Membrane-Active Peptides and Their Potential Biomedical Application
Abstract
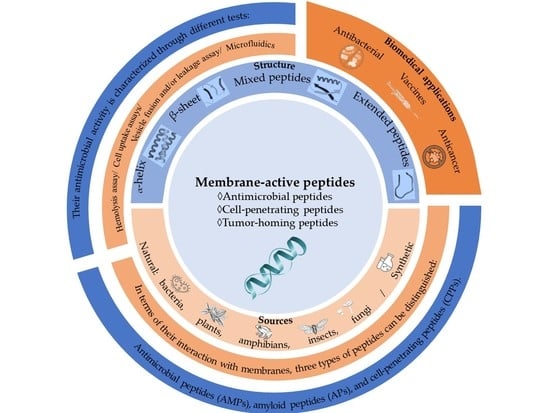
:1. Introduction
1.1. Membrane-Active Peptide Classes
1.1.1. Antimicrobial Peptides (AMPs)
1.1.2. Cell-Penetrating Peptides (CPPs)
1.1.3. Tumor-Homing Peptides
1.2. Methods of Obtaining Membrane-Active Peptides
1.2.1. Extraction of Membrane-Active Peptides from Natural Sources
1.2.2. Peptides Produced from Genetically Modified Organisms
1.2.3. Solid-Phase Peptide Synthesis
1.2.4. Supported Liquid Membranes for Peptide Extraction
1.3. Purification and Characterization of Membrane-Active Peptides
1.3.1. High-Performance Liquid Chromatography (HPLC)
1.3.2. Ultrafiltration
1.3.3. Mass Spectrometry
1.3.4. Edman Degradation
1.3.5. Circular Dichroism (CD)
1.4. Physicochemical Properties
2. Mechanisms of Action
2.1. Interactions of Membrane-Active Peptides with a Model or Biological Membranes
2.2. Antimicrobial Activity
2.3. Haemolysis Assay
2.4. Cell Uptake Assays
2.5. Vesicle Fusion and/or Leakage Assay
2.6. Microfluidics
3. Biomedical Application
3.1. Antibacterial
3.1.1. Systemic Infections
3.1.2. Food Preservation
3.1.3. Oral Care
3.2. Anticancer
3.2.1. Peptides Used for Lung Cancer Treatment
3.2.2. Peptide Treatment for Multiple Myeloma Tumors
3.2.3. Peptide Treatment for Neuroendocrine Cancer
3.2.4. Peptide-Based Drugs for Ovarian Cancer
3.2.5. Peptide-Based Drugs Used for the Treatment of Prostate Cancer
3.3. Vaccines
3.3.1. Antitumoral Vaccines
Heregulin (HER)-2/neu Peptide
3.3.2. Antiviral Vaccines
3.3.3. Vaccines for Neurodegenerative Diseases
3.3.4. Arginine-Rich Peptides as Neuroprotectants in Ischemic Stroke
| Peptide | Commercial Drug Name | Applications | Trial Phase | References |
|---|---|---|---|---|
| Antibacterial | ||||
| Gramicidin | Neosporin® | Treating bacterial conjunctivitis | Admitted to the market | [22] |
| Vancomycin | Vancocin®HCl | Treatment of Gram-positive bacterial infections | Admitted to the market | [22] |
| Daptomycin | Cubicin® | Treatment for skin infections and Staphylococcus aureus infections | Admitted to the market | [22] |
| Telavancin | Orbactiv® | Treatment of Gram-positive bacterial infections | Admitted to the market | [22] |
| Nisin | Nisaplin®, Chrisin® and Delvo®Nis | Biopreservative | Admitted to the market | [114] |
| Streptococcus salivarius K12 | BLIS K12 | Protection against oral pathogenic bacteria in humans | Admitted to the market | [142] |
| Antitumoral | ||||
| Microcin E492 | - | Cervical adenocarcinoma, acute T-cell leukemia, Burkitt’s lymphoma, B-lymphoblastoid cells | Preclinical | [143,144] |
| Colicin A and E1 | - | Breast carcinoma, osteosarcoma, leiomyosarcoma, fibrosarcoma | Preclinical | [143,144] |
| Azurin-Derived Peptide p28 | - | Breast cancer | Phase I of clinical trials | [145] |
| Nisin A | - | Head and neck squamous cell carcinoma, breast adenocarcinoma, liver hepatocellular carcinoma, acute T-cell leukemia | Preclinical | [143,144] |
| Pediocin CP2 | - | Mammary gland adenocarcinoma, hepatocarcinoma, cervical adenocarcinoma | Preclinical | [143,144] |
| Peptide | Commercial Drug Name | Used for | Trial Phase | References |
| Vaccines | ||||
| Combination of 25 amino acids from several immune mutations of the repetitive region of MUC1 combined with immunoadjuvant monophosphoryl lipid A | Liposomal BLP-25 | Anticancer | Phase III of clinical trials | [146] |
| Combination of 16 peptides | ISA101 | HPV immunization | Phase II of clinical trials | [147] |
| Combination of nucleoprotein, matrix 1, and both B- and T-cell linear epitopes from HA into a single recombinantly synthesized polypeptide | Multimeric-001 | Influenza and HIV-1 immunization | Phase I of clinical trials | [133] |
| Synthesized peptide immunogens of the SARS-CoV-2 S protein conjugated to a carrier protein and absorbed on aluminum hydroxide | EpiVacCorona | Coronavirus disease 19 (COVID-19) immunization | Phase II of clinical trials | [148,149] |
| Short C-terminal fragments of Aβ40 and an aluminum hydroxide adjuvant | ABvac40 | Treatment for Alzheimer’s disease | Phase II of clinical trials | [150] |
4. Current Limitations and Future Perspectives
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Sani, M.A.; Separovic, F. How Membrane-Active Peptides Get into Lipid Membranes. Acc. Chem. Res. 2016, 49, 1130–1138. [Google Scholar] [CrossRef] [PubMed]
- Avci, F.G.; Akbulut, B.S.; Ozkirimli, E. Membrane Active Peptides and Their Biophysical Characterization. Biomolecules 2018, 8, 77. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, L.; Wang, N.; Zhang, W.; Cheng, X.; Yan, Z.; Shao, G.; Wang, X.; Wang, R.; Fu, C. Therapeutic peptides: Current applications and future directions. Signal Transduct. Target. Ther. 2022, 7, 48. [Google Scholar] [CrossRef] [PubMed]
- Sachon, E.; Walrant, A.; Sagan, S.; Cribier, S.; Rodriguez, N. Binding and crossing: Methods for the characterization of membrane-active peptides interactions with membranes at the molecular level. Arch. Biochem. Biophys. 2021, 699, 108751. [Google Scholar] [CrossRef]
- Erdem Büyükkiraz, M.; Kesmen, Z. Antimicrobial peptides (AMPs): A promising class of antimicrobial compounds. J. Appl. Microbiol. 2022, 132, 1573–1596. [Google Scholar] [CrossRef]
- Fjell, C.D.; Hiss, J.A.; Hancock, R.E.W.; Schneider, G. Designing antimicrobial peptides: Form follows function. Nat. Rev. Drug Discov. 2012, 11, 37–51. [Google Scholar] [CrossRef]
- Merrifield, R.B.; Vizioli, L.D.; Boman, H.G. Synthesis of the antibacterial peptide cecropin A(1-33). Biochemistry 1982, 21, 5020–5031. [Google Scholar] [CrossRef]
- Spitznagel, J.K. Antibiotic proteins of human neutrophils. J. Clin. Investig. 1990, 86, 1381–1386. [Google Scholar] [CrossRef]
- Blondelle, S.E.; Houghten, R.A. Design of model amphipathic peptides having potent antimicrobial activities. Biochemistry 1992, 31, 12688–12694. [Google Scholar] [CrossRef]
- Huang, Y.; Huang, J.; Chen, Y. Alpha-helical cationic antimicrobial peptides: Relationships of structure and function. Protein Cell 2010, 1, 143–152. [Google Scholar] [CrossRef] [Green Version]
- Shori, A.B.; Baba, A.S. The Effect of Refrigerated Storage on Anti-Diabetic and Antioxidant Potency of Probiotic Yogurt Treated with Some Medicinal Plants. Fermentation 2023, 9, 427. [Google Scholar] [CrossRef]
- Patra, A.; Das, J.; Agrawal, N.R.; Kushwaha, G.S.; Ghosh, M.; Son, Y.-O. Marine Antimicrobial Peptides-Based Strategies for Tackling Bacterial Biofilm and Biofouling Challenges. Molecules 2022, 27, 7546. [Google Scholar]
- Saucedo-Vázquez, J.P.; Gushque, F.; Vispo, N.S.; Rodriguez, J.; Gudiño-Gomezjurado, M.E.; Albericio, F.; Tellkamp, M.P.; Alexis, F. Marine Arthropods as a Source of Antimicrobial Peptides. Mar. Drugs 2022, 20, 501. [Google Scholar] [CrossRef] [PubMed]
- Baindara, P.; Mandal, S.M. Plant-Derived Antimicrobial Peptides: Novel Preservatives for the Food Industry. Foods 2022, 11, 2415. [Google Scholar] [PubMed]
- Balls, A.; Hale, W.; Harris, T. A crystalline protein obtained from a lipoprotein of wheat flour. Cereal Chem. 1942, 19, 279–288. [Google Scholar]
- Yi, H.-Y.; Chowdhury, M.; Huang, Y.-D.; Yu, X.-Q. Insect antimicrobial peptides and their applications. Appl. Microbiol. Biotechnol. 2014, 98, 5807–5822. [Google Scholar] [CrossRef] [Green Version]
- Stączek, S.; Cytryńska, M.; Zdybicka-Barabas, A. Unraveling the Role of Antimicrobial Peptides in Insects. Int. J. Mol. Sci. 2023, 24, 5753. [Google Scholar] [CrossRef]
- Wu, Y.; Williams, J.; Calder, E.D.D.; Walport, L.J. Strategies to expand peptide functionality through hybridisation with a small molecule component. RSC Chem. Biol. 2021, 2, 151–165. [Google Scholar] [CrossRef]
- Han, Y.; Zhang, M.; Lai, R.; Zhang, Z. Chemical modifications to increase the therapeutic potential of antimicrobial peptides. Peptides 2021, 146, 170666. [Google Scholar] [CrossRef]
- Röckendorf, N.; Nehls, C.; Gutsmann, T. Design of Membrane Active Peptides Considering Multi-Objective Optimization for Biomedical Application. Membranes 2022, 12, 180. [Google Scholar]
- Khairkhah, N.; Namvar, A.; Bolhassani, A. Application of Cell Penetrating Peptides as a Promising Drug Carrier to Combat Viral Infections. Mol. Biotechnol. 2023. [Google Scholar] [CrossRef]
- Chen, C.H.; Lu, T.K. Development and Challenges of Antimicrobial Peptides for Therapeutic Applications. Antibiotics 2020, 9, 24. [Google Scholar] [CrossRef] [Green Version]
- Frankel, A.D.; Pabo, C.O. Cellular uptake of the tat protein from human immunodeficiency virus. Cell 1988, 55, 1189–1193. [Google Scholar] [CrossRef]
- Joliot, A.; Pernelle, C.; Deagostini-Bazin, H.; Prochiantz, A. Antennapedia homeobox peptide regulates neural morphogenesis. Proc. Natl. Acad. Sci. USA 1991, 88, 1864–1868. [Google Scholar] [CrossRef]
- Pooga, M.; Hällbrink, M.; Zorko, M.; Langel, U. Cell penetration by transportan. FASEB J. Off. Publ. Fed. Am. Soc. Exp. Biol. 1998, 12, 67–77. [Google Scholar] [CrossRef]
- Gautam, A.; Singh, H.; Tyagi, A.; Chaudhary, K.; Kumar, R.; Kapoor, P.; Raghava, G.P.S. CPPsite: A curated database of cell penetrating peptides. Database 2012, 2012, bas015. [Google Scholar] [CrossRef] [Green Version]
- Kalafatovic, D.; Giralt, E. Cell-Penetrating Peptides: Design Strategies beyond Primary Structure and Amphipathicity. Molecules 2017, 22, 1929. [Google Scholar] [CrossRef] [Green Version]
- Stalmans, S.; Bracke, N.; Wynendaele, E.; Gevaert, B.; Peremans, K.; Burvenich, C.; Polis, I.; De Spiegeleer, B. Cell-Penetrating Peptides Selectively Cross the Blood-Brain Barrier In Vivo. PLoS ONE 2015, 10, e0139652. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, J.; Khan, A.R.; Fu, M.; Wang, R.; Ji, J.; Zhai, G. Cell-penetrating peptide: A means of breaking through the physiological barriers of different tissues and organs. J. Control. Release 2019, 309, 106–124. [Google Scholar] [CrossRef] [PubMed]
- Derakhshankhah, H.; Jafari, S. Cell penetrating peptides: A concise review with emphasis on biomedical applications. Biomed. Pharmacother. 2018, 108, 1090–1096. [Google Scholar] [CrossRef] [PubMed]
- Taylor, R.E.; Zahid, M. Cell Penetrating Peptides, Novel Vectors for Gene Therapy. Pharmaceutics 2020, 12, 225. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ruseska, I.; Zimmer, A. Internalization mechanisms of cell-penetrating peptides. Beilstein J. Nanotechnol. 2020, 11, 101–123. [Google Scholar] [CrossRef] [PubMed]
- Sharma, A.; Kapoor, P.; Gautam, A.; Chaudhary, K.; Kumar, R.; Chauhan, J.S.; Tyagi, A.; Raghava, G.P.S. Computational approach for designing tumor homing peptides. Sci. Rep. 2013, 3, 1607. [Google Scholar] [CrossRef] [Green Version]
- Zuo, H. iRGD: A Promising Peptide for Cancer Imaging and a Potential Therapeutic Agent for Various Cancers. J. Oncol. 2019, 2019, 9367845. [Google Scholar] [CrossRef]
- Svensen, N.; Walton, J.G.A.; Bradley, M. Peptides for cell-selective drug delivery. Trends Pharmacol. Sci. 2012, 33, 186–192. [Google Scholar] [CrossRef]
- Islam, M.M.; Zaman, S.U.; Asif, F.; Hasan Arnab, M.K.; Rahman, M.M.; Hasan, M. Isolation of Antimicrobial Peptides (AMPs) from Different Sources: A Review. Bangladesh Pharm. J. 2023, 26, 99–111. [Google Scholar] [CrossRef]
- Pingitore, E.V.; Salvucci, E.; Sesma, F.; Nader-Macias, M. Different strategies for purification of antimicrobial peptides from lactic acid bacteria (LAB). Commun. Curr. Res. Educ. Top. Trends Appl. Microbiol. 2007, 1, 557–568. [Google Scholar]
- Ramazi, S.; Mohammadi, N.; Allahverdi, A.; Khalili, E.; Abdolmaleki, P. A review on antimicrobial peptides databases and the computational tools. Database 2022, 2022, baac011. [Google Scholar] [CrossRef] [PubMed]
- Kleiner-Grote, G.R.M.; Risse, J.M.; Friehs, K. Secretion of recombinant proteins from E. coli. Eng. Life Sci. 2018, 18, 532–550. [Google Scholar] [CrossRef] [Green Version]
- Grewal, A.S.; Kumar, K.; Redhu, S.; Bhardwaj, S. Microwave assisted synthesis: A green chemistry approach. Int. Res. J. Pharm. Appl. Sci. 2013, 3, 278–285. [Google Scholar]
- Karas, J.A.; Boland, M.; Haigh, C.; Johanssen, V.; Hill, A.; Barnham, K.; Collins, S.; Scanlon, D. Microwave Synthesis of Prion Protein Fragments up to 111 Amino Acids in Length Generates Biologically Active Peptides. Int. J. Pept. Res. Ther. 2012, 18, 21–29. [Google Scholar] [CrossRef]
- Drapala, A.; Wieczorek, P. Extraction of short peptides using supported liquid membranes. Desalination 2002, 148, 235–239. [Google Scholar] [CrossRef]
- Michaels, A.S.; Nelsen, L.; Porter, M.C. Ultrafiltration. In Membrane Processes in Industry and Biomedicine: Proceedings of a Symposium Held at the 160th National Meeting of the American Chemical Society, under the Sponsorship of the Division of Industrial and Engineering Chemistry, Chicago, IL, USA, 16–17 September 1970; Bier, M., Ed.; Springer: Boston, MA, USA, 1971; pp. 197–232. [Google Scholar]
- Beaubier, S.; Przybylski, R.; Bodin, A.; Nedjar, N.; Dhulster, P.; Kapel, R. Ultrafiltration Fractionation of Bovine Hemoglobin Hydrolysates: Prediction of Separation Performances for Optimal Enrichment in Antimicrobial Peptide. Membranes 2021, 11, 73. [Google Scholar] [CrossRef]
- Hofstadler, S.A.; Sannes-Lowery, K.A. Applications of ESI-MS in drug discovery: Interrogation of noncovalent complexes. Nat. Rev. Drug Discov. 2006, 5, 585–595. [Google Scholar] [CrossRef] [PubMed]
- Laursen, R.A. Solid-phase Edman degradation. An automatic peptide sequencer. Eur. J. Biochem. 1971, 20, 89–102. [Google Scholar] [CrossRef] [PubMed]
- Gopal, R.; Park, J.S.; Seo, C.H.; Park, Y. Applications of Circular Dichroism for Structural Analysis of Gelatin and Antimicrobial Peptides. Int. J. Mol. Sci. 2012, 13, 3229–3244. [Google Scholar] [CrossRef] [PubMed]
- Kang, S.-J.; Kim, D.-H.; Mishig-Ochir, T.; Lee, B.-J. Antimicrobial peptides: Their physicochemical properties and therapeutic application. Arch. Pharmacal. Res. 2012, 35, 409–413. [Google Scholar] [CrossRef]
- Jindal, M.; Le, C.; Mohd Yusof, M.; Sekaran, S. Net charge, hydrophobicity and specific amino acids contribute to the activity of antimicrobial peptides. J. Health Transl. Med. 2014, 17, 1–7. [Google Scholar]
- Kim, S.; Chen, J.; Cheng, T.; Gindulyte, A.; He, J.; He, S.; Li, Q.; Shoemaker, B.A.; Thiessen, P.A.; Yu, B.; et al. PubChem 2023 update. Nucleic Acids Res. 2022, 51, D1373–D1380. [Google Scholar] [CrossRef]
- Mabrouk, D.M. Antimicrobial peptides: Features, applications and the potential use against covid-19. Mol. Biol. Rep. 2022, 49, 10039–10050. [Google Scholar] [CrossRef]
- Wang, G. Human Antimicrobial Peptides and Proteins. Pharmaceuticals 2014, 7, 545–594. [Google Scholar]
- Lei, J.; Sun, L.; Huang, S.; Zhu, C.; Li, P.; He, J.; Mackey, V.; Coy, D.H.; He, Q. The antimicrobial peptides and their potential clinical applications. Am. J. Transl. Res. 2019, 11, 3919–3931. [Google Scholar]
- de Oliveira, E.C.L.; Santana, K.; Josino, L.; Lima e Lima, A.H.; de Souza de Sales Júnior, C. Predicting cell-penetrating peptides using machine learning algorithms and navigating in their chemical space. Sci. Rep. 2021, 11, 7628. [Google Scholar] [CrossRef] [PubMed]
- Böhmová, E.; Machová, D.; Pechar, M.; Pola, R.; Venclíková, K.; Janoušková, O.; Etrych, T. Cell-penetrating peptides: A useful tool for the delivery of various cargoes into cells. Physiol. Res. 2018, 67, S267–S279. [Google Scholar] [CrossRef] [PubMed]
- Last, N.B.; Schlamadinger, D.E.; Miranker, A.D. A common landscape for membrane-active peptides. Protein Sci. 2013, 22, 870–882. [Google Scholar] [PubMed] [Green Version]
- Benfield, A.H.; Henriques, S.T. Mode-of-action of antimicrobial peptides: Membrane disruption vs. intracellular mechanisms. Front. Med. Technol. 2020, 2, 610997. [Google Scholar] [CrossRef] [PubMed]
- Hollmann, A.; Martinez, M.; Maturana, P.; Semorile, L.C.; Maffia, P.C. Antimicrobial peptides: Interaction with model and biological membranes and synergism with chemical antibiotics. Front. Chem. 2018, 6, 204. [Google Scholar] [CrossRef] [Green Version]
- Berthelot, K.; Cullin, C.; Lecomte, S. What does make an amyloid toxic: Morphology, structure or interaction with membrane? Biochimie 2013, 95, 12–19. [Google Scholar] [CrossRef]
- Milletti, F. Cell-penetrating peptides: Classes, origin, and current landscape. Drug Discov. Today 2012, 17, 850–860. [Google Scholar] [CrossRef]
- Bischetti, M.; Alaimo, N.; Nardelli, F.; Punzi, P.; Amariei, C.; Ingenito, R.; Musco, G.; Gallo, M.; Cicero, D.O. Structural insights on the selective interaction of the histidine-rich piscidin antimicrobial peptide Of-Pis1 with membranes. Biochim. Biophys. Acta (BBA)-Biomembr. 2023, 1865, 184080. [Google Scholar] [CrossRef]
- Aguilar, S.; Brunetti, A.E.; Garay, A.V.; Santos, L.C.; Perez, L.O.; Moreira, D.; Cancelarich, N.L.; Barbosa, E.A.; Basso, N.G.; de Freitas, S.M.; et al. Structure and function of cationic hylin bioactive peptides from the tree frog Boana pulchella in interaction with lipid membranes. Peptides 2023, 159, 170900. [Google Scholar] [CrossRef] [PubMed]
- Syryamina, V.N.; Afanasyeva, E.F.; Dzuba, S.A.; Formaggio, F.; De Zotti, M. Peptide-membrane binding is not enough to explain bioactivity: A case study. Biochim. Biophys. Acta (BBA)-Biomembr. 2022, 1864, 183978. [Google Scholar] [CrossRef] [PubMed]
- Mohid, S.A.; Biswas, K.; Won, T.; Mallela, L.S.; Gucchait, A.; Butzke, L.; Sarkar, R.; Barkham, T.; Reif, B.; Leipold, E.; et al. Structural insights into the interaction of antifungal peptides and ergosterol containing fungal membrane. Biochim. Et Biophys. Acta (BBA)-Biomembr. 2022, 1864, 183996. [Google Scholar] [CrossRef]
- Guha, S.; Ferrie, R.P.; Ghimire, J.; Ventura, C.R.; Wu, E.; Sun, L.; Kim, S.Y.; Wiedman, G.R.; Hristova, K.; Wimley, W.C. Applications and evolution of melittin, the quintessential membrane active peptide. Biochem. Pharmacol. 2021, 193, 114769. [Google Scholar] [CrossRef]
- Viana de Freitas, T.; Karmakar, U.; Vasconcelos, A.; Santos, M.; Oliveira do Vale Lira, B.; Costa, S.; Barbosa, E.; Cardozo-Fh, J.; Correa, R.; Ribeiro, D.S.; et al. Release of immunomodulatory peptides at bacterial membrane interfaces as a novel strategy to fight microorganisms. J. Biol. Chem. 2023, 299, 103056. [Google Scholar] [CrossRef]
- Kumar, S.; Balayaa, R.D.A.; Kanekar, S.; Raju, R.; Prasad, T.S.K.; Kandasamy, R.K. Computational Tools for Exploring Peptide-Membrane Interactions in Gram-Positive Bacteria. Comput. Struct. Biotechnol. J. 2023, 21, 1995–2008. [Google Scholar] [CrossRef]
- Sebastiao, M.; Babych, M.; Quittot, N.; Kumar, K.; Arnold, A.A.; Marcotte, I.; Bourgault, S. Development of a novel fluorescence assay for studying lipid bilayer perturbation induced by amyloidogenic peptides using cell plasma membrane vesicles. Biochim. Biophys. Acta (BBA)-Biomembr. 2023, 1865, 184118. [Google Scholar] [CrossRef]
- Sani, M.-A.; Le Brun, A.P.; Rajput, S.; Attard, T.; Separovic, F. The membrane activity of the antimicrobial peptide caerin 1.1 is pH dependent. Biophys. J. 2023, 122, 1058–1067. [Google Scholar] [CrossRef] [PubMed]
- Hazam, P.K.; Cheng, C.-C.; Lin, W.-C.; Hsieh, C.-Y.; Hsu, P.-H.; Chen, Y.-R.; Li, C.-C.; Hsueh, P.-R.; Chen, J.-Y. Strategic modification of low-activity natural antimicrobial peptides confers antibacterial potential in vitro and in vivo. Eur. J. Med. Chem. 2023, 249, 115131. [Google Scholar] [CrossRef]
- Peng, K.-C.; Lee, S.-H.; Hour, A.-L.; Pan, C.-Y.; Lee, L.-H.; Chen, J.-Y. Five different piscidins from Nile tilapia, Oreochromis niloticus: Analysis of their expressions and biological functions. PLoS ONE 2012, 7, e50263. [Google Scholar] [CrossRef]
- da Silva, B.S.; Díaz-Roa, A.; Yamane, E.S.; Hayashi, M.A.F.; Silva Junior, P.I. Doderlin: Isolation and characterization of a broad-spectrum antimicrobial peptide from Lactobacillus acidophilus. Res. Microbiol. 2023, 174, 103995. [Google Scholar] [CrossRef]
- Zhou, J.; Cha, R.; Wu, Z.; Zhang, C.; He, Y.; Zhang, H.; Liu, K.; Fareed, M.S.; Wang, Z.; Yang, C.; et al. An injectable, natural peptide hydrogel with potent antimicrobial activity and excellent wound healing-promoting effects. Nano Today 2023, 49, 101801. [Google Scholar] [CrossRef]
- Cai, S.; Liu, P.; Liu, S.; Cao, X.; Peng, J.; Meng, K.; Qu, Y. Characterization and functional analysis of three novel liver-expressed antimicrobial peptide 2 (LEAP-2) in Glyptothorax zanaensis. Aquac. Rep. 2023, 28, 101455. [Google Scholar] [CrossRef]
- Zhang, H.; Cao, Z.; Diao, Q.; Zhou, Y.; Ao, J.; Liu, C.; Sun, Y. Antimicrobial activity and mechanisms of a derived antimicrobial peptide TroNKL-27 from golden pompano (Trachinotus ovatus) NK-lysin. Fish Shellfish Immunol. 2022, 126, 357–369. [Google Scholar] [CrossRef]
- Park, H.J.; Kang, H.K.; Park, E.; Kim, M.K.; Park, Y. Bactericidal activities and action mechanism of the novel antimicrobial peptide Hylin a1 and its analog peptides against Acinetobacter baumannii infection. Eur. J. Pharm. Sci. 2022, 175, 106205. [Google Scholar] [CrossRef]
- Taveira, G.B.; de Oliveira Mello, É.; Simão, T.L.B.V.; Cherene, M.B.; de Oliveira Carvalho, A.; Muzitano, M.F.; Lassounskaia, E.; Pireda, S.; de Castro Miguel, E.; Basso, L.G.M.; et al. A new bioinspired peptide on defensin from C. annuum fruits: Antimicrobial activity, mechanisms of action and therapeutical potential. Biochim. Biophys. Acta (BBA)-Gen. Subj. 2022, 1866, 130218. [Google Scholar] [CrossRef]
- Luo, X.; Song, Y.; Cao, Z.; Qin, Z.; Dessie, W.; He, N.; Wang, Z.; Tan, Y. Evaluation of the antimicrobial activities and mechanisms of synthetic antimicrobial peptide against food-borne pathogens. Food Biosci. 2022, 49, 101903. [Google Scholar] [CrossRef]
- Won, T.; Mohid, S.A.; Choi, J.; Kim, M.; Krishnamoorthy, J.; Biswas, I.; Bhunia, A.; Lee, D. The role of hydrophobic patches of de novo designed MSI-78 and VG16KRKP antimicrobial peptides on fragmenting model bilayer membranes. Biophys. Chem. 2023, 296, 106981. [Google Scholar] [CrossRef]
- Horváti, K.; Bacsa, B.; Mlinkó, T.; Szabó, N.; Hudecz, F.; Zsila, F.; Bősze, S. Comparative analysis of internalisation, haemolytic, cytotoxic and antibacterial effect of membrane-active cationic peptides: Aspects of experimental setup. Amino Acids 2017, 49, 1053–1067. [Google Scholar] [CrossRef]
- Tenore, G.C.; Ritieni, A.; Campiglia, P.; Stiuso, P.; Di Maro, S.; Sommella, E.; Pepe, G.; D’Urso, E.; Novellino, E. Antioxidant peptides from “Mozzarella di Bufala Campana DOP” after simulated gastrointestinal digestion: In vitro intestinal protection, bioavailability, and anti-haemolytic capacity. J. Funct. Foods 2015, 15, 365–375. [Google Scholar] [CrossRef]
- Gluvić, A.; Ulrih, N.P. Peptides derived from food sources: Antioxidative activities and interactions with model lipid membranes. Food Chem. 2019, 287, 324–332. [Google Scholar] [CrossRef] [PubMed]
- Jafar, S.; Kamal, H.; Mudgil, P.; Hassan, H.M.; Maqsood, S. Camel whey protein hydrolysates displayed enhanced cholesteryl esterase and lipase inhibitory, anti-hypertensive and anti-haemolytic properties. LWT 2018, 98, 212–218. [Google Scholar] [CrossRef]
- Edwards, A.B.; Mastaglia, F.L.; Knuckey, N.W.; Yip, K.-H.; Meloni, B. Assessment of the safety of the cationic arginine-rich peptides (CARPs) poly-arginine-18 (R18 and R18D) in ex vivo models of mast cell degranulation and red blood cell hemolysis. Biochem. Biophys. Rep. 2022, 31, 101305. [Google Scholar] [CrossRef]
- Dennison, S.R.; Reddy, S.M.; Morton, L.H.G.; Harris, F.; Badiani, K.; Phoenix, D.A. PEGylation enhances the antibacterial and therapeutic potential of amphibian host defence peptides. Biochim. Biophys. Acta (BBA)-Biomembr. 2022, 1864, 183806. [Google Scholar] [CrossRef] [PubMed]
- Langer, M.K.; Rahman, A.; Dey, H.; Anderssen, T.; Blencke, H.-M.; Haug, T.; Stensvåg, K.; Strøm, M.B.; Bayer, A. Investigation of tetrasubstituted heterocycles reveals hydantoins as a promising scaffold for development of novel antimicrobials with membranolytic properties. Eur. J. Med. Chem. 2023, 249, 115147. [Google Scholar] [CrossRef] [PubMed]
- Zou, W.; Zhang, Y.; Zhou, M.; Chen, X.; Ma, C.; Wang, T.; Jiang, Y.; Chen, T.; Shaw, C.; Wang, L. Exploring the active core of a novel antimicrobial peptide, palustrin-2LTb, from the Kuatun frog, Hylarana latouchii, using a bioinformatics-directed approach. Comput. Struct. Biotechnol. J. 2022, 20, 6192–6205. [Google Scholar] [CrossRef] [PubMed]
- Bhat, R.A.H.; Khangembam, V.C.; Thakuria, D.; Pant, V.; Tandel, R.S.; Tripathi, G.; Sarma, D. Antimicrobial activity of an artificially designed peptide against fish pathogens. Microbiol. Res. 2022, 260, 127039. [Google Scholar] [CrossRef] [PubMed]
- Ponnappan, N.; Budagavi, D.P.; Chugh, A. CyLoP-1: Membrane-active peptide with cell-penetrating and antimicrobial properties. Biochim. Biophys. Acta-Biomembr. BBA 2017, 1859, 167–176. [Google Scholar] [CrossRef] [PubMed]
- Plaza, J.G.R.; Morales-Nava, R.; Diener, C.; Schreiber, G.; Gonzalez, Z.D.; Ortiz, M.T.L.; Blake, I.O.; Pantoja, O.; Volkmer, R.; Klipp, E. Cell penetrating peptides and cationic antibacterial peptides: Two sides of the same coin. J. Biol. Chem. 2014, 289, 14448–14457. [Google Scholar] [CrossRef] [Green Version]
- Ascoët, S.; Touchard, A.; Téné, N.; Lefranc, B.; Leprince, J.; Paquet, F.; Jouvensal, L.; Barassé, V.; Treilhou, M.; Billet, A.; et al. The mechanism underlying toxicity of a venom peptide against insects reveals how ants are master at disrupting membranes. iScience 2023, 26, 106157. [Google Scholar] [CrossRef]
- Sharma, K.; Aaghaz, S.; Maurya, I.K.; Sharma, K.K.; Singh, S.; Rudramurthy, S.M.; Kumar, V.; Tikoo, K.; Jain, R. Synthetic amino acids-derived peptides target Cryptococcus neoformans by inducing cell membrane disruption. Bioorganic Chem. 2023, 130, 106252. [Google Scholar] [CrossRef]
- Wade, H.M.; Darling, L.E.O.; Elmore, D.E. Hybrids made from antimicrobial peptides with different mechanisms of action show enhanced membrane permeabilization. Biochim. Biophys. Acta (BBA)-Biomembr. 2019, 1861, 182980. [Google Scholar] [CrossRef]
- Tang, Q.; Tan, P.; Dai, Z.; Wang, T.; Xu, S.; Ding, Y.; Jin, J.; Zhang, X.; Zhang, Y.; Zhou, C.; et al. Hydrophobic modification improves the delivery of cell-penetrating peptides to eliminate intracellular pathogens in animals. Acta Biomater. 2023, 157, 210–224. [Google Scholar] [CrossRef] [PubMed]
- Parra, A.L.C.; Freitas, C.D.T.; Souza, P.F.N.; von Aderkas, P.; Borchers, C.H.; Beattie, G.A.; Silva, F.D.A.; Thornburg, R.W. Ornamental tobacco floral nectar is a rich source of antimicrobial peptides. Plant Sci. 2022, 324, 111427. [Google Scholar] [CrossRef] [PubMed]
- Muchintala, D.; Suresh, V.; Raju, D.; Sashidhar, R.B. Synthesis and characterization of cecropin peptide-based silver nanocomposites: Its antibacterial activity and mode of action. Mater. Sci. Eng. C 2020, 110, 110712. [Google Scholar] [CrossRef]
- Wichmann, N.; Lund, P.M.; Hansen, M.B.; Hjørringgaard, C.U.; Larsen, J.B.; Kristensen, K.; Andresen, T.L.; Simonsen, J.B. Applying flow cytometry to identify the modes of action of membrane-active peptides in a label-free and high-throughput fashion. Biochim. Biophys. Acta (BBA)-Biomembr. 2022, 1864, 183820. [Google Scholar] [CrossRef]
- Sun, S.; Xia, Y.; Liu, J.; Dou, Y.; Yang, K.; Yuan, B.; Kang, Z. Real-time monitoring the interfacial dynamic processes at model cell membranes: Taking cell penetrating peptide TAT as an example. J. Colloid Interface Sci. 2022, 609, 707–717. [Google Scholar] [CrossRef]
- Basso, L.G.M.; Zeraik, A.E.; Felizatti, A.P.; Costa-Filho, A.J. Membranotropic and biological activities of the membrane fusion peptides from SARS-CoV spike glycoprotein: The importance of the complete internal fusion peptide domain. Biochim. Et Biophys. Acta (BBA)-Biomembr. 2021, 1863, 183697. [Google Scholar] [CrossRef]
- Shekunov, E.V.; Zlodeeva, P.D.; Efimova, S.S.; Muryleva, A.A.; Zarubaev, V.V.; Slita, A.V.; Ostroumova, O.S. Cyclic lipopeptides as membrane fusion inhibitors against SARS-CoV-2: New tricks for old dogs. Antivir. Res. 2023, 212, 105575. [Google Scholar] [CrossRef]
- Reuter, M.; Schwieger, C.; Meister, A.; Karlsson, G.; Blume, A. Poly-l-lysines and poly-l-arginines induce leakage of negatively charged phospholipid vesicles and translocate through the lipid bilayer upon electrostatic binding to the membrane. Biophys. Chem. 2009, 144, 27–37. [Google Scholar] [CrossRef] [Green Version]
- Cummings, J.E.; Vanderlick, T.K. Aggregation and hemi-fusion of anionic vesicles induced by the antimicrobial peptide cryptdin-4. Biochim. Biophys. Acta (BBA)-Biomembr. 2007, 1768, 1796–1804. [Google Scholar] [CrossRef] [Green Version]
- Rice, A.; Zimmerberg, J.; Pastor, R.W. Initiation and evolution of pores formed by influenza fusion peptides probed by lysolipid inclusion. Biophys. J. 2022. [Google Scholar] [CrossRef]
- Suárez, T.; Gómara, M.a.J.; Goñi, F.M.; Mingarro, I.; Muga, A.; Pérez-Payá, E.; Nieva, J.L. Calcium-dependent conformational changes of membrane-bound Ebola fusion peptide drive vesicle fusion. FEBS Lett. 2003, 535, 23–28. [Google Scholar] [CrossRef] [Green Version]
- Chiou, P.-C.; Hsu, W.-W.; Chang, Y.; Chen, Y.-F. Molecular packing of lipid membranes and action mechanisms of membrane-active peptides. Colloids Surf. B Biointerfaces 2022, 213, 112384. [Google Scholar] [CrossRef]
- Vitorino, R.; Guedes, S.; da Costa, J.P.; Kašička, V. Microfluidics for peptidomics, proteomics, and cell analysis. Nanomaterials 2021, 11, 1118. [Google Scholar] [CrossRef] [PubMed]
- Xun, X.-M.; Zhang, Z.-A.; Yuan, Z.-X.; Tuhong, K.; Yan, C.-H.; Zhan, Y.-F.; Kang, G.-P.; Wu, Q.-Y.; Wang, J. Novel caffeic acid grafted chitosan-sodium alginate microcapsules generated by microfluidic technique for the encapsulation of bioactive peptides from silkworm pupae. Sustain. Chem. Pharm. 2023, 32, 100974. [Google Scholar] [CrossRef]
- Al Nahas, K.; Cama, J.; Schaich, M.; Hammond, K.; Deshpande, S.; Dekker, C.; Ryadnov, M.; Keyser, U. A microfluidic platform for the characterisation of membrane active antimicrobials. Lab A Chip 2019, 19, 837–844. [Google Scholar] [CrossRef] [Green Version]
- Gómez, S.C.; Quezada, V.; Quiroz, I.; Muñoz-Camargo, C.; Osma, J.F.; Reyes, L.H.; Cruz, J.C. Design and Manufacture of a Low-Cost Microfluidic System for the Synthesis of Giant Liposomes for the Encapsulation of Yeast Homologues: Applications in the Screening of Membrane-Active Peptide Libraries. Micromachines 2021, 12, 1377. [Google Scholar] [CrossRef]
- Cama, J.; Al Nahas, K.; Fletcher, M.; Hammond, K.; Ryadnov, M.G.; Keyser, U.F.; Pagliara, S. An ultrasensitive microfluidic approach reveals correlations between the physico-chemical and biological activity of experimental peptide antibiotics. Sci. Rep. 2022, 12, 4005. [Google Scholar] [CrossRef]
- Al Nahas, K.; Fletcher, M.; Hammond, K.; Nehls, C.; Cama, J.; Ryadnov, M.G.; Keyser, U.F. Measuring Thousands of Single-Vesicle Leakage Events Reveals the Mode of Action of Antimicrobial Peptides. Anal. Chem. 2022, 94, 9530–9539. [Google Scholar] [CrossRef] [PubMed]
- Moretta, A.; Scieuzo, C.; Petrone, A.M.; Salvia, R.; Manniello, M.D.; Franco, A.; Lucchetti, D.; Vassallo, A.; Vogel, H.; Sgambato, A.; et al. Antimicrobial Peptides: A New Hope in Biomedical and Pharmaceutical Fields. Front. Cell. Infect. Microbiol. 2021, 11, 668632. [Google Scholar] [CrossRef]
- Bahar, A.A.; Ren, D. Antimicrobial Peptides. Pharmaceuticals 2013, 6, 1543–1575. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Soltani, S.; Hammami, R.; Cotter, P.D.; Rebuffat, S.; Said, L.B.; Gaudreau, H.; Bédard, F.; Biron, E.; Drider, D.; Fliss, I. Bacteriocins as a new generation of antimicrobials: Toxicity aspects and regulations. FEMS Microbiol. Rev. 2020, 45, fuaa039. [Google Scholar] [CrossRef]
- Favaro, L.; Campanaro, S.; Fugaban, J.I.I.; Treu, L.; Jung, E.S.; d’Ovidio, L.; de Oliveira, D.P.; Liong, M.T.; Ivanova, I.V.; Todorov, S.D. Genomic, metabolomic, and functional characterisation of beneficial properties of Pediococcus pentosaceus ST58, isolated from human oral cavity. Benef. Microbes 2023, 14, 57–72. [Google Scholar] [CrossRef] [PubMed]
- da Silva, A.R.S.; de Souza de Azevedo, P.O.; Converti, A.; de Souza Oliveira, R.P. Cultivation of Lactic Acid Bacteria and Evaluation of the Antimicrobial Potential of Partially Purified Bacteriocin-like Inhibitory Substances against Cariogenic and Food Pathogens. Fermentation 2022, 8, 400. [Google Scholar] [CrossRef]
- Stiltner, J.; McCandless, K.; Zahid, M. Cell-Penetrating Peptides: Applications in Tumor Diagnosis and Therapeutics. Pharmaceutics 2021, 13, 890. [Google Scholar] [CrossRef]
- Kondo, E.; Iioka, H.; Saito, K. Tumor-homing peptide and its utility for advanced cancer medicine. Cancer Sci. 2021, 112, 2118–2125. [Google Scholar] [CrossRef]
- Kaur, S.; Kaur, S. Bacteriocins as Potential Anticancer Agents. Front. Pharmacol. 2015, 6, 272. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, T.; Li, J.; Jia, Q.; Zhan, S.; Zhang, Q.; Wang, Y.; Wang, X. Antimicrobial peptide 17BIPHE2 inhibits the proliferation of lung cancer cells in vitro and in vivo by regulating the ERK signaling pathway. Oncol. Lett. 2021, 22, 501. [Google Scholar] [CrossRef]
- Xiao, Y.-F.; Jie, M.-M.; Li, B.-S.; Hu, C.-J.; Xie, R.; Tang, B.; Yang, S.-M. Peptide-Based Treatment: A Promising Cancer Therapy. J. Immunol. Res. 2015, 2015, 761820. [Google Scholar] [CrossRef] [Green Version]
- Fu, C.; Yu, L.; Miao, Y.; Liu, X.; Yu, Z.; Wei, M. Peptide–drug conjugates (PDCs): A novel trend of research and development on targeted therapy, hype or hope? Acta Pharm. Sin. B 2023, 13, 498–516. [Google Scholar] [CrossRef] [PubMed]
- Chavda, V.P.; Solanki, H.K.; Davidson, M.; Apostolopoulos, V.; Bojarska, J. Peptide-Drug Conjugates: A New Hope for Cancer Management. Molecules 2022, 27, 7232. [Google Scholar]
- Zhang, J.; Song, Q.; Cai, L.; Xie, Y.; Chen, Y. The efficacy of 177Lu-DOTATATE peptide receptor radionuclide therapy (PRRT) in patients with metastatic neuroendocrine tumours: A systematic review and meta-analysis. J. Cancer Res. Clin. Oncol. 2020, 146, 1533–1543. [Google Scholar] [CrossRef] [PubMed]
- Flanagan, K.; Kumari, R.; Miettinen, J.J.; Haney, S.L.; Varney, M.L.; Williams, J.T.; Majumder, M.M.; Suvela, M.; Slipicevic, A.; Lehmann, F.; et al. The Peptide-Drug Conjugate Melflufen Modulates the Unfolded Protein Response of Multiple Myeloma and Amyloidogenic Plasma Cells and Induces Cell Death. Hemasphere 2022, 6, e687. [Google Scholar] [CrossRef] [PubMed]
- Carlier, C.; Strese, S.; Viktorsson, K.; Velander, E.; Nygren, P.; Uustalu, M.; Juntti, T.; Lewensohn, R.; Larsson, R.; Spira, J.; et al. Preclinical activity of melflufen (J1) in ovarian cancer. Oncotarget 2016, 7, 59322–59335. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huynh, T.T.; van Dam, E.M.; Sreekumar, S.; Mpoy, C.; Blyth, B.J.; Muntz, F.; Harris, M.J.; Rogers, B.E. Copper-67-Labeled Bombesin Peptide for Targeted Radionuclide Therapy of Prostate Cancer. Pharmaceuticals 2022, 15, 728. [Google Scholar] [CrossRef]
- Ita, K. Coronavirus Disease (COVID-19): Current Status and Prospects for Drug and Vaccine Development. Arch. Med. Res. 2021, 52, 15–24. [Google Scholar] [CrossRef] [PubMed]
- Naz, R.K.; Dabir, P. Peptide vaccines against cancer, infectious diseases, and conception. Front. Biosci.-Landmark 2007, 12, 1833–1844. [Google Scholar] [CrossRef] [Green Version]
- Sangha, R.; Butts, C. L-BLP25: A Peptide Vaccine Strategy in Non–Small Cell Lung Cancer. Clin. Cancer Res. 2007, 13, 4652s–4654s. [Google Scholar] [CrossRef] [Green Version]
- Zhou, X.; Jiang, X.; Qu, M.; Aninwene, G.E., II; Jucaud, V.; Moon, J.J.; Gu, Z.; Sun, W.; Khademhosseini, A. Engineering Antiviral Vaccines. ACS Nano 2020, 14, 12370–12389. [Google Scholar] [CrossRef]
- Zhou, J.Z.; Jou, J.; Cohen, E. Vaccine Strategies for Human Papillomavirus-Associated Head and Neck Cancers. Cancers 2022, 14, 33. [Google Scholar] [CrossRef] [PubMed]
- Malonis, R.J.; Lai, J.R.; Vergnolle, O. Peptide-Based Vaccines: Current Progress and Future Challenges. Chem. Rev. 2020, 120, 3210–3229. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tsilibary, E.-P.; Charonis, S.A.; Georgopoulos, A.P. Vaccines for Influenza. Vaccines 2021, 9, 47. [Google Scholar] [CrossRef] [PubMed]
- Barchuk, A.; Bulina, A.; Cherkashin, M.; Berezina, N.; Rakova, T.; Kuplevatskaya, D.; Skougarevskiy, D.; Okhotin, A. Gam-COVID-Vac, EpiVacCorona, and CoviVac effectiveness against lung injury during Delta and Omicron variant surges in St. Petersburg, Russia: A test-negative case–control study. Respir. Res. 2022, 23, 276. [Google Scholar] [CrossRef] [PubMed]
- Bayani, F.; Hashkavaei, N.S.; Arjmand, S.; Rezaei, S.; Uskoković, V.; Alijanianzadeh, M.; Uversky, V.N.; Ranaei Siadat, S.O.; Mozaffari-Jovin, S.; Sefidbakht, Y. An overview of the vaccine platforms to combat COVID-19 with a focus on the subunit vaccines. Prog. Biophys. Mol. Biol. 2023, 178, 32–49. [Google Scholar] [CrossRef] [PubMed]
- Bravo-Vázquez, L.A.; Mora-Hernández, E.O.; Rodríguez, A.L.; Sahare, P.; Bandyopadhyay, A.; Duttaroy, A.K.; Paul, S. Current Advances of Plant-Based Vaccines for Neurodegenerative Diseases. Pharmaceutics 2023, 15, 711. [Google Scholar] [CrossRef]
- Vassilakopoulou, V.; Karachaliou, C.-E.; Evangelou, A.; Zikos, C.; Livaniou, E. Peptide-Based Vaccines for Neurodegenerative Diseases: Recent Endeavors and Future Perspectives. Vaccines 2021, 9, 1278. [Google Scholar] [CrossRef]
- Meloni, B.P.; Brookes, L.M.; Clark, V.W.; Cross, J.L.; Edwards, A.B.; Anderton, R.S.; Hopkins, R.M.; Hoffmann, K.; Knuckey, N.W. Poly-Arginine and Arginine-Rich Peptides are Neuroprotective in Stroke Models. J. Cereb. Blood Flow Metab. 2015, 35, 993–1004. [Google Scholar] [CrossRef]
- Dergunova, L.V.; Filippenkov, I.B.; Limborska, S.A.; Myasoedov, N.F. Neuroprotective Peptides and New Strategies for Ischemic Stroke Drug Discoveries. Genes 2023, 14, 953. [Google Scholar] [CrossRef]
- Eskandari, S.; Rezayof, A.; Asghari, S.M.; Hashemizadeh, S. Neurobiochemical characteristics of arginine-rich peptides explain their potential therapeutic efficacy in neurodegenerative diseases. Neuropeptides 2023, 101, 102356. [Google Scholar] [CrossRef]
- Laws, G.L.; Hale, J.D.F.; Kemp, R.A. Human Systemic Immune Response to Ingestion of the Oral Probiotic Streptococcus salivarius BLIS K12. Probiotics Antimicrob. Proteins 2021, 13, 1521–1529. [Google Scholar] [CrossRef]
- Karpiński, T.M.; Adamczak, A. Anticancer Activity of Bacterial Proteins and Peptides. Pharmaceutics 2018, 10, 54. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Goh, K.S.; Ng, Z.J.; Halim, M.; Oslan, S.N.; Oslan, S.N.H.; Tan, J.S. A Comprehensive Review on the Anticancer Potential of Bacteriocin: Preclinical and Clinical Studies. Int. J. Pept. Res. Ther. 2022, 28, 75. [Google Scholar] [CrossRef]
- Arsenio, M.F.; Nuno, B.; Ananda, M.C. Exploring the anticancer potential of the bacterial protein azurin. AIMS Microbiol. 2016, 2, 292–303. [Google Scholar] [CrossRef]
- Dahal, T. Immuno-Therapy in Lung Cancer-How Does Immuno-therapy for Lung Cancer Change Patients’ Vision. Int. J. Immunol. Immunother. 2022, 9, 062. [Google Scholar]
- Sousa, L.G.; Rajapakshe, K.; Rodriguez Canales, J.; Chin, R.L.; Feng, L.; Wang, Q.; Barrese, T.Z.; Massarelli, E.; William, W.; Johnson, F.M.; et al. ISA101 and nivolumab for HPV-16(+) cancer: Updated clinical efficacy and immune correlates of response. J. Immunother. Cancer 2022, 10, e004232. [Google Scholar] [CrossRef] [PubMed]
- Ryzhikov, A.; Ryzhikov, E.; Bogryantseva, M.; Usova, S.; Danilenko, E.; Nechaeva, E.; Pyankov, O.; Pyankova, O.; Gudymo, A.; Bodnev, S. A single blind, placebo-controlled randomized study of the safety, reactogenicity and immunogenicity of the “EpiVacCorona” Vaccine for the prevention of COVID-19, in volunteers aged 18–60 years (phase I-II). Russ. J. Infect. Immun. 2021, 11, 283–296. [Google Scholar] [CrossRef]
- Basheeruddin Asdaq, S.M.; Jomah, S.; Rabbani, S.I.; Alamri, A.M.; Salem Alshammari, S.K.; Duwaidi, B.S.; Alshammari, M.S.; Alamri, A.S.; Alsanie, W.F.; Alhomrani, M.; et al. Insight into the Advances in Clinical Trials of SARS-CoV-2 Vaccines. Can. J. Infect. Dis. Med. Microbiol. 2022, 2022, 6913772. [Google Scholar] [CrossRef]
- Zieneldien, T.; Kim, J.; Sawmiller, D.; Cao, C. The Immune System as a Therapeutic Target for Alzheimer’s Disease. Life 2022, 12, 1440. [Google Scholar] [CrossRef]
- Fosgerau, K.; Hoffmann, T. Peptide therapeutics: Current status and future directions. Drug Discov. Today 2015, 20, 122–128. [Google Scholar] [CrossRef] [Green Version]
- Sharma, K.; Sharma, K.K.; Sharma, A.; Jain, R. Peptide-based drug discovery: Current status and recent advances. Drug Discov. Today 2023, 28, 103464. [Google Scholar] [CrossRef] [PubMed]
- Xie, J.; Bi, Y.; Zhang, H.; Dong, S.; Teng, L.; Lee, R.J.; Yang, Z. Cell-Penetrating Peptides in Diagnosis and Treatment of Human Diseases: From Preclinical Research to Clinical Application. Front. Pharmacol. 2020, 11, 697. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Koh, J.J.; Liu, S.; Lakshminarayanan, R.; Verma, C.S.; Beuerman, R.W. Membrane Active Antimicrobial Peptides: Translating Mechanistic Insights to Design. Front. Neurosci. 2017, 11, 73. [Google Scholar] [CrossRef] [Green Version]
- Kmeck, A.; Tancer, R.J.; Ventura, C.R.; Wiedman, G.R. Synergies with and Resistance to Membrane-Active Peptides. Antibiotics 2020, 9, 620. [Google Scholar] [CrossRef] [PubMed]
- De Moraes, L.F.R.N.; Silva, P.S.e.; Pereira, T.C.P.L.; Almeida Rodrigues, T.A.; Farias Frihling, B.E.; da Costa, R.A.; Torquato, H.F.V.; Lima, C.S.; Paredes-Gamero, E.J.; Migliolo, L. First generation of multifunctional peptides derived from latarcin-3a from Lachesana tarabaevi spider toxin. Front. Microbiol. 2022, 13, 965621. [Google Scholar] [CrossRef]


| Membrane-Active Peptides | Peptide Sequence | Aminoacids | Net Charge (at pH = 7) | Hydrophobicity (%) | Peptide Origin |
|---|---|---|---|---|---|
| AMPs | |||||
| α-Defensin | DCYCRIPACIAGERRYGTCIYQGRLWAFCC | 30 | 1.73 | 23 | Human |
| BuCATHL4C (CAT) | RIRFPWPWRWPWWRRVRG | 18 | 5 | 33 | Buffalo |
| Indolicin | ILPWKWPWWPWRR-NH2 | 13 | 4 | 23 | Cow |
| Lactoferricin B | KCRRWQWRMKKLGAPSITCV | 20 | 5.91 | 40 | Cow milk |
| Piscidin 3 | FFHHIFRGIVHVGKTIHRLVTG | 22 | 3.36 | 14 | Fish |
| Parasin | KGRGKQGKVRAKAKTRSS | 18 | 8 | 61 | Fish |
| Drosocin | GKPRPTSPRPTSHPRPIRV-NH2 | 19 | 6.09 | 37 | Fly |
| Magainin 1 | GIGKFLHSAGKFGKAFVGEIMKS | 23 | 3.09 | 30 | Frog |
| Histatin 5 | DSHAKRHHGYKRKFHEKHHSHRGY | 24 | 5.63 | 46 | Human |
| Cathelicidin | HLLGDFFRKSKEKIGKEFKRIVQRIKDFLRNLVPRTES | 37 | 6 | 54 | Humans and other mammals |
| Protegrin-1 | RGGRLC1YC2RRRFC2VC1VGR-NH2 | 18 | 6.82 | 33 | Pig |
| PR-39 | RRRPRPPYLPRPRPPPFFPPRLPPRIPPGFPPRFPPRFP | 39 | 10 | 26 | Pig |
| Cecropin B | KWKVFKKIEKMGRNIRNGIVKAGPAIAVLGEAKAL-NH2 | 35 | 8 | 37 | The giant silkmoth, Hyalophora cecropia |
| CPPs | |||||
| TP10 | AGYLLGKINLKALAALAKKIL | 21 | 4 | 24 | Analogue of transportan |
| Penetratin 1 | RQIKIWFQNRRMKWKK-NH2 | 16 | 8 | 63 | Fly |
| HIV-1 Tat | YGRKKRRQRRR | 11 | 8 | 82 | HIV infected cells |
| Bactenecin (Bac7) | RRIRPRPPRLPRPRPRPLPFPRPG | 24 | 9 | 38 | Mammals |
| Maurocalcine | GDCLPHLKLCKENKDCCSKKCKRRGTNIEKRCR | 33 | 6.82 | 55 | Scorpion |
| Transportan | GWTLNSAGYLLGKINLKALAALAKKIL-NH2 | 27 | 5 | 26 | Scorpion venom |
| Nona-arginine (R9) | RRRRRRRRR | 9 | 9 | 100 | Synthetic |
| R6/W3 | RRWWRRWRR | 9 | 6 | 67 | Synthetic |
| CyLoP-1 | CRWRWKCCKK | 10 | 4.86 | 50 | Synthetic |
| CB5005 | KLKLALALALAVQRKRQKLMP | 21 | 6 | 38 | Synthetic |
| Mastoparan | INLKALAALAKKIL-NH2 | 14 | 4 | 29 | Wasp venom |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gostaviceanu, A.; Gavrilaş, S.; Copolovici, L.; Copolovici, D.M. Membrane-Active Peptides and Their Potential Biomedical Application. Pharmaceutics 2023, 15, 2091. https://doi.org/10.3390/pharmaceutics15082091
Gostaviceanu A, Gavrilaş S, Copolovici L, Copolovici DM. Membrane-Active Peptides and Their Potential Biomedical Application. Pharmaceutics. 2023; 15(8):2091. https://doi.org/10.3390/pharmaceutics15082091
Chicago/Turabian StyleGostaviceanu, Andreea, Simona Gavrilaş, Lucian Copolovici, and Dana Maria Copolovici. 2023. "Membrane-Active Peptides and Their Potential Biomedical Application" Pharmaceutics 15, no. 8: 2091. https://doi.org/10.3390/pharmaceutics15082091