Polysarcosine-Functionalized mRNA Lipid Nanoparticles Tailored for Immunotherapy
Abstract
1. Introduction
2. Materials and Methods
2.1. Materials
2.2. RNA Constructs and In Vitro Transcription
2.3. LNP Preparation
2.4. Dynamic Light Scattering and Zeta Potential Measurements
2.5. Accessible mRNA
2.6. pKa Fluorescence Assay
2.7. Small-Angle X-ray Scattering
2.8. Data Treatment
2.9. In Vitro Thy1.1 Transfection Assay
2.10. In Vitro Cy5 Cell Binding Assay
2.11. Flow Cytometry
2.12. Statistical Analysis
3. Results
3.1. Particle Size, Zeta Potential and mRNA Incorporation
3.2. Fluorescence-Based pKa Determination
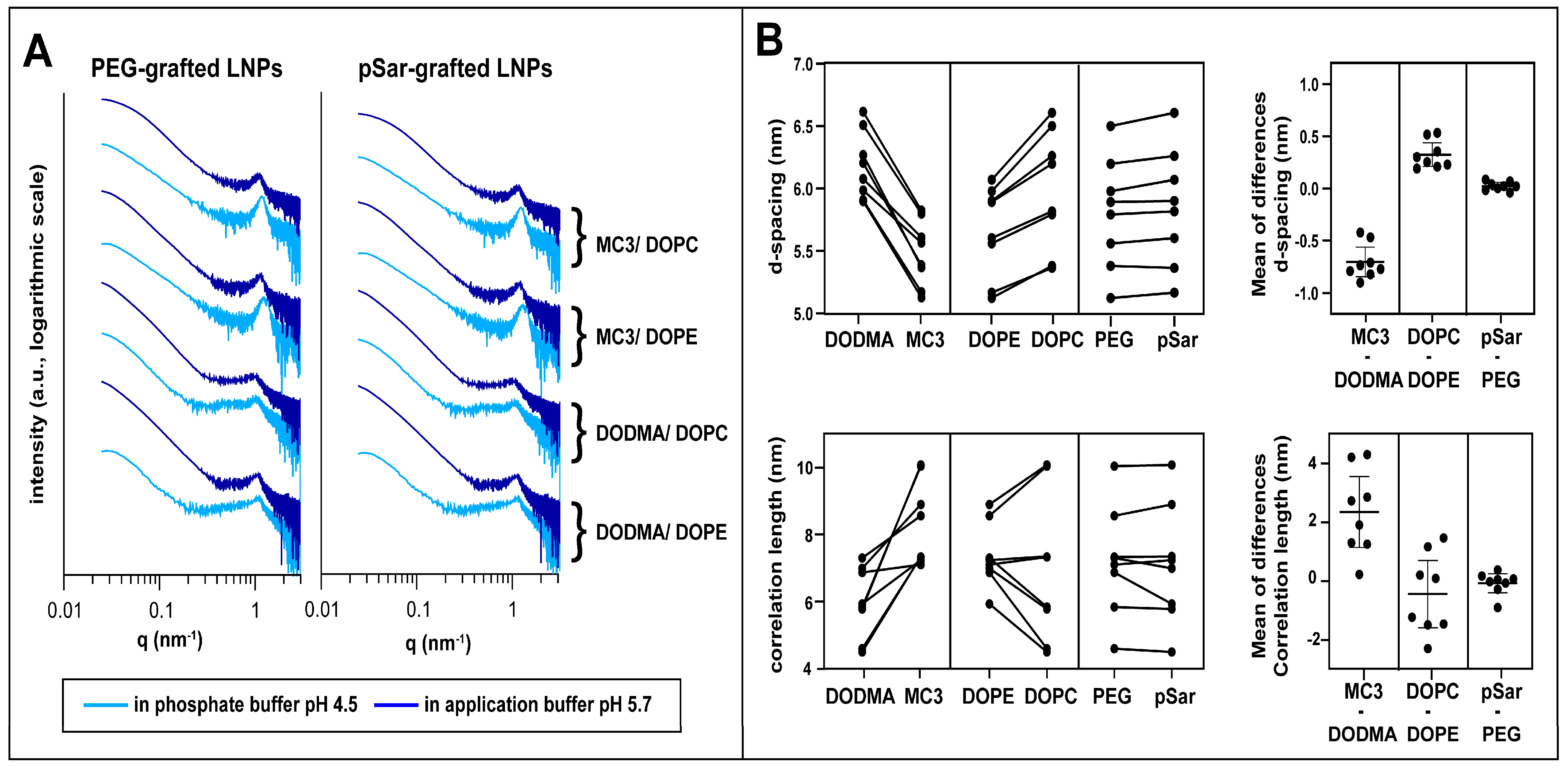
3.3. Small-Angle X-ray Scattering
3.4. In Vitro Transfection Studies
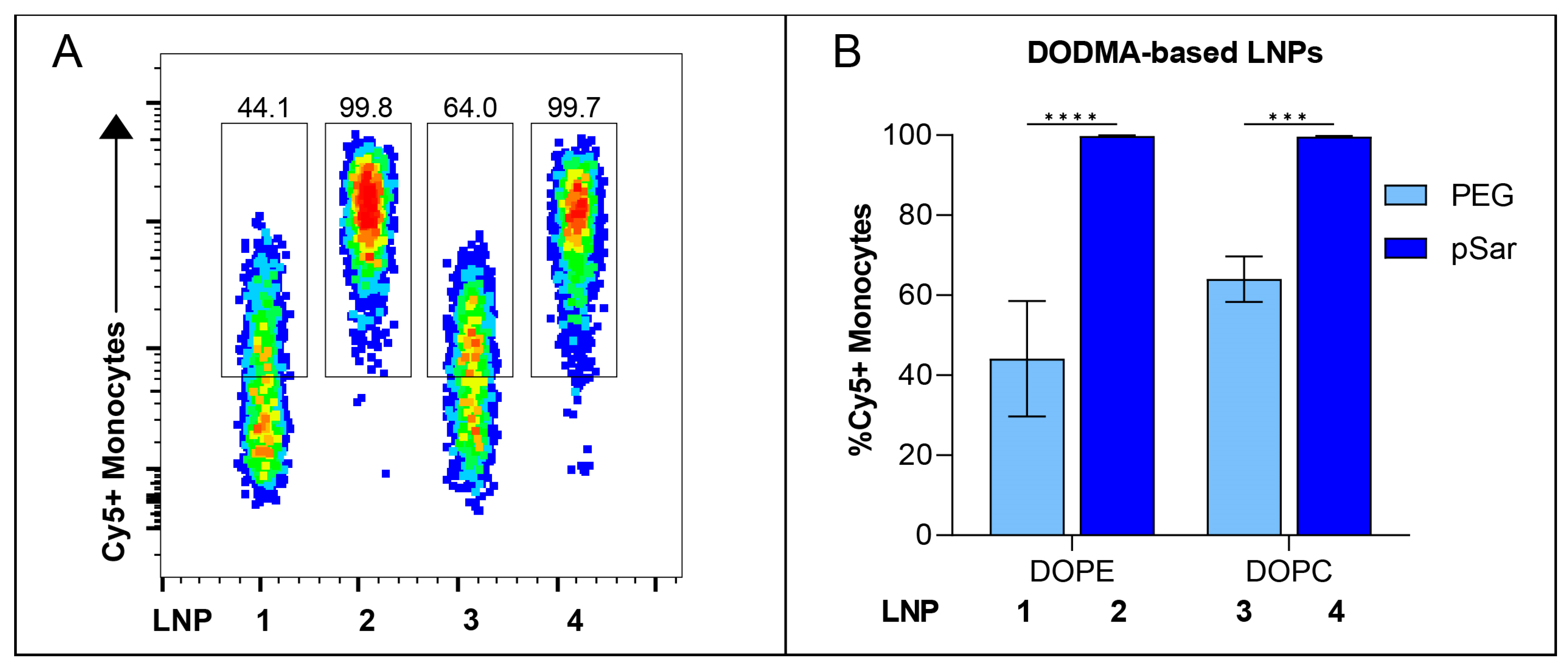
3.5. In Vitro Cell-Binding Studies
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sahin, U.; Karikó, K.; Türeci, Ö. mRNA-based therapeutics--developing a new class of drugs. Nat. Rev. Drug Discov. 2014, 13, 759–780. [Google Scholar] [CrossRef] [PubMed]
- Akinc, A.; Maier, M.A.; Manoharan, M.; Fitzgerald, K.; Jayaraman, M.; Barros, S.; Ansell, S.; Du, X.; Hope, M.J.; Madden, T.D.; et al. The Onpattro story and the clinical translation of nanomedicines containing nucleic acid-based drugs. Nat. Nanotechnol. 2019, 14, 1084–1087. [Google Scholar] [CrossRef]
- Hou, X.; Zaks, T.; Langer, R.; Dong, Y. Lipid nanoparticles for mRNA delivery. Nat. Rev. Mater. 2021, 6, 1078–1094. [Google Scholar] [CrossRef]
- Tenchov, R.; Bird, R.; Curtze, A.E.; Zhou, Q. Lipid Nanoparticles—From Liposomes to mRNA Vaccine Delivery, a Landscape of Research Diversity and Advancement. ACS Nano 2021, 15, 16982–17015. [Google Scholar] [CrossRef] [PubMed]
- Sahin, U.; Oehm, P.; Derhovanessian, E.; Jabulowsky, R.A.; Vormehr, M.; Gold, M.; Maurus, D.; Schwarck-Kokarakis, D.; Kuhn, A.N.; Omokoko, T.; et al. An RNA vaccine drives immunity in checkpoint-inhibitor-treated melanoma. Nature 2020, 585, 107–112. [Google Scholar] [CrossRef]
- Sahin, U.; Türeci, Ö. Personalized vaccines for cancer immunotherapy. Science 2018, 359, 1355–1360. [Google Scholar] [CrossRef]
- Sahin, U.; Derhovanessian, E.; Miller, M.; Kloke, B.-P.; Simon, P.; Löwer, M.; Bukur, V.; Tadmor, A.D.; Luxemburger, U.; Schrörs, B.; et al. Personalized RNA mutanome vaccines mobilize poly-specific therapeutic immunity against cancer. Nature 2017, 547, 222–226. [Google Scholar] [CrossRef]
- Walsh, E.E.; Frenck, R.W.; Falsey, A.R.; Kitchin, N.; Absalon, J.; Gurtman, A.; Lockhart, S.; Neuzil, K.; Mulligan, M.J.; Bailey, R.; et al. Safety and Immunogenicity of Two RNA-Based COVID-19 Vaccine Candidates. N. Engl. J. Med. 2020, 383, 2439–2450. [Google Scholar] [CrossRef]
- Corbett, K.S.; Edwards, D.K.; Leist, S.R.; Abiona, O.M.; Boyoglu-Barnum, S.; Gillespie, R.A.; Himansu, S.; Schäfer, A.; Ziwawo, C.T.; DiPiazza, A.T.; et al. SARS-CoV-2 mRNA vaccine design enabled by prototype pathogen preparedness. Nature 2020, 586, 567–571. [Google Scholar] [CrossRef] [PubMed]
- Hald Albertsen, C.; Kulkarni, J.A.; Witzigmann, D.; Lind, M.; Petersson, K.; Simonsen, J.B. The role of lipid components in lipid nanoparticles for vaccines and gene therapy. Adv. Drug Deliv. Rev. 2022, 188, 114416. [Google Scholar] [CrossRef]
- Cheng, X.; Lee, R.J. The role of helper lipids in lipid nanoparticles (LNPs) designed for oligonucleotide delivery. Adv. Drug Deliv. Rev. 2016, 99, 129–137. [Google Scholar] [CrossRef] [PubMed]
- Rietwyk, S.; Peer, D. Next-Generation Lipids in RNA Interference Therapeutics. ACS Nano 2017, 11, 7572–7586. [Google Scholar] [CrossRef]
- Torchilin, V.P.; Zhou, F.; Huang, L. pH-Sensitive Liposomes. J. Liposome Res. 1993, 3, 201–255. [Google Scholar] [CrossRef]
- Ramezanpour, M.; Schmidt, M.L.; Bodnariuc, I.; Kulkarni, J.A.; Leung, S.S.W.; Cullis, P.R.; Thewalt, J.L.; Tieleman, D.P. Ionizable amino lipid interactions with POPC: Implications for lipid nanoparticle function. Nanoscale 2019, 11, 14141–14146. [Google Scholar] [CrossRef] [PubMed]
- Semple, S.C.; Akinc, A.; Chen, J.; Sandhu, A.P.; Mui, B.L.; Cho, C.K.; Sah, D.W.Y.; Stebbing, D.; Crosley, E.J.; Yaworski, E.; et al. Rational design of cationic lipids for siRNA delivery. Nat. Biotechnol. 2010, 28, 172–176. [Google Scholar] [CrossRef] [PubMed]
- Jayaraman, M.; Ansell, S.M.; Mui, B.L.; Tam, Y.K.; Chen, J.; Du, X.; Butler, D.; Eltepu, L.; Matsuda, S.; Narayanannair, J.K.; et al. Maximizing the Potency of siRNA Lipid Nanoparticles for Hepatic Gene Silencing In Vivo. Angew. Chem. 2012, 124, 8657–8661. [Google Scholar] [CrossRef]
- Uebbing, L.; Ziller, A.; Siewert, C.; Schroer, M.A.; Blanchet, C.E.; Svergun, D.I.; Ramishetti, S.; Peer, D.; Sahin, U.; Haas, H.; et al. Investigation of pH-Responsiveness inside Lipid Nanoparticles for Parenteral mRNA Application Using Small-Angle X-ray Scattering. Langmuir 2020, 36, 13331–13341. [Google Scholar] [CrossRef]
- Kranz, L.M.; Diken, M.; Haas, H.; Kreiter, S.; Loquai, C.; Reuter, K.C.; Meng, M.; Fritz, D.; Vascotto, F.; Hefesha, H.; et al. Systemic RNA delivery to dendritic cells exploits antiviral defence for cancer immunotherapy. Nature 2016, 534, 396–401. [Google Scholar] [CrossRef]
- Rosigkeit, S.; Meng, M.; Grunwitz, C.; Gomes, P.; Kreft, A.; Hayduk, N.; Heck, R.; Pickert, G.; Ziegler, K.; Abassi, Y.; et al. Monitoring Translation Activity of mRNA-Loaded Nanoparticles in Mice. Mol. Pharm. 2018, 15, 3909–3919. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Q.; Wei, T.; Farbiak, L.; Johnson, L.T.; Dilliard, S.A.; Siegwart, D.J. Selective organ targeting (SORT) nanoparticles for tissue-specific mRNA delivery and CRISPR-Cas gene editing. Nat. Nanotechnol. 2020, 15, 313–320. [Google Scholar] [CrossRef]
- Segalla, G. Chemical-physical criticality and toxicological potential of lipid nanomaterials contained in a COVID-19 mRNA vaccine. Int. J. Vaccine Theory Pract. Res. 2023, 3, 787–817. [Google Scholar] [CrossRef]
- Jörgensen, A.M.; Wibel, R.; Bernkop-Schnürch, A. Biodegradable Cationic and Ionizable Cationic Lipids: A Roadmap for Safer Pharmaceutical Excipients. Small 2023, 19, e2206968. [Google Scholar] [CrossRef] [PubMed]
- Ishida, T.; Masuda, K.; Ichikawa, T.; Ichihara, M.; Irimura, K.; Kiwada, H. Accelerated clearance of a second injection of PEGylated liposomes in mice. Int. J. Pharm. 2003, 255, 167–174. [Google Scholar] [CrossRef] [PubMed]
- Garay, R.P.; Labaune, J.P. Immunogenicity of Polyethylene Glycol (PEG). Open Conf. Proc. J. 2011, 2, 104–107. [Google Scholar] [CrossRef][Green Version]
- Semple, K.T.; Reid, B.J.; Fermor, T.R. Impact of composting strategies on the treatment of soils contaminated with organic pollutants. Environ. Pollut. 2001, 112, 269–283. [Google Scholar] [CrossRef]
- Estapé Senti, M.; de Jongh, C.A.; Dijkxhoorn, K.; Verhoef, J.J.F.; Szebeni, J.; Storm, G.; Hack, C.E.; Schiffelers, R.M.; Fens, M.H.; Boross, P. Anti-PEG antibodies compromise the integrity of PEGylated lipid-based nanoparticles via complement. J. Control. Release 2022, 341, 475–486. [Google Scholar] [CrossRef]
- Secker, C.; Brosnan, S.M.; Luxenhofer, R.; Schlaad, H. Poly(α-Peptoid)s Revisited: Synthesis, Properties, and Use as Biomaterial. Macromol. Biosci. 2015, 15, 881–891. [Google Scholar] [CrossRef] [PubMed]
- Kidchob, T.; Kimura, S.; Imanishi, Y. Amphiphilic poly(Ala)-b-poly(Sar) microspheres loaded with hydrophobic drug. J. Control. Release 1998, 51, 241–248. [Google Scholar] [CrossRef]
- Birke, A.; Ling, J.; Barz, M. Polysarcosine-containing copolymers: Synthesis, characterization, self-assembly, and applications. Prog. Polym. Sci. 2018, 81, 163–208. [Google Scholar] [CrossRef]
- Bleher, S.; Buck, J.; Muhl, C.; Sieber, S.; Barnert, S.; Witzigmann, D.; Huwyler, J.; Barz, M.; Süss, R. Poly(Sarcosine) Surface Modification Imparts Stealth-Like Properties to Liposomes. Small 2019, 15, e1904716. [Google Scholar] [CrossRef]
- Maurer, P.H.; Subrahmanyam, D.; Katchalski, E.; Blout, E.R. Antigenicity of Polypeptides (Poly Alpha Amino Acids). J. Immunol. 1959, 83, 193–197. [Google Scholar] [CrossRef]
- Fenaroli, F.; Repnik, U.; Xu, Y.; Johann, K.; van Herck, S.; Dey, P.; Skjeldal, F.M.; Frei, D.M.; Bagherifam, S.; Kocere, A.; et al. Enhanced Permeability and Retention-like Extravasation of Nanoparticles from the Vasculature into Tuberculosis Granulomas in Zebrafish and Mouse Models. ACS Nano 2018, 12, 8646–8661. [Google Scholar] [CrossRef] [PubMed]
- Nogueira, S.S.; Schlegel, A.; Maxeiner, K.; Weber, B.; Barz, M.; Schroer, M.A.; Blanchet, C.E.; Svergun, D.I.; Ramishetti, S.; Peer, D.; et al. Polysarcosine-Functionalized Lipid Nanoparticles for Therapeutic mRNA Delivery. ACS Appl. Nano Mater. 2020, 3, 10634–10645. [Google Scholar] [CrossRef]
- US Food and Drug Administration. Liposome Drug Products: Chemistry, Manufacturing, and Controls; Human Pharmacokinetics and Bioavailability; and Labeling Documentation. In Guidance for Industry.(CDER); Food and Drug Administration: Rockville, MD, USA, 2018. [Google Scholar]
- Ziller, A.; Nogueira, S.S.; Hühn, E.; Funari, S.S.; Brezesinski, G.; Hartmann, H.; Sahin, U.; Haas, H.; Langguth, P. Incorporation of mRNA in Lamellar Lipid Matrices for Parenteral Administration. Mol. Pharm. 2018, 15, 642–651. [Google Scholar] [CrossRef] [PubMed]
- Siewert, C.; Haas, H.; Nawroth, T.; Ziller, A.; Nogueira, S.S.; Schroer, M.A.; Blanchet, C.E.; Svergun, D.I.; Radulescu, A.; Bates, F.; et al. Investigation of charge ratio variation in mRNA—DEAE-dextran polyplex delivery systems. Biomaterials 2019, 192, 612–620. [Google Scholar] [CrossRef]
- Siewert, C.D.; Haas, H.; Cornet, V.; Nogueira, S.S.; Nawroth, T.; Uebbing, L.; Ziller, A.; Al-Gousous, J.; Radulescu, A.; Schroer, M.A.; et al. Hybrid Biopolymer and Lipid Nanoparticles with Improved Transfection Efficacy for mRNA. Cells 2020, 9, 2034. [Google Scholar] [CrossRef] [PubMed]
- Felgner, P.L.; Gadek, T.R.; Holm, M.; Roman, R.; Chan, H.W.; Wenz, M.; Northrop, J.P.; Ringold, G.M.; Danielsen, M. Lipofection: A highly efficient, lipid-mediated DNA-transfection procedure. Proc. Natl. Acad. Sci. USA 1987, 84, 7413–7417. [Google Scholar] [CrossRef]
- Muhl, C.; Conrad, M.; Unthan, D.; Barz, M. Synthesis and characterization of bisalkylated polysarcosine-based lipopolymers. Eur. Polym. J. 2019, 120, 109223. [Google Scholar] [CrossRef]
- Kuhn, A.N.; Diken, M.; Kreiter, S.; Selmi, A.; Kowalska, J.; Jemielity, J.; Darzynkiewicz, E.; Huber, C.; Türeci, O.; Sahin, U. Phosphorothioate cap analogs increase stability and translational efficiency of RNA vaccines in immature dendritic cells and induce superior immune responses in vivo. Gene Ther. 2010, 17, 961–971. [Google Scholar] [CrossRef]
- Karikó, K.; Muramatsu, H.; Welsh, F.A.; Ludwig, J.; Kato, H.; Akira, S.; Weissman, D. Incorporation of pseudouridine into mRNA yields superior nonimmunogenic vector with increased translational capacity and biological stability. Mol. Ther. 2008, 16, 1833–1840. [Google Scholar] [CrossRef]
- Baiersdörfer, M.; Boros, G.; Muramatsu, H.; Mahiny, A.; Vlatkovic, I.; Sahin, U.; Karikó, K. A Facile Method for the Removal of dsRNA Contaminant from In Vitro-Transcribed mRNA. Mol. Ther. Nucleic Acids 2019, 15, 26–35. [Google Scholar] [CrossRef] [PubMed]
- Leung, A.K.K.; Hafez, I.M.; Baoukina, S.; Belliveau, N.M.; Zhigaltsev, I.V.; Afshinmanesh, E.; Tieleman, D.P.; Hansen, C.L.; Hope, M.J.; Cullis, P.R. Lipid Nanoparticles Containing siRNA Synthesized by Microfluidic Mixing Exhibit an Electron-Dense Nanostructured Core. J. Phys. Chem. C Nanomater. Interfaces 2012, 116, 18440–18450. [Google Scholar] [CrossRef] [PubMed]
- Heyes, J.; Palmer, L.; Bremner, K.; MacLachlan, I. Cationic lipid saturation influences intracellular delivery of encapsulated nucleic acids. J. Control. Release 2005, 107, 276–287. [Google Scholar] [CrossRef]
- Romeis, B. Taschenbuch der Mikroskopischen Technik; De Gruyter: Berlin, Germany, 1943; ISBN 9783486764369. [Google Scholar]
- Blanchet, C.E.; Spilotros, A.; Schwemmer, F.; Graewert, M.A.; Kikhney, A.; Jeffries, C.M.; Franke, D.; Mark, D.; Zengerle, R.; Cipriani, F.; et al. Versatile sample environments and automation for biological solution X-ray scattering experiments at the P12 beamline (PETRA III, DESY). J. Appl. Crystallogr. 2015, 48, 431–443. [Google Scholar] [CrossRef]
- Manalastas-Cantos, K.; Konarev, P.V.; Hajizadeh, N.R.; Kikhney, A.G.; Petoukhov, M.V.; Molodenskiy, D.S.; Panjkovich, A.; Mertens, H.D.T.; Gruzinov, A.; Borges, C.; et al. ATSAS 3.0: Expanded functionality and new tools for small-angle scattering data analysis. J. Appl. Crystallogr. 2021, 54, 343–355. [Google Scholar] [CrossRef]
- Franke, D.; Kikhney, A.G.; Svergun, D.I. Automated acquisition and analysis of small angle X-ray scattering data. Nucl. Instrum. Methods Phys. Res. Sect. A Accel. Spectrometers Detect. Assoc. Equip. 2012, 689, 52–59. [Google Scholar] [CrossRef]
- Bragg, W.H.; Bragg William, L. The reflection of X-rays by crystals. Proc. R. Soc. Lond. A 1913, 88, 428–438. [Google Scholar] [CrossRef]
- Goodby, J.W.; Tschierske, C.; Raynes, P.; Gleeson, H.; Kato, T.; Collings, P.J. (Eds.) Handbook of Liquid Crystals; Wiley-VCH Verlag GmbH & Co. KGaA: Weinheim, Germany, 2014; ISBN 9783527671403. [Google Scholar]
- Ciccariello, S.; Goodisman, J.; Brumberger, H. On the Porod law. J. Appl. Crystallogr. 1988, 21, 117–128. [Google Scholar] [CrossRef]
- Carrasco, M.J.; Alishetty, S.; Alameh, M.-G.; Said, H.; Wright, L.; Paige, M.; Soliman, O.; Weissman, D.; Cleveland, T.E.; Grishaev, A.; et al. Ionization and structural properties of mRNA lipid nanoparticles influence expression in intramuscular and intravascular administration. Commun. Biol. 2021, 4, 956. [Google Scholar] [CrossRef]
- Belliveau, N.M.; Huft, J.; Lin, P.J.; Chen, S.; Leung, A.K.; Leaver, T.J.; Wild, A.W.; Lee, J.B.; Taylor, R.J.; Tam, Y.K.; et al. Microfluidic Synthesis of Highly Potent Limit-size Lipid Nanoparticles for In Vivo Delivery of siRNA. Mol. Ther. Nucleic Acids 2012, 1, e37. [Google Scholar] [CrossRef]
- Yanez Arteta, M.; Kjellman, T.; Bartesaghi, S.; Wallin, S.; Wu, X.; Kvist, A.J.; Dabkowska, A.; Székely, N.; Radulescu, A.; Bergenholtz, J.; et al. Successful reprogramming of cellular protein production through mRNA delivered by functionalized lipid nanoparticles. Proc. Natl. Acad. Sci. USA 2018, 115, E3351–E3360. [Google Scholar] [CrossRef] [PubMed]
- Bale, H.D.; Schmidt, P.W. Small-Angle X-Ray-Scattering Investigation of Submicroscopic Porosity with Fractal Properties. Phys. Rev. Lett. 1984, 53, 596–599. [Google Scholar] [CrossRef]
- Teixeira, J. Small-angle scattering by fractal systems. J. Appl. Crystallogr. 1988, 21, 781–785. [Google Scholar] [CrossRef]
- Evers, M.J.W.; Kulkarni, J.A.; van der Meel, R.; Cullis, P.R.; Vader, P.; Schiffelers, R.M. State-of-the-Art Design and Rapid-Mixing Production Techniques of Lipid Nanoparticles for Nucleic Acid Delivery. Small Methods 2018, 2, 1700375. [Google Scholar] [CrossRef]
- Wan, C.; Allen, T.M.; Cullis, P.R. Lipid nanoparticle delivery systems for siRNA-based therapeutics. Drug Deliv. Transl. Res. 2014, 4, 74–83. [Google Scholar] [CrossRef]
- Ramishetti, S.; Kedmi, R.; Goldsmith, M.; Leonard, F.; Sprague, A.G.; Godin, B.; Gozin, M.; Cullis, P.R.; Dykxhoorn, D.M.; Peer, D. Systemic Gene Silencing in Primary T Lymphocytes Using Targeted Lipid Nanoparticles. ACS Nano 2015, 9, 6706–6716. [Google Scholar] [CrossRef]
- Kozma, G.T.; Shimizu, T.; Ishida, T.; Szebeni, J. Anti-PEG antibodies: Properties, formation, testing and role in adverse immune reactions to PEGylated nano-biopharmaceuticals. Adv. Drug Deliv. Rev. 2020, 154–155, 163–175. [Google Scholar] [CrossRef]
- Ju, Y.; Lee, W.S.; Pilkington, E.H.; Kelly, H.G.; Li, S.; Selva, K.J.; Wragg, K.M.; Subbarao, K.; Nguyen, T.H.O.; Rowntree, L.C.; et al. Anti-PEG Antibodies Boosted in Humans by SARS-CoV-2 Lipid Nanoparticle mRNA Vaccine. ACS Nano 2022, 16, 11769–11780. [Google Scholar] [CrossRef] [PubMed]
- Weber, B.; Seidl, C.; Schwiertz, D.; Scherer, M.; Bleher, S.; Süss, R.; Barz, M. Polysarcosine-Based Lipids: From Lipopolypeptoid Micelles to Stealth-Like Lipids in Langmuir Blodgett Monolayers. Polymers 2016, 8, 427. [Google Scholar] [CrossRef]
- Son, K.; Ueda, M.; Taguchi, K.; Maruyama, T.; Takeoka, S.; Ito, Y. Evasion of the accelerated blood clearance phenomenon by polysarcosine coating of liposomes. J. Control. Release 2020, 322, 209–216. [Google Scholar] [CrossRef]
- Barz, M.; Weber, B.; Haas, H.; Heller, P.; Nogueira, S.; Schlegel, A. RNA Particles Comprising Polysarcosine. WO2020069718A1, 1 October 2018. [Google Scholar]
- Patel, S.; Ashwanikumar, N.; Robinson, E.; Xia, Y.; Mihai, C.; Griffith, J.P.; Hou, S.; Esposito, A.A.; Ketova, T.; Welsher, K.; et al. Naturally-occurring cholesterol analogues in lipid nanoparticles induce polymorphic shape and enhance intracellular delivery of mRNA. Nat. Commun. 2020, 11, 983. [Google Scholar] [CrossRef] [PubMed]
- Sebastiani, F.; Yanez Arteta, M.; Lerche, M.; Porcar, L.; Lang, C.; Bragg, R.A.; Elmore, C.S.; Krishnamurthy, V.R.; Russell, R.A.; Darwish, T.; et al. Apolipoprotein E Binding Drives Structural and Compositional Rearrangement of mRNA-Containing Lipid Nanoparticles. ACS Nano 2021, 15, 6709–6722. [Google Scholar] [CrossRef] [PubMed]

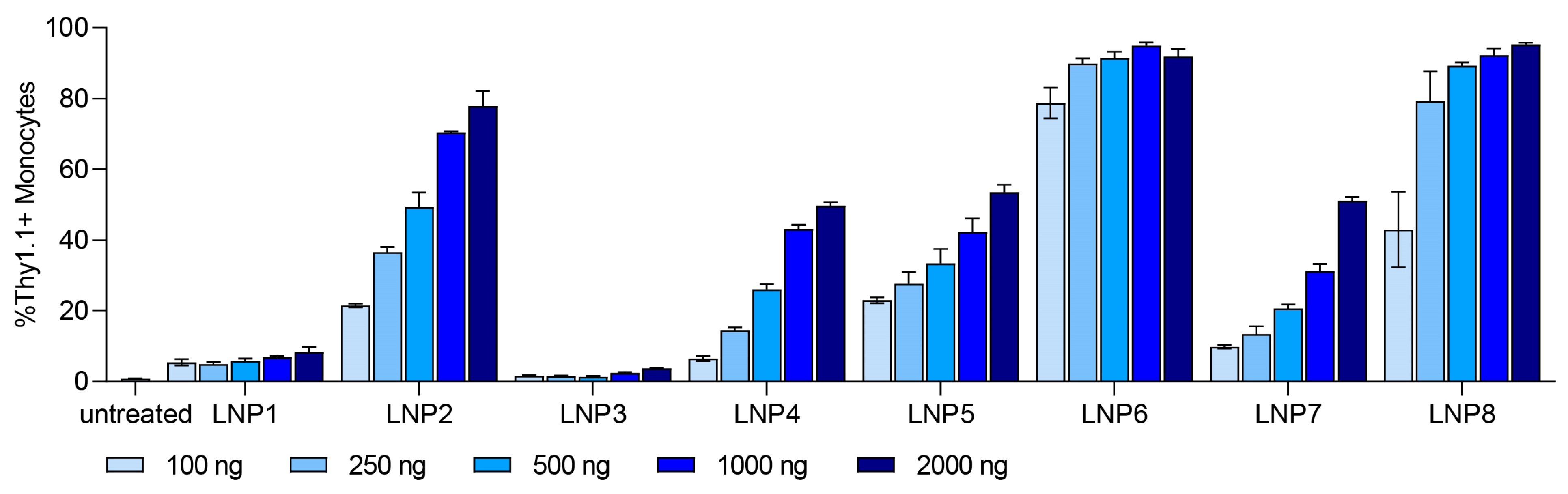
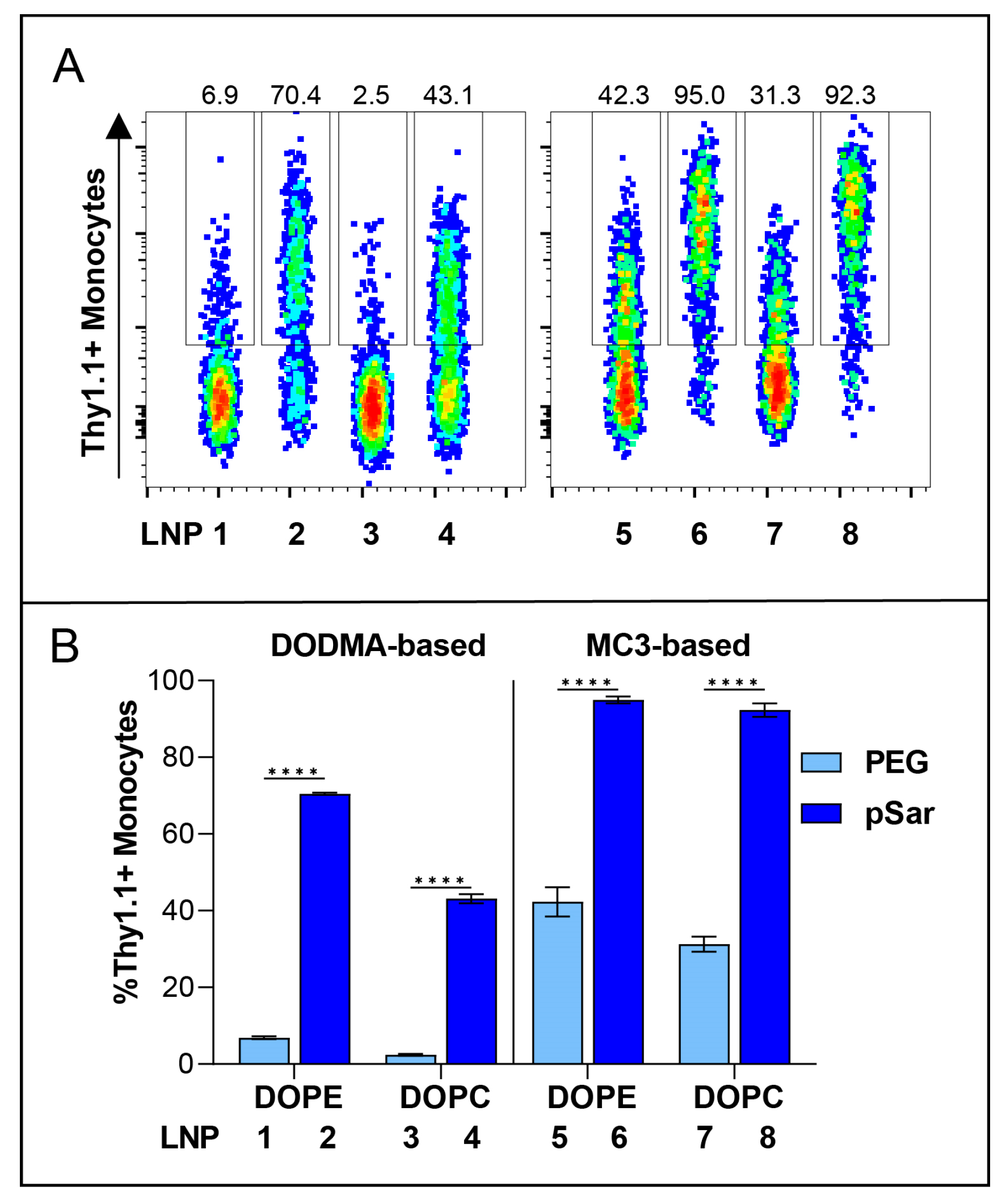
| Formulation | Helper Lipid | Helper Lipid (mol%) | Ionizable Lipid | Ionizable Lipid (mol%) | Cholesterol (mol%) | Stealth Moiety | Stealth Moiety (mol%) | mRNA (mol%) |
|---|---|---|---|---|---|---|---|---|
| LNP 1 | DOPE | 10 | DODMA | 40 | 48 | PEG | 2 | 8 |
| LNP 2 | DOPE | 10 | DODMA | 40 | 48 | pSar | 2 | 8 |
| LNP 3 | DOPC | 10 | DODMA | 40 | 48 | PEG | 2 | 8 |
| LNP 4 | DOPC | 10 | DODMA | 40 | 48 | pSar | 2 | 8 |
| LNP 5 | DOPE | 10 | MC3 | 40 | 48 | PEG | 2 | 8 |
| LNP 6 | DOPE | 10 | MC3 | 40 | 48 | pSar | 2 | 8 |
| LNP 7 | DOPC | 10 | MC3 | 40 | 48 | PEG | 2 | 8 |
| LNP 8 | DOPC | 10 | MC3 | 40 | 48 | pSar | 2 | 8 |
| Formulation | Composition | Diameter (nm) | PDI | Zeta Potential (mV) | Inaccessible mRNA (%) |
|---|---|---|---|---|---|
| LNP 1 | DODMA/DOPE/PEG | 201 ± 15 | 0.202 ± 0.043 | 27 ± 7 | 95 ± 3 |
| LNP 2 | DODMA/DOPE/pSar | 239 ± 21 | 0.173 ± 0.028 | 34 ± 6 | 91 ± 4 |
| LNP 3 | DODMA/DOPC/PEG | 221 ± 15 | 0.205 ± 0.021 | 28 ± 5 | 92 ± 4 |
| LNP 4 | DODMA/DOPC/pSar | 242 ± 33 | 0.193 ± 0.040 | 35 ± 3 | 90 ± 4 |
| LNP 5 | MC3/DOPE/PEG | 174 ± 07 | 0.124 ± 0.021 | 30 ± 3 | 96 ± 2 |
| LNP 6 | MC3/DOPE/pSar | 196 ±13 | 0.170 ± 0.015 | 33 ± 2 | 93 ± 2 |
| LNP 7 | MC3/DOPC/PEG | 159 ± 15 | 0.117 ±.0.024 | 27 ± 6 | 97 ± 1 |
| LNP 8 | MC3/DOPC/pSar | 197 ± 17 | 0.156 ± 0.028 | 34 ± 1 | 94 ± 2 |
| Formulation (DODMA) | Apparent pKa | Formulation (MC3) | Apparent pKa |
|---|---|---|---|
| LNP 1 | 6.5 ± 0.0 | LNP 5 | 6.7 ± 0.0 |
| LNP 2 | 6.5 ± 0.1 | LNP 6 | 6.6 ± 0.0 |
| LNP 3 | 6.5 ± 0.2 | LNP 7 | 6.5 ± 0.1 |
| LNP 4 | 6.6 ± 0.1 | LNP 8 | 6.5 ± 0.1 |
| Formulation | Application Buffer (pH 5.7) | Phosphate Buffer (pH 4.5) | ||||
|---|---|---|---|---|---|---|
| d-Spacing (nm) | Correlation Length (nm) | Porod Exponent | d-Spacing (nm) | Correlation Length (nm) | Porod Exponent | |
| LNP 1 | 6.0 ± 0.1 | 6.9 ± 0.3 | −3.9 ± 0.1 | 5.9 ± 0.1 | 7.3 ± 0.2 | −3.4 ± 0.1 |
| LNP 2 | 6.1 ± 0.1 | 4.6 ± 0.3 | −3.9 ± 0.1 | 5.9 ± 0.1 | 7.0 ± 0.2 | −3.4 ± 0.1 |
| LNP 3 | 6.5 ± 0.1 | 5.9 ± 0.2 | −3.9 ± 0.1 | 6.2 ± 0.1 | 5.8 ± 0.1 | −3.9 ± 0.1 |
| LNP 4 | 6.6 ± 0.1 | 4.5 ± 0.2 | −3.9 ± 0.1 | 6.3 ± 0.1 | 5.8 ± 0.2 | −3.9 ± 0.1 |
| LNP 5 | 5.6 ± 0.1 | 7.1 ± 0.2 | −3.8 ± 0.1 | 5.1 ± 0.1 | 8.6 ± 0.2 | −3.3 ± 0.1 |
| LNP 6 | 5.8 ± 0.1 | 7.3 ± 0.2 | −3.5 ± 0.1 | 5.2 ± 0.1 | 8.9 ± 0.2 | −2.6 ± 0.1 |
| LNP 7 | 5.6 ± 0.1 | 7.2 ± 0.3 | −3.7 ± 0.1 | 5.4 ± 0.1 | 10.0 ± 0.1 | −3.5 ± 0.1 |
| LNP 8 | 5.8 ± 0.1 | 7.3 ± 0.3 | −3.6 ± 0.1 | 5.4 ± 0.1 | 10.1 ± 0.2 | −2.7 ± 0.1 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wilhelmy, C.; Keil, I.S.; Uebbing, L.; Schroer, M.A.; Franke, D.; Nawroth, T.; Barz, M.; Sahin, U.; Haas, H.; Diken, M.; et al. Polysarcosine-Functionalized mRNA Lipid Nanoparticles Tailored for Immunotherapy. Pharmaceutics 2023, 15, 2068. https://doi.org/10.3390/pharmaceutics15082068
Wilhelmy C, Keil IS, Uebbing L, Schroer MA, Franke D, Nawroth T, Barz M, Sahin U, Haas H, Diken M, et al. Polysarcosine-Functionalized mRNA Lipid Nanoparticles Tailored for Immunotherapy. Pharmaceutics. 2023; 15(8):2068. https://doi.org/10.3390/pharmaceutics15082068
Chicago/Turabian StyleWilhelmy, Christoph, Isabell Sofia Keil, Lukas Uebbing, Martin A. Schroer, Daniel Franke, Thomas Nawroth, Matthias Barz, Ugur Sahin, Heinrich Haas, Mustafa Diken, and et al. 2023. "Polysarcosine-Functionalized mRNA Lipid Nanoparticles Tailored for Immunotherapy" Pharmaceutics 15, no. 8: 2068. https://doi.org/10.3390/pharmaceutics15082068
APA StyleWilhelmy, C., Keil, I. S., Uebbing, L., Schroer, M. A., Franke, D., Nawroth, T., Barz, M., Sahin, U., Haas, H., Diken, M., & Langguth, P. (2023). Polysarcosine-Functionalized mRNA Lipid Nanoparticles Tailored for Immunotherapy. Pharmaceutics, 15(8), 2068. https://doi.org/10.3390/pharmaceutics15082068