cCPE Fusion Proteins as Molecular Probes to Detect Claudins and Tight Junction Dysregulation in Gastrointestinal Cell Lines, Tissue Explants and Patient-Derived Organoids
Abstract
1. Introduction
2. Materials and Methods
2.1. Protein Preparations
2.2. Cell Culture
2.3. Plate Reader Binding Assay
2.4. Ex vivo Incubation of Tissue Specimens with GST-cCPE
2.5. Generation and Characterization of Patient-Derived Gastric Cancer Organoids
2.5.1. Gastric Tissue Processing
2.5.2. Gastric Organoid Culture
2.6. Live-Cell Imaging
2.6.1. Live-Cell Imaging of HT-29/B6 with AF647-GST-cCPE and YFP-cCPE
2.6.2. Live-Cell Imaging of Colonic Ulcers
2.6.3. Live-Cell Imaging of Gastric Organoids
2.7. Histology and Immunochemistry
2.8. Quantification of cCPE Binding in Whole Mount Samples
2.9. Flow Cytometry
2.9.1. Flow Cytometry for Assessing YFP-cCPE Binding
2.9.2. Flow Cytometry of Gastric Organoids
2.10. Targeted DNA Sequencing
3. Results
3.1. YFP-cCPE-wt and YFP-cCPE-SSS Bind Specifically to CLDN4 and Other Claudins on the Cell Surface
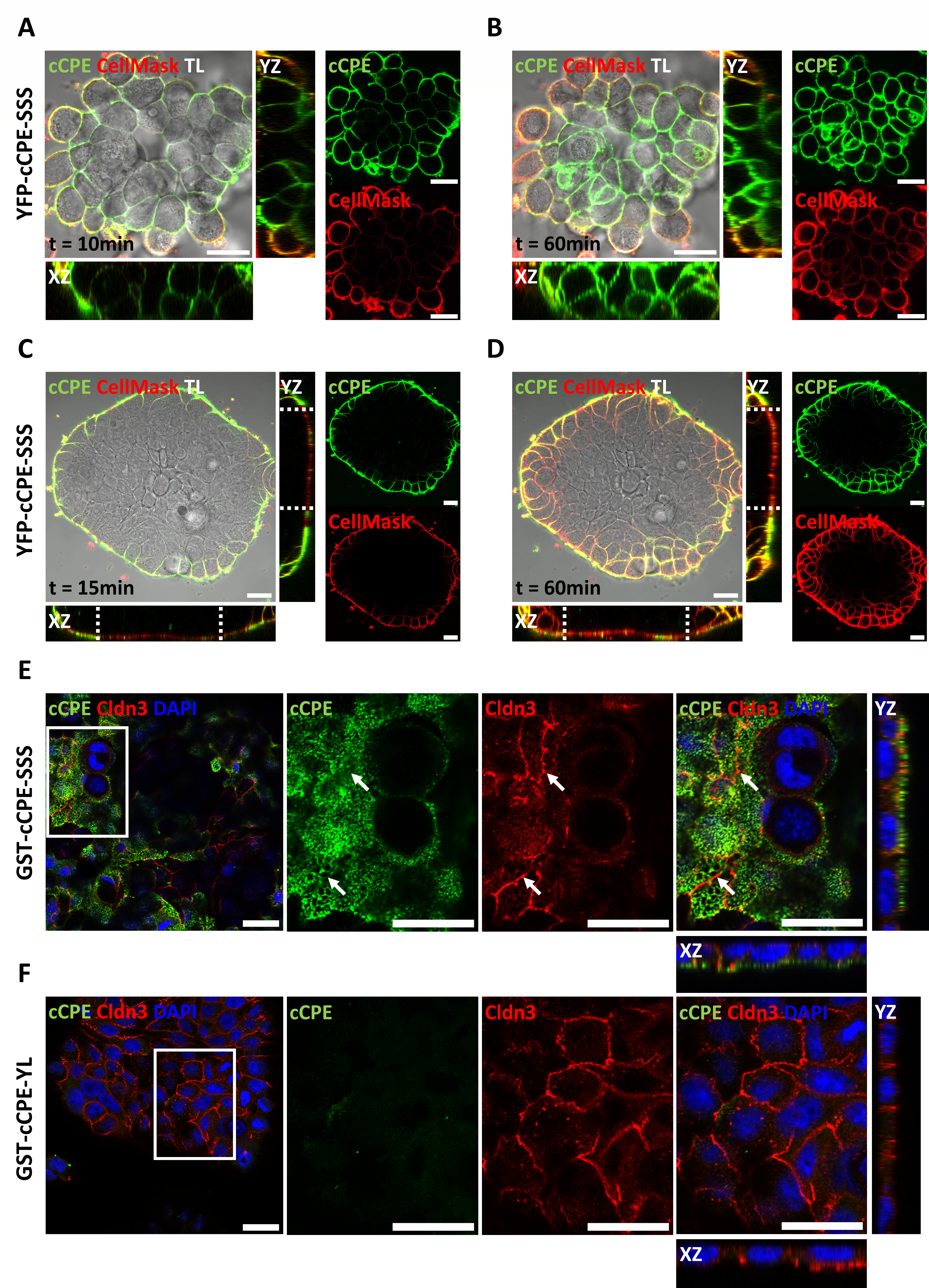
3.2. cCPE Fusion Proteins Visualize Non-Junctional Claudins and TJ Diffusion Barriers in Colonocyte Islands
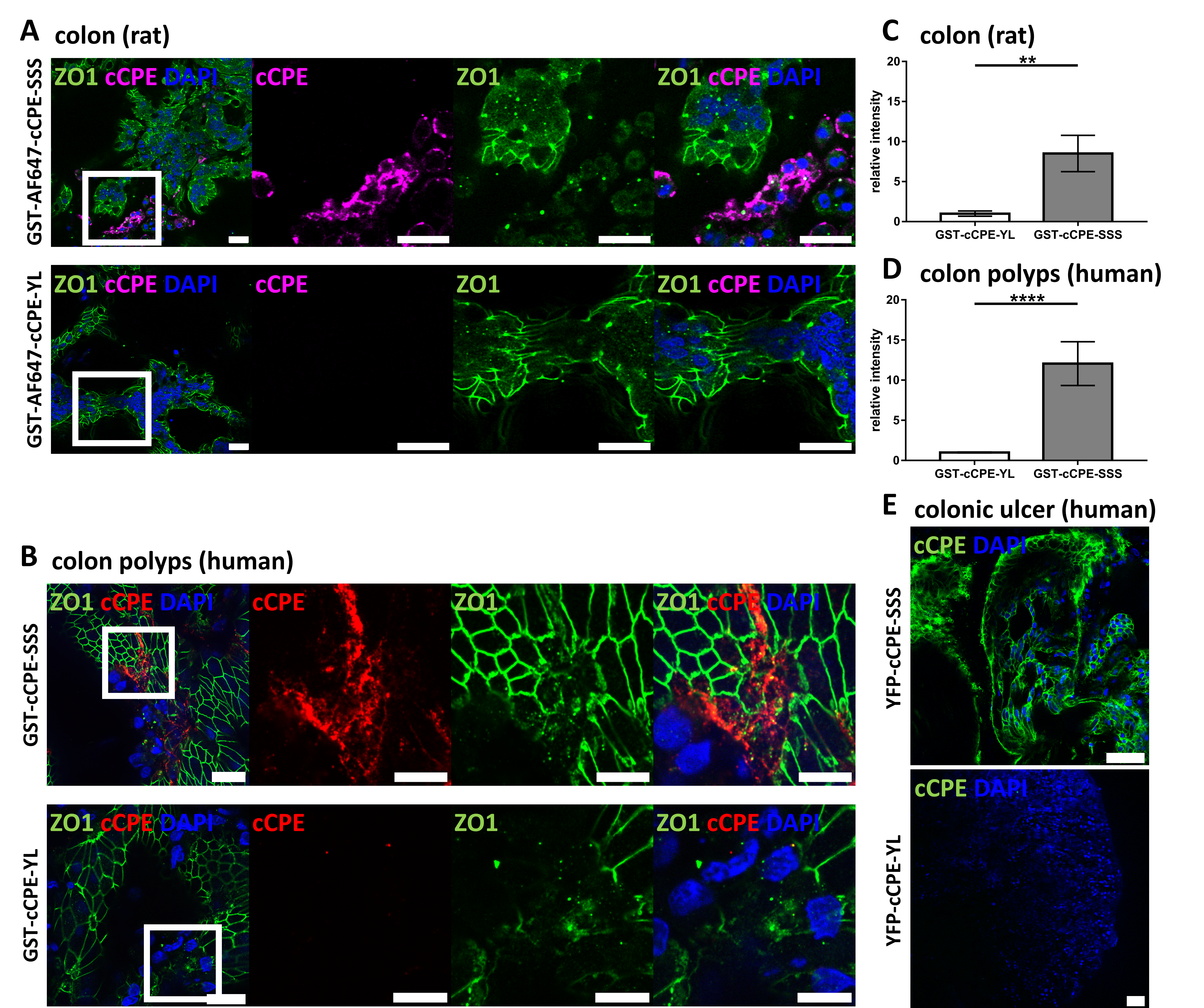
3.3. Use of cCPE Variants for Detection of Delocalized Non-Junctional Claudins in Ex Vivo Models of Rat and Human Colon Tissue
3.4. Use of YFP-cCPE to Probe the Expression, Polar Localization and Dysregulation of Claudins in Human Gastric Cancer Organoids
3.4.1. Establishment of Patient-Derived Gastric Organoids as a 3D Testing Platform for Cancer-Related Dedifferentiation
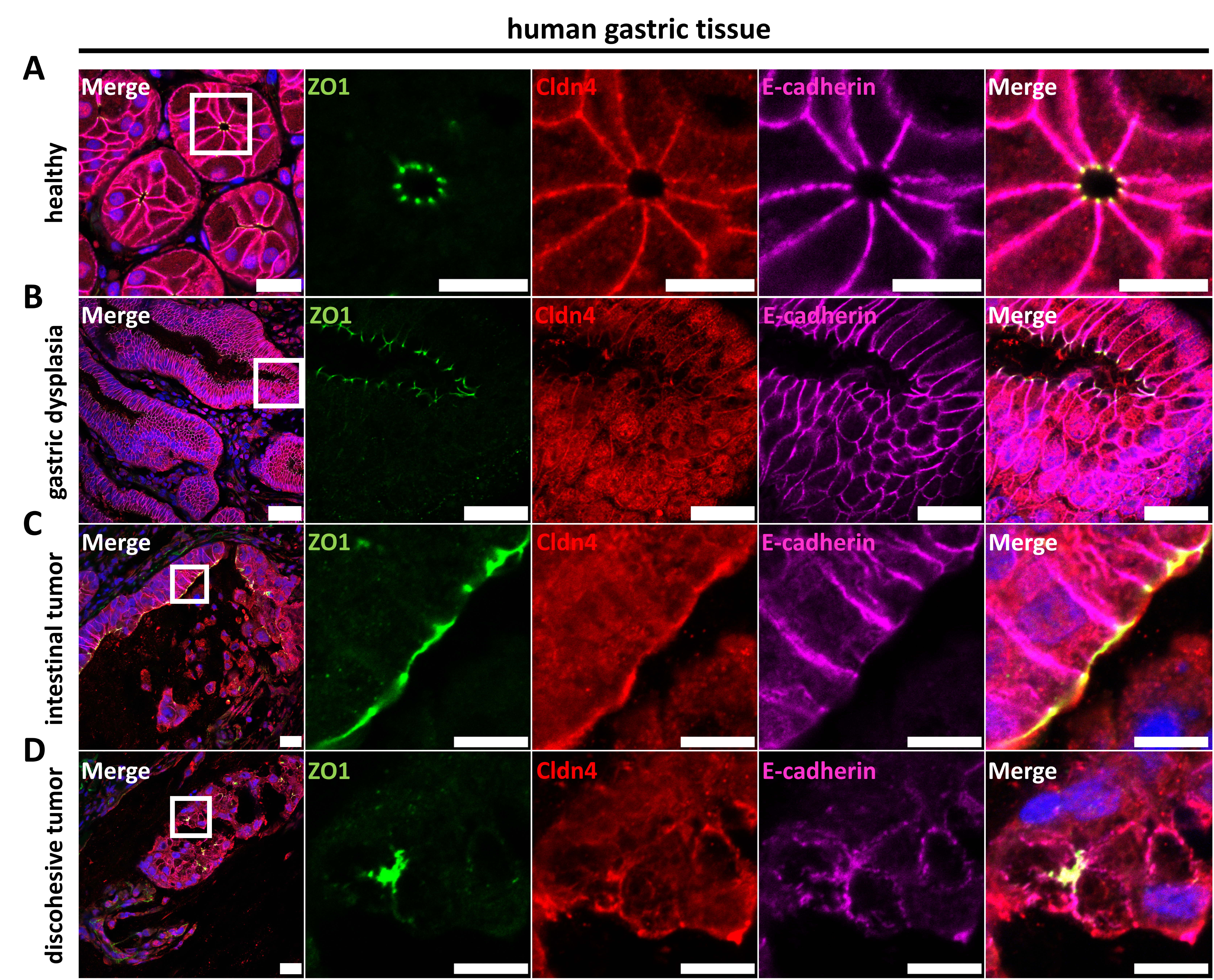
3.4.2. Characterization of Epithelial Differentiation in Gastric Tissue
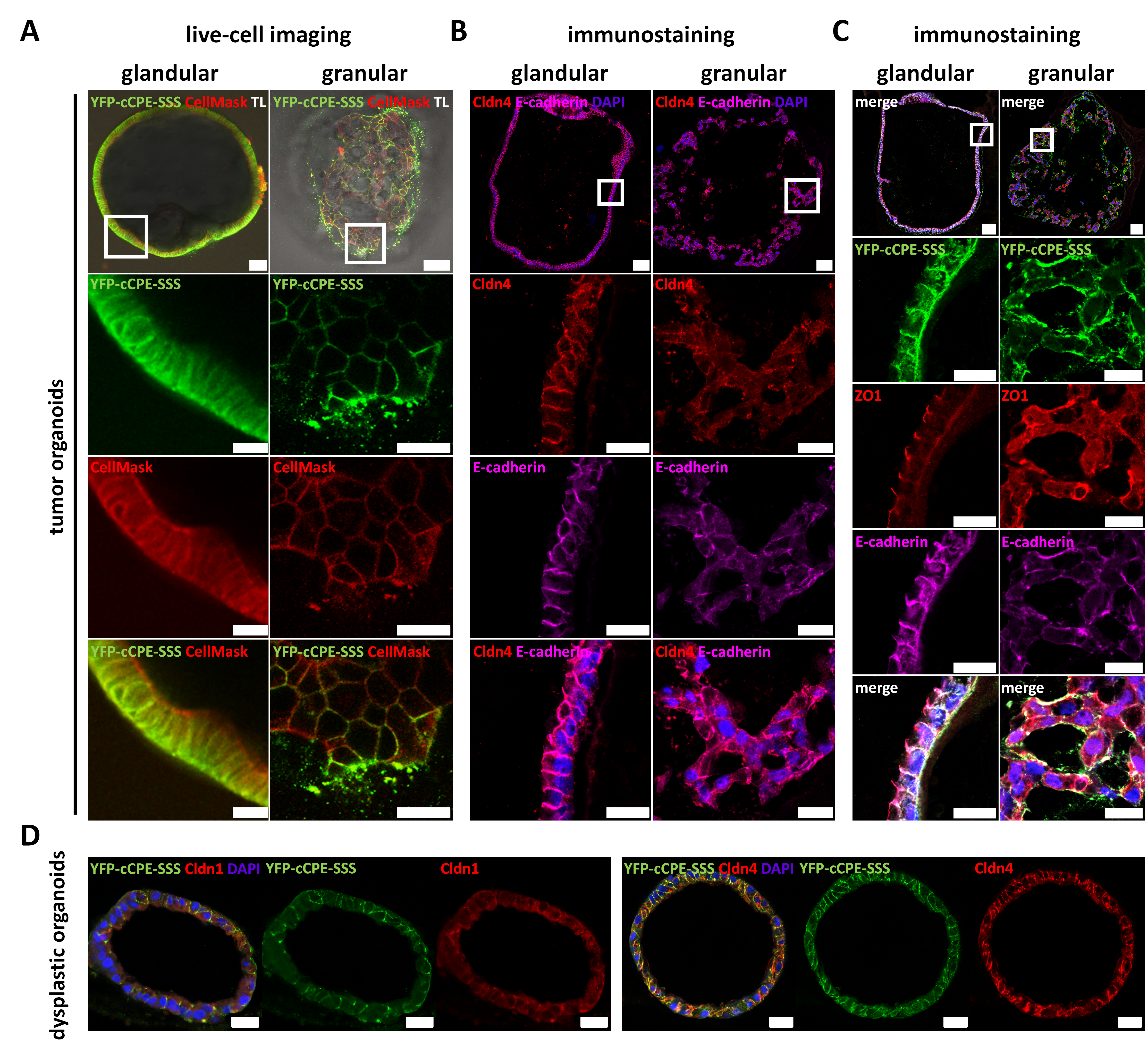
3.4.3. Claudin Expression, as Well as Differential Cell Polarity and Junctional Integrity, is Largely Conserved between Parental Tissues and Organoids
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Günzel, D.; Fromm, M. Claudins and other tight junction proteins. Compr. Physiol. 2012, 2, 1819–1852. [Google Scholar] [CrossRef] [PubMed]
- Hartsock, A.; Nelson, W.J. Adherens and tight junctions: Structure, function and connections to the actin cytoskeleton. Biochim. Biophys. Acta 2008, 1778, 660–669. [Google Scholar] [CrossRef] [PubMed]
- Schlegel, N.; Boerner, K.; Waschke, J. Targeting desmosomal adhesion and signalling for intestinal barrier stabilization in inflammatory bowel diseases-Lessons from experimental models and patients. Acta Physiol. 2021, 231, e13492. [Google Scholar] [CrossRef] [PubMed]
- Farquhar, M.; Palade, G. Junctional complexes in various epithelia. J. Cell. Biol. 1963, 17, 375–412. [Google Scholar] [CrossRef] [PubMed]
- Furuse, M.; Sasaki, H.; Fujimoto, K.; Tsukita, S. A single gene product, claudin-1 or -2, reconstitutes tight junction strands and recruits occludin in fibroblasts. J. Cell. Biol. 1998, 143, 391–401. [Google Scholar] [CrossRef]
- Morita, K.; Furuse, M.; Fujimoto, K.; Tsukita, S. Claudin multigene family encoding four-transmembrane domain protein components of tight junction strands. Proc. Natl. Acad. Sci. USA 1999, 96, 511–516. [Google Scholar] [CrossRef]
- Mineta, K.; Yamamoto, Y.; Yamazaki, Y.; Tanaka, H.; Tada, Y.; Saito, K.; Tamura, A.; Igarashi, M.; Endo, T.; Takeuchi, K.; et al. Predicted expansion of the claudin multigene family. FEBS Lett. 2011, 585, 606–612. [Google Scholar] [CrossRef]
- Piontek, J.; Krug, S.M.; Protze, J.; Krause, G.; Fromm, M. Molecular architecture and assembly of the tight junction backbone. Biochim. Biophys. Acta. Biomembr. 2020, 1862, 183279. [Google Scholar] [CrossRef]
- Claude, P.; Goodenough, D.A. Fracture faces of zonulae occludentes from “tight” and “leaky” epithelia. J. Cell Biol. 1973, 58, 390–400. [Google Scholar] [CrossRef]
- Otani, T.; Nguyen, T.P.; Tokuda, S.; Sugihara, K.; Sugawara, T.; Furuse, K.; Miura, T.; Ebnet, K.; Furuse, M. Claudins and JAM-A coordinately regulate tight junction formation and epithelial polarity. J. Cell Biol. 2019, 218, 3372–3396. [Google Scholar] [CrossRef]
- Barmeyer, C.; Schulzke, J.D.; Fromm, M. Claudin-related intestinal diseases. Semin. Cell Dev. Biol. 2015, 42, 30–38. [Google Scholar] [CrossRef] [PubMed]
- Hashimoto, I.; Oshima, T. Claudins and Gastric Cancer: An Overview. Cancers 2022, 14, 290. [Google Scholar] [CrossRef] [PubMed]
- Oshima, T.; Miwa, H.; Joh, T. Changes in the expression of claudins in active ulcerative colitis. J. Gastroenterol. Hepatol. 2008, 23 (Suppl. 2), S146–S150. [Google Scholar] [CrossRef]
- Prasad, S.; Mingrino, R.; Kaukinen, K.; Hayes, K.L.; Powell, R.M.; MacDonald, T.T.; Collins, J.E. Inflammatory processes have differential effects on claudins 2, 3 and 4 in colonic epithelial cells. Lab. Investig. 2005, 85, 1139–1162. [Google Scholar] [CrossRef] [PubMed]
- Zeissig, S.; Burgel, N.; Günzel, D.; Richter, J.; Mankertz, J.; Wahnschaffe, U.; Kroesen, A.J.; Zeitz, M.; Fromm, M.; Schulzke, J.D. Changes in expression and distribution of claudin 2, 5 and 8 lead to discontinuous tight junctions and barrier dysfunction in active Crohn’s disease. Gut 2007, 56, 61–72. [Google Scholar] [CrossRef]
- Weber, C.R.; Nalle, S.C.; Tretiakova, M.; Rubin, D.T.; Turner, J.R. Claudin-1 and claudin-2 expression is elevated in inflammatory bowel disease and may contribute to early neoplastic transformation. Lab. Investig. 2008, 88, 1110–1120. [Google Scholar] [CrossRef]
- Turpin, W.; Lee, S.H.; Raygoza Garay, J.A.; Madsen, K.L.; Meddings, J.B.; Bedrani, L.; Power, N.; Espin-Garcia, O.; Xu, W.; Smith, M.I.; et al. Increased Intestinal Permeability Is Associated With Later Development of Crohn’s Disease. Gastroenterology 2020, 159, 2092–2100.e5. [Google Scholar] [CrossRef]
- D’Inca, R.; Di Leo, V.; Corrao, G.; Martines, D.; D’Odorico, A.; Mestriner, C.; Venturi, C.; Longo, G.; Sturniolo, G.C. Intestinal permeability test as a predictor of clinical course in Crohn’s disease. Am. J. Gastroenterol. 1999, 94, 2956–2960. [Google Scholar] [CrossRef]
- Dhawan, P.; Singh, A.B.; Deane, N.G.; No, Y.; Shiou, S.R.; Schmidt, C.; Neff, J.; Washington, M.K.; Beauchamp, R.D. Claudin-1 regulates cellular transformation and metastatic behavior in colon cancer. J. Clin. Investig. 2005, 115, 1765–1776. [Google Scholar] [CrossRef]
- Philip, R.; Heiler, S.; Mu, W.; Buchler, M.W.; Zoller, M.; Thuma, F. Claudin-7 promotes the epithelial-mesenchymal transition in human colorectal cancer. Oncotarget 2015, 6, 2046–2063. [Google Scholar] [CrossRef]
- Dahiya, N.; Becker, K.G.; Wood, W.H., 3rd; Zhang, Y.; Morin, P.J. Claudin-7 is frequently overexpressed in ovarian cancer and promotes invasion. PLoS ONE 2011, 6, e22119. [Google Scholar] [CrossRef] [PubMed]
- Kwon, M.J.; Kim, S.H.; Jeong, H.M.; Jung, H.S.; Kim, S.S.; Lee, J.E.; Gye, M.C.; Erkin, O.C.; Koh, S.S.; Choi, Y.L.; et al. Claudin-4 overexpression is associated with epigenetic derepression in gastric carcinoma. Lab. Investig. 2011, 91, 1652–1667. [Google Scholar] [CrossRef] [PubMed]
- Resnick, M.B.; Gavilanez, M.; Newton, E.; Konkin, T.; Bhattacharya, B.; Britt, D.E.; Sabo, E.; Moss, S.F. Claudin expression in gastric adenocarcinomas: A tissue microarray study with prognostic correlation. Hum. Pathol. 2005, 36, 886–892. [Google Scholar] [CrossRef] [PubMed]
- Kim, W.S.; Kim, H.; Joo, M.K.; Choi, B.I.; Yoo, A.Y.; Park, J.J.; Lee, B.J.; Kim, S.H.; Chun, H.J. High Expression of Claudin-4 Is Associated with Synchronous Tumors in Patients with Early Gastric Cancer. J. Clin. Med. 2022, 11, 3550. [Google Scholar] [CrossRef] [PubMed]
- Matsuda, Y.; Semba, S.; Ueda, J.; Fuku, T.; Hasuo, T.; Chiba, H.; Sawada, N.; Kuroda, Y.; Yokozaki, H. Gastric and intestinal claudin expression at the invasive front of gastric carcinoma. Cancer. Sci. 2007, 98, 1014–1019. [Google Scholar] [CrossRef] [PubMed]
- Fujiwara-Tani, R.; Mori, S.; Ogata, R.; Sasaki, R.; Ikemoto, A.; Kishi, S.; Kondoh, M.; Kuniyasu, H. Claudin-4: A New Molecular Target for Epithelial Cancer Therapy. Int. J. Mol. Sci. 2023, 24, 5494. [Google Scholar] [CrossRef]
- Hempel, C.; Protze, J.; Altun, E.; Riebe, B.; Piontek, A.; Fromm, A.; Lee, I.M.; Saleh, T.; Günzel, D.; Krause, G.; et al. Assembly of Tight Junction Strands: Claudin-10b and Claudin-3 Form Homo-Tetrameric Building Blocks that Polymerise in a Channel-Independent Manner. J. Mol. Biol. 2020, 432, 2405–2427. [Google Scholar] [CrossRef]
- Boireau, S.; Buchert, M.; Samuel, M.S.; Pannequin, J.; Ryan, J.L.; Choquet, A.; Chapuis, H.; Rebillard, X.; Avances, C.; Ernst, M.; et al. DNA-methylation-dependent alterations of claudin-4 expression in human bladder carcinoma. Carcinogenesis 2007, 28, 246–258. [Google Scholar] [CrossRef]
- Kwon, M.J. Emerging roles of claudins in human cancer. Int. J. Mol. Sci. 2013, 14, 18148–18180. [Google Scholar] [CrossRef]
- Corsini, M.; Ravaggi, A.; Odicino, F.; Santin, A.D.; Ravelli, C.; Presta, M.; Romani, C.; Mitola, S. Claudin3 is localized outside the tight junctions in human carcinomas. Oncotarget 2018, 9, 18446–18453. [Google Scholar] [CrossRef]
- Eichner, M.; Augustin, C.; Fromm, A.; Piontek, A.; Walther, W.; Bucker, R.; Fromm, M.; Krause, G.; Schulzke, J.D.; Günzel, D.; et al. In Colon Epithelia, Clostridium perfringens Enterotoxin Causes Focal Leaks by Targeting Claudins Which are Apically Accessible Due to Tight Junction Derangement. J. Infect. Dis. 2017, 217, 147–157. [Google Scholar] [CrossRef] [PubMed]
- Berselli, A.; Alberini, G.; Benfenati, F.; Maragliano, L. The Impact of Pathogenic and Artificial Mutations on Claudin-5 Selectivity from Molecular Dynamics Simulations. bioRxiv 2023. [Google Scholar] [CrossRef] [PubMed]
- Winkler, L.; Gehring, C.; Wenzel, A.; Muller, S.L.; Piehl, C.; Krause, G.; Blasig, I.E.; Piontek, J. Molecular determinants of the interaction between Clostridium perfringens enterotoxin fragments and claudin-3. J. Biol. Chem. 2009, 284, 18863–18872. [Google Scholar] [CrossRef] [PubMed]
- Yamaguchi, H.; Kojima, T.; Ito, T.; Kyuno, D.; Kimura, Y.; Imamura, M.; Hirata, K.; Sawada, N. Effects of Clostridium perfringens enterotoxin via claudin-4 on normal human pancreatic duct epithelial cells and cancer cells. Cell Mol. Biol. Lett. 2011, 16, 385–397. [Google Scholar] [CrossRef] [PubMed]
- Krause, G.; Protze, J.; Piontek, J. Assembly and function of claudins: Structure-function relationships based on homology models and crystal structures. Semin. Cell Dev. Biol. 2015, 42, 3–12. [Google Scholar] [CrossRef] [PubMed]
- Fujita, K.; Katahira, J.; Horiguchi, Y.; Sonoda, N.; Furuse, M.; Tsukita, S. Clostridium perfringens enterotoxin binds to the second extracellular loop of claudin-3, a tight junction integral membrane protein. FEBS Lett. 2000, 476, 258–261. [Google Scholar] [CrossRef]
- Veshnyakova, A.; Piontek, J.; Protze, J.; Waziri, N.; Heise, I.; Krause, G. Mechanism of Clostridium perfringens enterotoxin interaction with claudin-3/-4 protein suggests structural modifications of the toxin to target specific claudins. J. Biol. Chem. 2012, 287, 1698–1708. [Google Scholar] [CrossRef] [PubMed]
- Ogbu, C.P.; Roy, S.; Vecchio, A.J. Disruption of Claudin-Made Tight Junction Barriers by Clostridium perfringens Enterotoxin: Insights from Structural Biology. Cells 2022, 11, 903. [Google Scholar] [CrossRef]
- Protze, J.; Eichner, M.; Piontek, A.; Dinter, S.; Rossa, J.; Blecharz, K.G.; Vajkoczy, P.; Piontek, J.; Krause, G. Directed structural modification of Clostridium perfringens enterotoxin to enhance binding to claudin-5. Cell. Mol. Life Sci. CMLS 2015, 72, 1417–1432. [Google Scholar] [CrossRef]
- Neuhaus, W.; Piontek, A.; Protze, J.; Eichner, M.; Mahringer, A.; Subileau, E.A.; Lee, I.M.; Schulzke, J.D.; Krause, G.; Piontek, J. Reversible opening of the blood-brain barrier by claudin-5-binding variants of Clostridium perfringens enterotoxin’s claudin-binding domain. Biomaterials 2018, 161, 129–143. [Google Scholar] [CrossRef]
- Piontek, A.; Eichner, M.; Zwanziger, D.; Beier, L.S.; Protze, J.; Walther, W.; Theurer, S.; Schmid, K.W.; Fuhrer-Sakel, D.; Piontek, J.; et al. Targeting claudin-overexpressing thyroid and lung cancer by modified Clostridium perfringens enterotoxin. Mol. Oncol. 2020, 14, 261–276. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, A.; Saito, Y.; Kondoh, M.; Matsushita, K.; Krug, S.M.; Suzuki, H.; Tsujino, H.; Li, X.; Aoyama, H.; Matsuhisa, K.; et al. Creation and biochemical analysis of a broad-specific claudin binder. Biomaterials 2012, 33, 3464–3474. [Google Scholar] [CrossRef] [PubMed]
- Hashimoto, Y.; Yagi, K.; Kondoh, M. Roles of the first-generation claudin binder, Clostridium perfringens enterotoxin, in the diagnosis and claudin-targeted treatment of epithelium-derived cancers. Pflug. Arch. 2017, 469, 45–53. [Google Scholar] [CrossRef] [PubMed]
- Beier, L.S.; Rossa, J.; Woodhouse, S.; Bergmann, S.; Kramer, H.B.; Protze, J.; Eichner, M.; Piontek, A.; Vidal, Y.S.S.; Brandner, J.M.; et al. Use of Modified Clostridium perfringens Enterotoxin Fragments for Claudin Targeting in Liver and Skin Cells. Int. J. Mol. Sci. 2019, 20, 4774. [Google Scholar] [CrossRef]
- Beier, L.S.; Waldow, A.; Khomeijani Farahani, S.; Mannweiler, R.; Vidal, Y.S.S.; Brandner, J.M.; Piontek, J.; Günzel, D. Claudin targeting as an effective tool for directed barrier modulation of the viable epidermis. Ann. N. Y. Acad. Sci. 2022, 1517, 251–265. [Google Scholar] [CrossRef]
- Takahashi, A.; Komiya, E.; Kakutani, H.; Yoshida, T.; Fujii, M.; Horiguchi, Y.; Mizuguchi, H.; Tsutsumi, Y.; Tsunoda, S.; Koizumi, N.; et al. Domain mapping of a claudin-4 modulator, the C-terminal region of C-terminal fragment of Clostridium perfringens enterotoxin, by site-directed mutagenesis. Biochem. Pharm. 2008, 75, 1639–1648. [Google Scholar] [CrossRef]
- Sonoda, N.; Furuse, M.; Sasaki, H.; Yonemura, S.; Katahira, J.; Horiguchi, Y.; Tsukita, S. Clostridium perfringens enterotoxin fragment removes specific claudins from tight junction strands: Evidence for direct involvement of claudins in tight junction barrier. J. Cell Biol. 1999, 147, 195–204. [Google Scholar] [CrossRef]
- Li, X.; Saeki, R.; Watari, A.; Yagi, K.; Kondoh, M. Tissue distribution and safety evaluation of a claudin-targeting molecule, the C-terminal fragment of Clostridium perfringens enterotoxin. Eur. J. Pharm. Sci. 2014, 52, 132–137. [Google Scholar] [CrossRef]
- Hashimoto, Y.; Shirakura, K.; Okada, Y.; Takeda, H.; Endo, K.; Tamura, M.; Watari, A.; Sadamura, Y.; Sawasaki, T.; Doi, T.; et al. Claudin-5-Binders Enhance Permeation of Solutes across the Blood-Brain Barrier in a Mammalian Model. J. Pharm. Exp. 2017, 363, 275–283. [Google Scholar] [CrossRef]
- Becker, A.; Leskau, M.; Schlingmann-Molina, B.L.; Hohmeier, S.C.; Alnajjar, S.; Murua Escobar, H.; Ngezahayo, A. Functionalization of gold-nanoparticles by the Clostridium perfringens enterotoxin C-terminus for tumor cell ablation using the gold nanoparticle-mediated laser perforation technique. Sci. Rep. 2018, 8, 14963. [Google Scholar] [CrossRef]
- Powley, I.R.; Patel, M.; Miles, G.; Pringle, H.; Howells, L.; Thomas, A.; Kettleborough, C.; Bryans, J.; Hammonds, T.; MacFarlane, M.; et al. Patient-derived explants (PDEs) as a powerful preclinical platform for anti-cancer drug and biomarker discovery. Br. J. Cancer 2020, 122, 735–744. [Google Scholar] [CrossRef] [PubMed]
- Seidlitz, T.; Merker, S.R.; Rothe, A.; Zakrzewski, F.; von Neubeck, C.; Grutzmann, K.; Sommer, U.; Schweitzer, C.; Scholch, S.; Uhlemann, H.; et al. Human gastric cancer modelling using organoids. Gut 2019, 68, 207–217. [Google Scholar] [CrossRef] [PubMed]
- Clevers, H. Modeling Development and Disease with Organoids. Cell 2016, 165, 1586–1597. [Google Scholar] [CrossRef]
- Kreusel, K.M.; Fromm, M.; Schulzke, J.D.; Hegel, U. Cl- secretion in epithelial monolayers of mucus-forming human colon cells (HT-29/B6). Am. J. Physiol. 1991, 261, C574–C582. [Google Scholar] [CrossRef] [PubMed]
- Piontek, A.; Witte, C.; May Rose, H.; Eichner, M.; Protze, J.; Krause, G.; Piontek, J.; Schroder, L. A cCPE-based xenon biosensor for magnetic resonance imaging of claudin-expressing cells. Ann. N. Y. Acad. Sci. 2017, 1397, 195–208. [Google Scholar] [CrossRef]
- Gekle, M.; Wunsch, S.; Oberleithner, H.; Silbernagl, S. Characterization of two MDCK-cell subtypes as a model system to study principal cell and intercalated cell properties. Pflug. Arch. 1994, 428, 157–162. [Google Scholar] [CrossRef] [PubMed]
- Steele, N.G.; Chakrabarti, J.; Wang, J.; Biesiada, J.; Holokai, L.; Chang, J.; Nowacki, L.M.; Hawkins, J.; Mahe, M.; Sundaram, N.; et al. An Organoid-Based Preclinical Model of Human Gastric Cancer. Cell Mol. Gastroenterol. Hepatol. 2019, 7, 161–184. [Google Scholar] [CrossRef] [PubMed]
- Miyoshi, H.; Stappenbeck, T.S. In vitro expansion and genetic modification of gastrointestinal stem cells in spheroid culture. Nat. Protoc. 2013, 8, 2471–2482. [Google Scholar] [CrossRef] [PubMed]
- Ehlen, L.; Arndt, J.; Treue, D.; Bischoff, P.; Loch, F.N.; Hahn, E.M.; Kotsch, K.; Klauschen, F.; Beyer, K.; Margonis, G.A.; et al. Novel methods for in vitro modeling of pancreatic cancer reveal important aspects for successful primary cell culture. BMC Cancer 2020, 20, 417. [Google Scholar] [CrossRef]
- Rosenthal, R.; Günzel, D.; Piontek, J.; Krug, S.M.; Ayala-Torres, C.; Hempel, C.; Theune, D.; Fromm, M. Claudin-15 forms a water channel through the tight junction with distinct function compared to claudin-2. Acta. Physiol. 2020, 228, e13334. [Google Scholar] [CrossRef]
- Epple, H.J.; Amasheh, S.; Mankertz, J.; Goltz, M.; Schulzke, J.D.; Fromm, M. Early aldosterone effect in distal colon by transcriptional regulation of ENaC subunits. Am. J. Physiol. Gastrointest. Liver. Physiol. 2000, 278, G718–G724. [Google Scholar] [CrossRef] [PubMed]
- Barmeyer, C.; Fromm, M.; Schulzke, J.D. Active and passive involvement of claudins in the pathophysiology of intestinal inflammatory diseases. Pflug. Arch. 2017, 469, 15–26. [Google Scholar] [CrossRef] [PubMed]
- Lauren, P. The Two Histological Main Types of Gastric Carcinoma: Diffuse and So-Called Intestinal-Type Carcinoma. An Attempt at a Histo-Clinical Classification. Acta. Pathol. Microbiol. Scand. 1965, 64, 31–49. [Google Scholar] [CrossRef] [PubMed]
- Nagtegaal, I.D.; Odze, R.D.; Klimstra, D.; Paradis, V.; Rugge, M.; Schirmacher, P.; Washington, K.M.; Carneiro, F.; Cree, I.A.; Board, W.H.O.C.O.T.E. The 2019 WHO classification of tumours of the digestive system. Histopathology 2020, 76, 182–188. [Google Scholar] [CrossRef]
- Becker, K.; Mueller, J.D.; Schulmacher, C.; Ott, K.; Fink, U.; Busch, R.; Bottcher, K.; Siewert, J.R.; Hofler, H. Histomorphology and grading of regression in gastric carcinoma treated with neoadjuvant chemotherapy. Cancer 2003, 98, 1521–1530. [Google Scholar] [CrossRef]
- Saitoh, Y.; Suzuki, H.; Tani, K.; Nishikawa, K.; Irie, K.; Ogura, Y.; Tamura, A.; Tsukita, S.; Fujiyoshi, Y. Tight junctions. Structural insight into tight junction disassembly by Clostridium perfringens enterotoxin. Science 2015, 347, 775–778. [Google Scholar] [CrossRef]
- Fujiwara-Tani, R.; Sasaki, T.; Luo, Y.; Goto, K.; Kawahara, I.; Nishiguchi, Y.; Kishi, S.; Mori, S.; Ohmori, H.; Kondoh, M.; et al. Anti-claudin-4 extracellular domain antibody enhances the antitumoral effects of chemotherapeutic and antibody drugs in colorectal cancer. Oncotarget 2018, 9, 37367–37378. [Google Scholar] [CrossRef]
- Hashimoto, Y.; Okada, Y.; Shirakura, K.; Tachibana, K.; Sawada, M.; Yagi, K.; Doi, T.; Kondoh, M. Anti-Claudin Antibodies as a Concept for Development of Claudin-Directed Drugs. J. Pharm. Exp. 2019, 368, 179–186. [Google Scholar] [CrossRef]
- Tachibana, K.; Iwashita, Y.; Wakayama, E.; Nishino, I.; Nishikaji, T.; Kondoh, M. Tight Junction Modulating Bioprobes for Drug Delivery System to the Brain: A Review. Pharmaceutics 2020, 12, 1236. [Google Scholar] [CrossRef]
- Tachibana, K.; Hashimoto, Y.; Shirakura, K.; Okada, Y.; Hirayama, R.; Iwashita, Y.; Nishino, I.; Ago, Y.; Takeda, H.; Kuniyasu, H.; et al. Safety and efficacy of an anti-claudin-5 monoclonal antibody to increase blood-brain barrier permeability for drug delivery to the brain in a non-human primate. J. Control. Release 2021, 336, 105–111. [Google Scholar] [CrossRef]
- Qi, C.; Gong, J.; Li, J.; Liu, D.; Qin, Y.; Ge, S.; Zhang, M.; Peng, Z.; Zhou, J.; Cao, Y.; et al. Claudin18.2-specific CAR T cells in gastrointestinal cancers: Phase 1 trial interim results. Nat. Med. 2022, 28, 1189–1198. [Google Scholar] [CrossRef] [PubMed]
- Moentenich, V.; Gebauer, F.; Comut, E.; Tuchscherer, A.; Bruns, C.; Schroeder, W.; Buettner, R.; Alakus, H.; Loeser, H.; Zander, T.; et al. Claudin 18.2 expression in esophageal adenocarcinoma and its potential impact on future treatment strategies. Oncol. Lett. 2020, 19, 3665–3670. [Google Scholar] [CrossRef] [PubMed]
- Shitara, K.; Lordick, F.; Bang, Y.J.; Enzinger, P.; Ilson, D.; Shah, M.A.; Van Cutsem, E.; Xu, R.H.; Aprile, G.; Xu, J.; et al. Zolbetuximab plus mFOLFOX6 in patients with CLDN18.2-positive, HER2-negative, untreated, locally advanced unresectable or metastatic gastric or gastro-oesophageal junction adenocarcinoma (SPOTLIGHT): A multicentre, randomised, double-blind, phase 3 trial. Lancet 2023. [Google Scholar] [CrossRef] [PubMed]
- Grizzi, G.; Venetis, K.; Denaro, N.; Bonomi, M.; Celotti, A.; Pagkali, A.; Hahne, J.C.; Tomasello, G.; Petrelli, F.; Fusco, N.; et al. Anti-Claudin Treatments in Gastroesophageal Adenocarcinoma: Mainstream and Upcoming Strategies. J. Clin. Med. 2023, 12, 2973. [Google Scholar] [CrossRef] [PubMed]
- Lohrberg, D.; Krause, E.; Schumann, M.; Piontek, J.; Winkler, L.; Blasig, I.E.; Haseloff, R.F. A strategy for enrichment of claudins based on their affinity to Clostridium perfringens enterotoxin. BMC Mol. Biol. 2009, 10, 61. [Google Scholar] [CrossRef]
- Gao, Z.; McClane, B.A. Use of Clostridium perfringens Enterotoxin and the Enterotoxin Receptor-Binding Domain (C-CPE) for Cancer Treatment: Opportunities and Challenges. J. Toxicol. 2012, 2012, 981626. [Google Scholar] [CrossRef]
- Mosley, M.; Knight, J.; Neesse, A.; Michl, P.; Iezzi, M.; Kersemans, V.; Cornelissen, B. Claudin-4 SPECT Imaging Allows Detection of Aplastic Lesions in a Mouse Model of Breast Cancer. J. Nucl. Med. 2015, 56, 745–751. [Google Scholar] [CrossRef]
- Alnajjar, S.; Nolte, I.; Becker, A.; Schille, J.T.; Trakooljul, N.; Frank, M.; Ngezahayo, A.; Murua Escobar, H. Ablation of Red Stable Transfected Claudin Expressing Canine Prostate Adenocarcinoma and Transitional Cell Carcinoma Cell Lines by C-CPE Gold-Nanoparticle-Mediated Laser Intervention. Int. J. Mol. Sci. 2021, 22, 2289. [Google Scholar] [CrossRef]
- Martin, D.T.; Lee, J.S.; Liu, Q.; Galiana, G.; Sprenkle, P.C.; Humphrey, P.A.; Petrylak, D.P.; Weinreb, J.C.; Schulam, P.G.; Weiss, R.M.; et al. Targeting prostate cancer with Clostridium perfringens enterotoxin functionalized nanoparticles co-encapsulating imaging cargo enhances magnetic resonance imaging specificity. Nanomedicine 2022, 40, 102477. [Google Scholar] [CrossRef]
- Neesse, A.; Hahnenkamp, A.; Griesmann, H.; Buchholz, M.; Hahn, S.A.; Maghnouj, A.; Fendrich, V.; Ring, J.; Sipos, B.; Tuveson, D.A.; et al. Claudin-4-targeted optical imaging detects pancreatic cancer and its precursor lesions. Gut 2013, 62, 1034–1043. [Google Scholar] [CrossRef]
- Cocco, E.; Shapiro, E.M.; Gasparrini, S.; Lopez, S.; Schwab, C.L.; Bellone, S.; Bortolomai, I.; Sumi, N.J.; Bonazzoli, E.; Nicoletti, R.; et al. Clostridium perfringens enterotoxin C-terminal domain labeled to fluorescent dyes for in vivo visualization of micrometastatic chemotherapy-resistant ovarian cancer. Int. J. Cancer 2015, 137, 2618–2629. [Google Scholar] [CrossRef] [PubMed]
- Liao, Z.; Yang, Z.; Piontek, A.; Eichner, M.; Krause, G.; Li, L.; Piontek, J.; Zhang, J. Specific binding of a mutated fragment of Clostridium perfringens enterotoxin to endothelial claudin-5 and its modulation of cerebral vascular permeability. Neuroscience 2016, 327, 53–63. [Google Scholar] [CrossRef] [PubMed]
- Ayala-Torres, C.; Krug, S.M.; Rosenthal, R.; Fromm, M. Angulin-1 (LSR) Affects Paracellular Water Transport, However Only in Tight Epithelial Cells. Int. J. Mol. Sci. 2021, 22, 7827. [Google Scholar] [CrossRef]
- Fuladi, S.; McGuinness, S.; Shen, L.; Weber, C.R.; Khalili-Araghi, F. Molecular mechanism of claudin-15 strand flexibility: A computational study. J. Gen. Physiol. 2022, 154, e202213116. [Google Scholar] [CrossRef]
- Nagarajan, S.K.; Klein, S.; Fadakar, B.S.; Piontek, J. Claudin-10b cation channels in tight junction strands: Octameric-interlocked pore barrels constitute paracellular channels with low water permeability. Comput. Struct. Biotechnol. J. 2023, 21, 1711–1727. [Google Scholar] [CrossRef] [PubMed]
- Berselli, A.; Benfenati, F.; Maragliano, L.; Alberini, G. Multiscale modelling of claudin-based assemblies: A magnifying glass for novel structures of biological interfaces. Comput. Struct. Biotechnol. J. 2022, 20, 5984–6010. [Google Scholar] [CrossRef]
- Schindelin, J.; Arganda-Carreras, I.; Frise, E.; Kaynig, V.; Longair, M.; Pietzsch, T.; Preibisch, S.; Rueden, C.; Saalfeld, S.; Schmid, B.; et al. Fiji: An open-source platform for biological-image analysis. Nat. Methods 2012, 9, 676–682. [Google Scholar] [CrossRef]
- Amasheh, M.; Grotjohann, I.; Amasheh, S.; Fromm, A.; Soderholm, J.D.; Zeitz, M.; Fromm, M.; Schulzke, J.D. Regulation of mucosal structure and barrier function in rat colon exposed to tumor necrosis factor alpha and interferon gamma in vitro: A novel model for studying the pathomechanisms of inflammatory bowel disease cytokines. Scand. J. Gastroenterol. 2009, 44, 1226–1235. [Google Scholar] [CrossRef]
- Fromm, M.; Schulzke, J.D.; Hegel, U. Control of electrogenic Na+ absorption in rat late distal colon by nanomolar aldosterone added in vitro. Am. J. Physiol. 1993, 264, E68–E73. [Google Scholar] [CrossRef]
- Studier, F.W.; Rosenberg, A.H.; Dunn, J.J.; Dubendorff, J.W. Use of T7 RNA polymerase to direct expression of cloned genes. Methods. Enzymol. 1990, 185, 60–89. [Google Scholar] [CrossRef]
- Pinto, M.P.; Jacobsen, B.M.; Horwitz, K.B. An immunohistochemical method to study breast cancer cell subpopulations and their growth regulation by hormones in three-dimensional cultures. Front. Endocrinol. 2011, 2, 15. [Google Scholar] [CrossRef] [PubMed]
- Huang, L.; Holtzinger, A.; Jagan, I.; BeGora, M.; Lohse, I.; Ngai, N.; Nostro, C.; Wang, R.; Muthuswamy, L.B.; Crawford, H.C.; et al. Ductal pancreatic cancer modeling and drug screening using human pluripotent stem cell- and patient-derived tumor organoids. Nat. Med. 2015, 21, 1364–1371. [Google Scholar] [CrossRef] [PubMed]


| Patient | Histology | Subtype | Morphology | T | N | M | G | R | L | V | Neoadjuvant Therapy | Tumor Regression |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| GC_1 | AC | Intestinal (Lauren) | Glandular, regressive changes | 3 | 1 | 1 | - | 0 | 0 | 0 | Yes | 1b |
| GC_2 | AC + high grade dysplasia | Mucinous (WHO 2019) | Mucinous | 1b | 0 | 0 | 3 | 0 | 0 | 0 | No | - |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Waldow, A.; Beier, L.-S.; Arndt, J.; Schallenberg, S.; Vollbrecht, C.; Bischoff, P.; Farrera-Sal, M.; Loch, F.N.; Bojarski, C.; Schumann, M.; et al. cCPE Fusion Proteins as Molecular Probes to Detect Claudins and Tight Junction Dysregulation in Gastrointestinal Cell Lines, Tissue Explants and Patient-Derived Organoids. Pharmaceutics 2023, 15, 1980. https://doi.org/10.3390/pharmaceutics15071980
Waldow A, Beier L-S, Arndt J, Schallenberg S, Vollbrecht C, Bischoff P, Farrera-Sal M, Loch FN, Bojarski C, Schumann M, et al. cCPE Fusion Proteins as Molecular Probes to Detect Claudins and Tight Junction Dysregulation in Gastrointestinal Cell Lines, Tissue Explants and Patient-Derived Organoids. Pharmaceutics. 2023; 15(7):1980. https://doi.org/10.3390/pharmaceutics15071980
Chicago/Turabian StyleWaldow, Ayk, Laura-Sophie Beier, Janine Arndt, Simon Schallenberg, Claudia Vollbrecht, Philip Bischoff, Martí Farrera-Sal, Florian N. Loch, Christian Bojarski, Michael Schumann, and et al. 2023. "cCPE Fusion Proteins as Molecular Probes to Detect Claudins and Tight Junction Dysregulation in Gastrointestinal Cell Lines, Tissue Explants and Patient-Derived Organoids" Pharmaceutics 15, no. 7: 1980. https://doi.org/10.3390/pharmaceutics15071980
APA StyleWaldow, A., Beier, L.-S., Arndt, J., Schallenberg, S., Vollbrecht, C., Bischoff, P., Farrera-Sal, M., Loch, F. N., Bojarski, C., Schumann, M., Winkler, L., Kamphues, C., Ehlen, L., & Piontek, J. (2023). cCPE Fusion Proteins as Molecular Probes to Detect Claudins and Tight Junction Dysregulation in Gastrointestinal Cell Lines, Tissue Explants and Patient-Derived Organoids. Pharmaceutics, 15(7), 1980. https://doi.org/10.3390/pharmaceutics15071980








