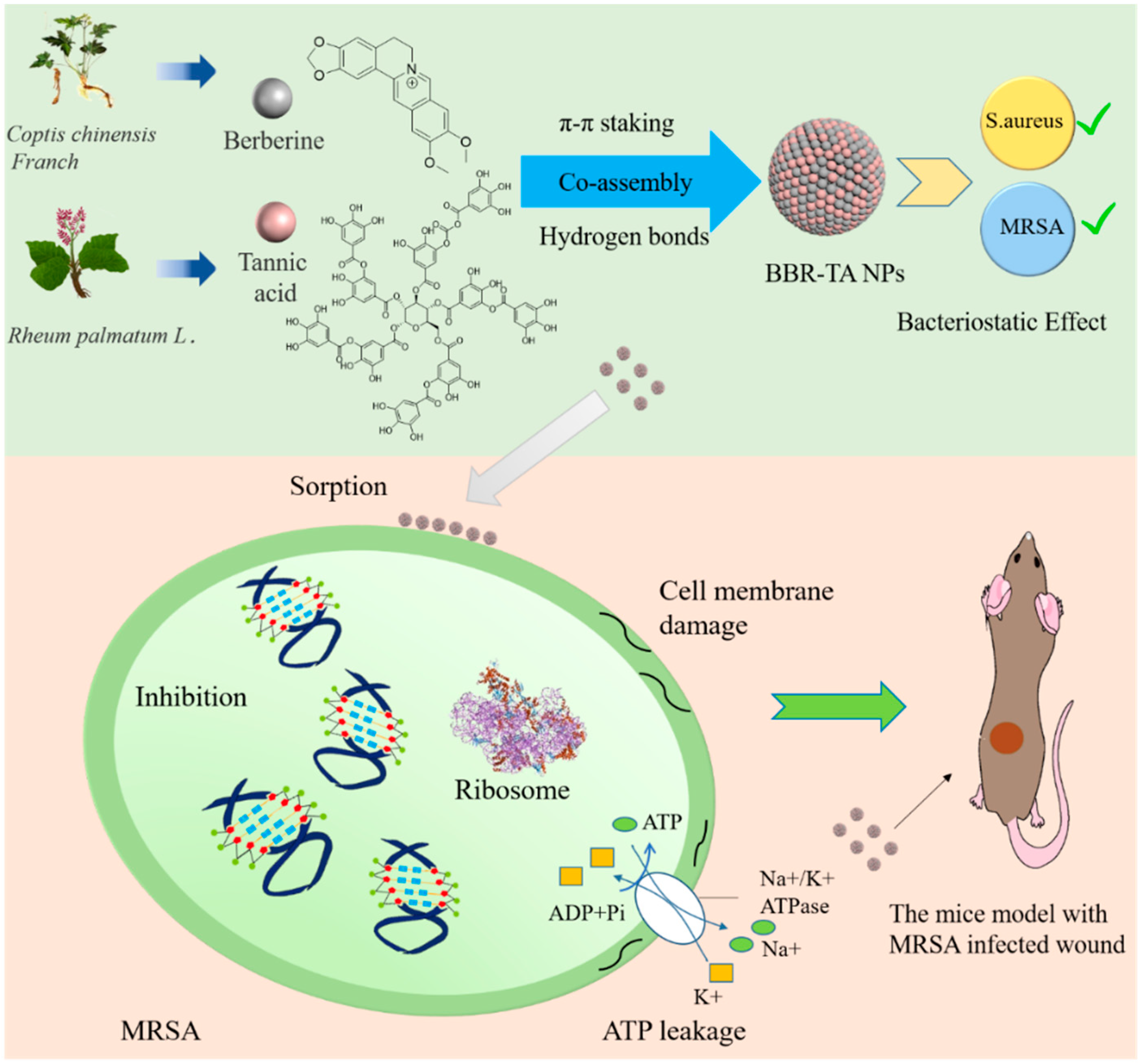
3.1. Preparation and Characterization of BBR-TA NPs
3.2. Co-Assembly Mechanism of BBR-TA NPs
3.3. Antibacterial Effect of BBR-TA NPs
3.4. Antibacterial Mechanism of BBR-TA NPs
3.5. Therapeutic Effect of BBR-TA NPs on MRSA-Infected Wounds in Mice
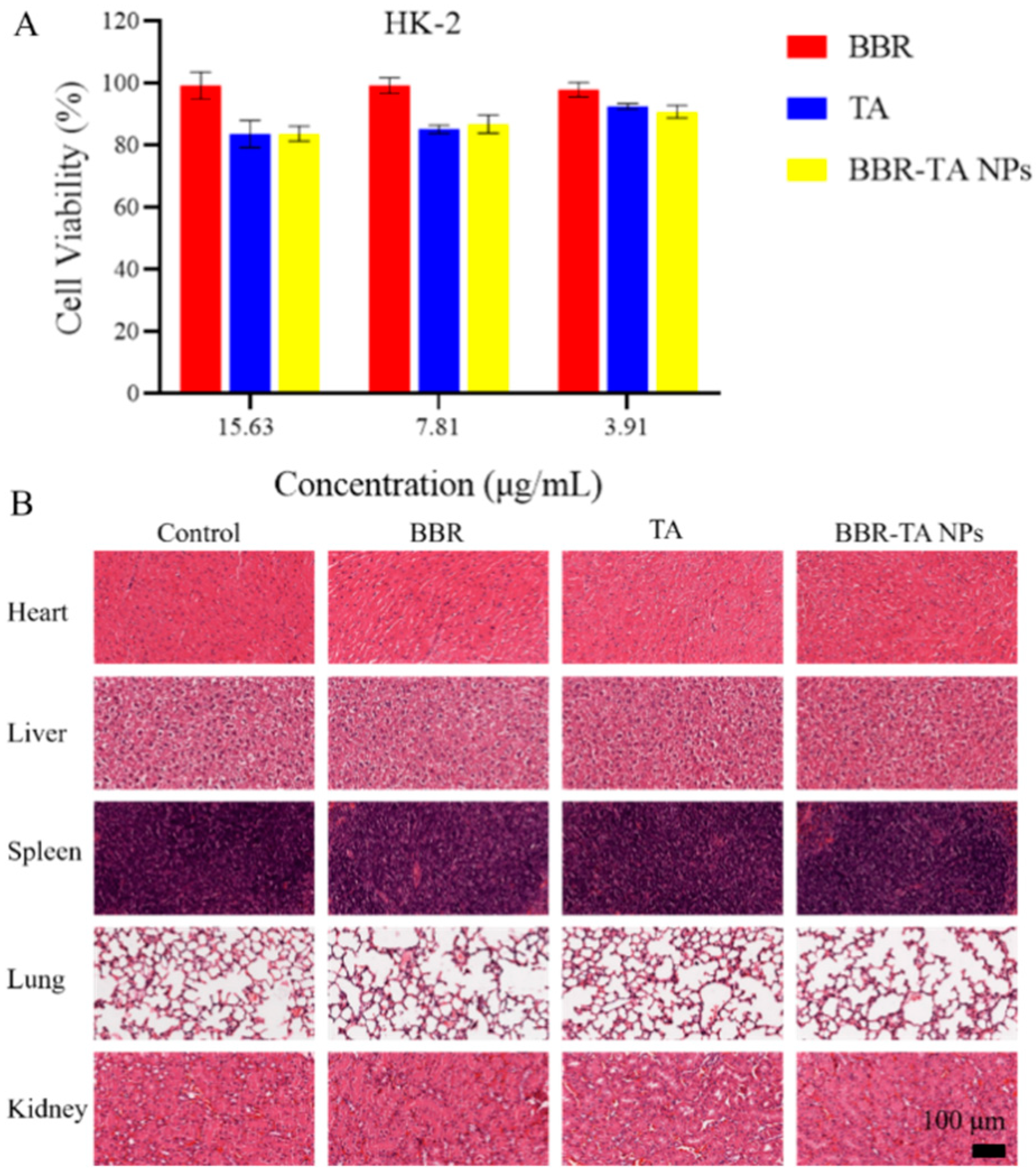
3.6. Biocompatibility Evaluation of BBR-TA NPs
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Darboe, S.; Okomo, U.; Muhammad, A.K.; Ceesay, B.; Jallow, M.; Usuf, E.; Tweed, S.; Akpalu, E.; Kwambana-Adams, B.; Kariuki, S.; et al. Community-acquired Invasive Bacterial Disease in Urban Gambia, 2005–2015: A Hospital-based Surveillance. Clin. Infect. Dis. 2019, 69, S105–S113. [Google Scholar] [CrossRef]
- Vrancianu, O.; Gheorghe, I.; Dobre, E.G.; Czobor, I.; Chifiriuc, M.C. Emerging Strategies to Combat β-Lactamase Producing ESKAPE Pathogens. Int. J. Mol. Sci. 2020, 21, 8527. [Google Scholar] [CrossRef] [PubMed]
- Hampton, H.G.; Watson, B.; Fineran, P.C. The arms race between bacteria and their phage foes. Nature 2020, 577, 327–336. [Google Scholar] [CrossRef] [PubMed]
- Ventola, C.L. The antibiotic resistance crisis: Part 1: Causes and threats. Pharm. Ther. 2015, 40, 277–283. [Google Scholar]
- Michael, C.A.; Dominey-Howes, D.; Labbate, M. The Antimicrobial Resistance Crisis: Causes, Consequences, and Management. Front. Public Health 2014, 2, 145. [Google Scholar] [CrossRef]
- Kim, M.; Park, J.; Kang, M.; Yang, J.; Park, W. Gain and loss of antibiotic resistant genes in multidrug resistant bacteria: One Health perspective. J. Microbiol. 2021, 59, 535–545. [Google Scholar] [CrossRef]
- Jansen, K.U.; Gruber, W.C.; Simon, R.; Wassil, J.; Anderson, A.S. The impact of human vaccines on bacterial antimicrobial resistance. A review. Environ. Chem. Lett. 2021, 19, 4031–4062. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.X. Global concern: Strategies for antibiotic development and risk of resistance. Chin. J. Antibiot. 2019, 44, 1–8. [Google Scholar]
- Asenjo, A.; Oteo-Iglesias, J.; Alós, J. What’s new in mechanisms of antibiotic resistance in bacteria of clinical origin? Enferm. Infecc. Microbiol. Clin. 2020, 39, 291–299. [Google Scholar] [CrossRef]
- Tang, S.; Zheng, J. Antibacterial Activity of Silver Nanoparticles: Structural Effects. Adv. Healthc. Mater. 2018, 7, e1701503. [Google Scholar] [CrossRef]
- Yin, I.X.; Zhang, J.; Zhao, I.S.; Mei, M.L.; Chu, C.H. The Antibacterial Mechanism of Silver Nanoparticles and Its Application in Dentistry. Int. J. Nanomed. 2020, 15, 2555–2562. [Google Scholar] [CrossRef]
- Liu, Y.; Shi, L.; Su, L.; Van, D.M.; Henny, C.; Jutte, P.C.; Ren, Y.; Busscher, H.J. Nanotechnology-based antimicrobials and delivery systems for biofilm-infection control. Chem. Soc. Rev. 2019, 48, 428–446. [Google Scholar] [CrossRef] [PubMed]
- Huh, A.J.; Kwon, Y.J. “Nanoantibiotics”: A new paradigm for treating infectious diseases using nanomaterials in the antibiotics resistant era. J. Control. Release 2011, 156, 128–145. [Google Scholar] [CrossRef] [PubMed]
- Kandi, V.; Kandi, S. Antimicrobial properties of nanomolecules: Potential candidates as antibiotics in the era of multi-drug resistance. Epidemiol. Health 2015, 37, e2015020. [Google Scholar] [CrossRef] [PubMed]
- Blecher, K.; Nasir, A.; Friedman, A. The growing role of nanotechnology in combating infectious disease. Virulence 2011, 2, 395–401. [Google Scholar] [CrossRef] [PubMed]
- Nisar, P.; Ali, N.; Rahman, L.; Ali, M.; Shinwari, Z.K. Antimicrobial activities of biologically synthesized metal nanoparticles: An insight into the mechanism of action. J. Biol. Inorg. Chem. 2019, 24, 929–941. [Google Scholar] [CrossRef]
- Napierska, D.; Thomassen, L.; Lison, D.; Martens, J.A.; Hoet, P.H. The nanosilica hazard: Another variable entity. Part. Fibre Toxicol. 2010, 7, 39. [Google Scholar] [CrossRef]
- Liu, J.; Zeng, F.; Allen, C. In vivo fate of unimers and micelles of a poly(ethylene glycol)-block-poly(caprolactone) copolymer in mice following intravenous administration. Eur. J. Pharm. Biopharm. 2007, 65, 309–319. [Google Scholar] [CrossRef]
- Elsabahy, M.; Wooley, K.L. Data Mining as a Guide for the Construction of Cross-Linked Nanoparticles with Low Immunotoxicity via Control of Polymer Chemistry and Supramolecular Assembly. Acc. Chem. Res. 2015, 48, 1620–1630. [Google Scholar] [CrossRef]
- Zheng, J.; Fan, R.; Wu, H.; Yao, H.; Yan, Y.; Liu, J.; Ran, L.; Sun, Z.; Yi, L.; Dang, L.; et al. Directed self-assembly of herbal small molecules into sustained release hydrogels for treating neural inflammation. Nat. Commun. 2019, 10, 1604. [Google Scholar] [CrossRef]
- Chen, M.H.; Lian, Y.Y.; Fang, D.S.; Chen, L.; Jia, J.; Zhang, W.L.; Lin, R.; Xie, Y.; Bi, H.K.; Jiang, H. Identification and antimicrobial properties of a new alkaloid produced by marine-derived Verrucosispora sp. FIM06-0036. Nat. Prod. Res. 2021, 35, 4211–4217. [Google Scholar] [CrossRef]
- Manosalva, L.; Mutis, A.; Urzúa, A.; Fajardo, V.; Quiroz, A. Antibacterial Activity of Alkaloid Fractions from Berberis microphylla G. Forst and Study of Synergism with Ampicillin and Cephalothin. Molecules 2016, 21, 76. [Google Scholar] [CrossRef] [PubMed]
- Torres, Y.R.; Berlinck, R.G.S.; Nascimento, G.G.F.; Fortier, S.C.; Pessoa, C.; Moraes, M.O.D. Antibacterial activity against resistant bacteria and cytotoxicity of four alkaloid toxins isolated from the marine sponge Arenosclera brasiliensis. Toxicon 2002, 40, 885–891. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Lam, A. Epigallocatechin gallate and gallic acid affect colonization of abiotic surfaces by oral bacteria. Arch. Oral Biol. 2020, 120, 104922. [Google Scholar] [CrossRef] [PubMed]
- Lu, J.; Wang, Z.; Ren, M.; Huang, G.; Fang, B.; Bu, X.; Liu, Y.; Guan, S. Antibacterial Effect of Gallic Acid against Aeromonas Hydrophila and Aeromonas Sobria through Damaging Membrane Integrity. Curr. Pharm. Biotechnol. 2016, 17, 1154–1158. [Google Scholar] [CrossRef]
- Gallique, M.; Wei, K.; Maisuria, V.B.; Okshevsky, M.; Tufenkji, N. Cranberry-Derived Proanthocyanidins Potentiate β-Lactam Antibiotics Against Resistant Bacteria. Appl. Environ. Microbiol. 2021, 87, e00127-21. [Google Scholar] [CrossRef]
- Rane, H.S.; Bernardo, S.M.; Howell, A.B.; Lee, S.A. Cranberry-derived proanthocyanidins prevent formation of Candida albicans biofilms in artificial urine through biofilm- and adherence-specific mechanisms. J. Antimicrob. Chemother. 2014, 69, 428–436. [Google Scholar] [CrossRef]
- Jing, W.; Xiaolan, C.; Yu, C.; Feng, Q.; Haifeng, Y. Pharmacological effects and mechanisms of tannic acid. Biomed. Pharmacother. 2022, 154, 113561. [Google Scholar] [CrossRef]
- Belhaoues, S.; Amri, S.; Bensouilah, M. Major phenolic compounds, antioxidant and antibacterial activities of Anthemis praecox Link aerial parts. S. Afr. J. Bot. 2020, 131, 200–205. [Google Scholar] [CrossRef]
- Pandey, A.; Negi, P.S. Phytochemical composition, in vitro antioxidant activity and antibacterial mechanisms of Neolamarckia cadamba fruits extracts. Nat. Prod. Res. 2018, 32, 1189–1192. [Google Scholar] [CrossRef]
- Aashique, M.; Roy, A.; Kosuru, R.Y.; Bera, S. Membrane Depolarization Sensitizes Pseudomonas aeruginosa Against Tannic Acid. Curr. Microbiol. 2021, 78, 713–717. [Google Scholar] [CrossRef]
- Li, H.; Fan, C.; Lu, H.; Feng, C.; He, P.; Yang, X.; Xiang, C.; Zuo, J.; Tang, W. Protective role of berberine on ulcerative colitis through modulating enteric glial cells-intestinal epithelial cells-immune cells interactions. Acta Pharm. Sin. B 2020, 10, 447–461. [Google Scholar] [CrossRef] [PubMed]
- Xiao, L.; Poudel, A.J.; Huang, L.X.; Wang, Y.; Abdalla, A.M.E.; Yang, G. Nanocellulose hyperfine network achieves sustained release of berberine hydrochloride solubilized with β-cyclodextrin for potential anti-infection oral administration. Int. J. Biol. Macromol. 2020, 153, 633–640. [Google Scholar] [CrossRef] [PubMed]
- Hu, H.; Zhou, S.; Sun, X.; Hao, L. A Potent antiarrhythmic drug N-methyl berbamine extends the action potential through inhibiting both calcium and potassium currents. J. Pharmacol. Sci. 2019, 142, 131–139. [Google Scholar] [CrossRef] [PubMed]
- Aleksandra, B.; Kamil, L.; Przemysaw, G. Berberine in the Treatment of Diabetes Mellitus: A Review. Endocr. Metab. Immune Disord. Drug Targets 2021, 21, 1379–1386. [Google Scholar]
- Hao, W.; Che, S.; Li, J.; Luo, J.; Zhang, W.; Chen, Y.; Zhao, Z.; Wei, H.; Xie, W. Synthesis of Berberine and Canagliflozin Chimera and Investigation into New Antibacterial Activity and Mechanisms. Molecules 2022, 27, 2948. [Google Scholar] [CrossRef]
- Tian, X.; Wang, P.; Li, T.; Huang, X.; Guo, W.; Yang, Y.; Yan, M.; Zhang, H.; Cai, D.; Jia, X. Self-assembled natural phytochemicals for synergistically antibacterial application from the enlightenment of traditional Chinese medicine combination. Acta Pharm. Sin. B 2020, 131, 200–205. [Google Scholar] [CrossRef]
- Xia, S.; Ma, L.; Wang, G.; Yang, J.; Zhang, M.; Wang, X.; Su, J.; Xie, M. In vitro Antimicrobial Activity and the Mechanism of Berberine Against Methicillin-Resistant Staphylococcus aureus Isolated from Bloodstream Infection Patients. Infect. Drug Resist 2022, 15, 1933–1944. [Google Scholar] [CrossRef]
- Wang, D.; Yu, L.; Xiang, H.; Fan, J.; He, L.; Guo, N.; Feng, H.; Deng, X. Global transcriptional profiles of Staphylococcus aureus treated with berberine chloride. FEMS Microbiol. Lett. 2008, 279, 217–225. [Google Scholar] [CrossRef]
- Malachowa, N.; Kobayashi, S.D.; Braughton, K.R.; DeLeo, F.R. Mouse Model of Staphylococcus aureus Skin Infection. Methods Mol. Biol. 2019, 1960, 139–147. [Google Scholar]
- Wan, Y.; Wang, X.; Yang, L.; Li, Q.; Zheng, X.; Bai, T.; Wang, X. Antibacterial Activity of Juglone Revealed in a Wound Model of Staphylococcus aureus Infection. Int. J. Mol. Sci. 2023, 24, 3931. [Google Scholar] [CrossRef]
- Sun, B.; Wu, F.; Zhang, Q.; Chu, X.; Wang, Z.; Huang, X.; Li, J.; Yao, C.; Zhou, N.; Shen, J. Insight into the effect of particle size distribution differences on the antibacterial activity of carbon dots. J. Colloid Interface Sci. 2021, 584, 505–519. [Google Scholar] [CrossRef]
- Zhao, Y.; Chen, F.; Pan, Y.; Li, Z.; Xue, X.; Okeke, C.I.; Wang, F.; Li, C.; Peng, L.; Wang, P.C.; et al. Nanodrug Formed by Coassembly of Dual Anticancer Drugs to Inhibit Cancer Cell Drug Resistance. ACS Appl. Mater. Interfaces 2015, 7, 19295–19305. [Google Scholar] [CrossRef]
- Huang, X.; Wang, P.; Li, T.; Tian, X.; Lei, H. Self-Assemblies Based on Traditional Medicine Berberine and Cinnamic Acid for Adhesion-Induced Inhibition Multidrug-Resistant Staphylococcus aureus. ACS Appl. Mater. Interfaces 2019, 12, 227–237. [Google Scholar] [CrossRef]
- Wiseman, T.; Williston, S.; Brandts, J.F.; Lin, L.N. Rapid measurement of binding constants and heats of binding using a new titration calorimeter. Anal. Biochem. 1989, 179, 131–137. [Google Scholar] [CrossRef]
- Patel, R.; Patel, M. Solid-State Characterization and Dissolution Properties of Lovastatin Hydroxylpropyl-β-Cyclodextrin Inclusion Complex. Pharm. Technol. 2007, 31, 72–82. [Google Scholar]
- Sahibzada, M.U.K.; Sadiq, A.; Faidah, H.S.; Khurram, M.; Amin, M.U.; Haseeb, A.; Kakar, M. Berberine nanoparticles with enhanced in vitro bioavailability: Characterization and antimicrobial activity. Drug Des. Dev. Ther. 2018, 12, 303–312. [Google Scholar] [CrossRef]
- Shi, C.; Zhang, X.; Sun, Y.; Yang, M.; Xia, X. Antimicrobial Activity of Ferulic Acid Against Cronobacter sakazakii and Possible Mechanism of Action. Foodborne Pathog. Dis. 2016, 13, 196. [Google Scholar] [CrossRef] [PubMed]
- Liu, M.; Yang, K.; Jing, W.; Jie, Z.; Qi, Y.; Wei, X.; Fan, M. Young astringent persimmon tannin inhibits methicillin-resistant Staphylococcus aureus isolated from pork-ScienceDirect. LWT 2019, 100, 48–55. [Google Scholar] [CrossRef]
- Roy, R.; Tiwari, M.; Donelli, G.; Tiwari, V. Strategies for combating bacterial biofilms: A focus on anti-biofilm agents and their mechanisms of action. Virulence 2018, 9, 522–554. [Google Scholar] [CrossRef] [PubMed]
- Li, T.; Wang, P.; Guo, W.; Huang, X.; Lei, H. Natural Berberine-Based Chinese Herb Medicine Assembled Nanostructures with Modified Antibacterial Application. ACS Nano 2019, 13, 6770–6781. [Google Scholar] [CrossRef] [PubMed]
- Gaglione, R.; Cesaro, A.; Dell’Olmo, E.; Ventura, B.D.; Casillo, A.; Girolamo, R.D.; Velotta, R.; Notomista, E.; Veldhuizen, E.; Corsaro, M.M. Effects of human antimicrobial cryptides identified in apolipoprotein B depend on specific features of bacterial strains. Sci. Rep. 2019, 9, 6728. [Google Scholar] [CrossRef] [PubMed]
- Arakha, M.; Saleem, M.; Mallick, B.C.; Jha, S. The effects of interfacial potential on antimicrobial propensity of ZnO nanoparticle. Sci. Rep. 2015, 5, 9578. [Google Scholar] [CrossRef] [PubMed]
- Li, R.; Fei, P.; Man, C.X.; Lou, B.B.; Niu, J.T.; Feng, J.; Sun, L.H.; Li, M.Y.; Jiang, Y.J. Tea polyphenols inactivate Cronobacter sakazakii isolated from powdered infant formula. J. Dairy Sci. 2016, 99, 1019–1028. [Google Scholar] [CrossRef]
- Zhu, Y.; Xu, J.; Wang, Y.; Chen, C.; Gu, H.; Chai, Y.; Wang, Y. Silver nanoparticles-decorated and mesoporous silica coated single-walled carbon nanotubes with an enhanced antibacterial activity for killing drug-resistant bacteria. Nano Res. 2020, 13, 389–400. [Google Scholar] [CrossRef]
- Wang, W.; Li, B.; Yang, H.; Lin, Z.; Chen, L.; Li, Z.; Ge, J.; Zhang, T.; Xia, H.; Li, L.; et al. Efficient elimination of multidrug-resistant bacteria using copper sulfide nanozymes anchored to graphene oxide nanosheets. Nano Res. 2020, 13, 2156–2164. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zheng, T.; Chen, H.; Wu, C.; Wang, J.; Cui, M.; Ye, H.; Feng, Y.; Li, Y.; Dong, Z. Fabrication of Co-Assembly from Berberine and Tannic Acid for Multidrug-Resistant Bacteria Infection Treatment. Pharmaceutics 2023, 15, 1782. https://doi.org/10.3390/pharmaceutics15071782
Zheng T, Chen H, Wu C, Wang J, Cui M, Ye H, Feng Y, Li Y, Dong Z. Fabrication of Co-Assembly from Berberine and Tannic Acid for Multidrug-Resistant Bacteria Infection Treatment. Pharmaceutics. 2023; 15(7):1782. https://doi.org/10.3390/pharmaceutics15071782
Chicago/Turabian StyleZheng, Tingting, Huan Chen, Chenyang Wu, Jinrui Wang, Mengyao Cui, Hanyi Ye, Yifan Feng, Ying Li, and Zhengqi Dong. 2023. "Fabrication of Co-Assembly from Berberine and Tannic Acid for Multidrug-Resistant Bacteria Infection Treatment" Pharmaceutics 15, no. 7: 1782. https://doi.org/10.3390/pharmaceutics15071782
APA StyleZheng, T., Chen, H., Wu, C., Wang, J., Cui, M., Ye, H., Feng, Y., Li, Y., & Dong, Z. (2023). Fabrication of Co-Assembly from Berberine and Tannic Acid for Multidrug-Resistant Bacteria Infection Treatment. Pharmaceutics, 15(7), 1782. https://doi.org/10.3390/pharmaceutics15071782





