Recent Advances and Innovations in the Preparation and Purification of In Vitro-Transcribed-mRNA-Based Molecules
Abstract
:1. Introduction
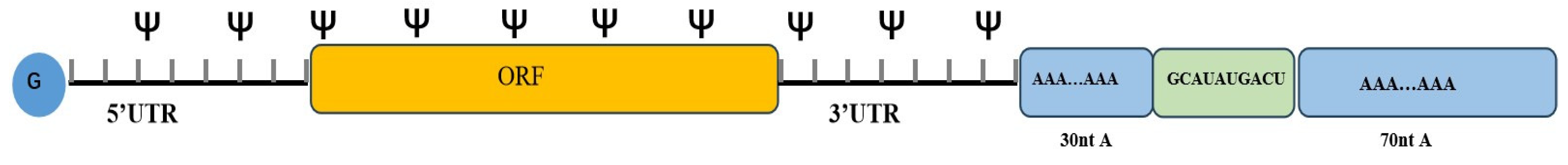
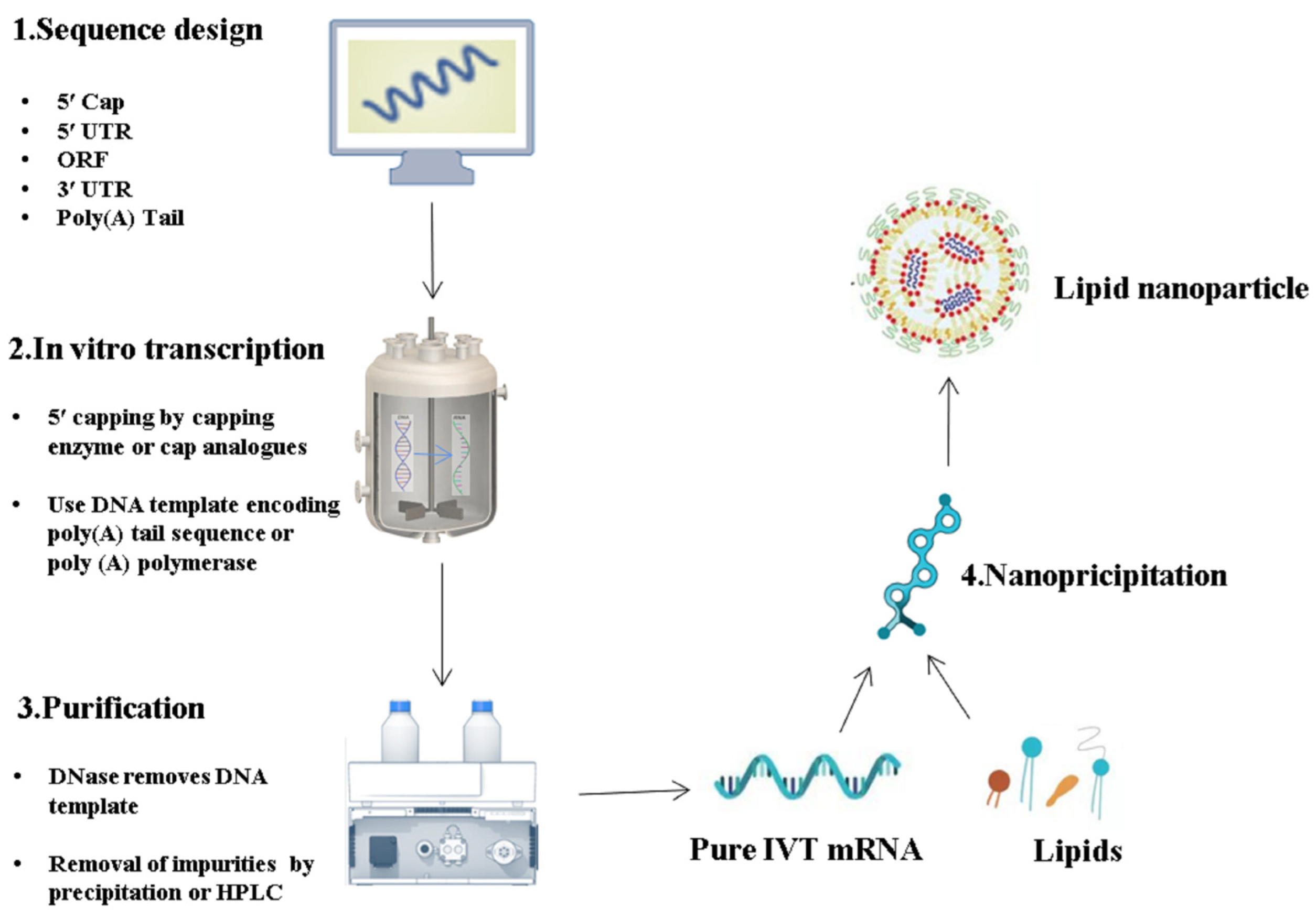
2. Latest Optimization Strategies for IVT mRNA Sequence Design
2.1. ORF
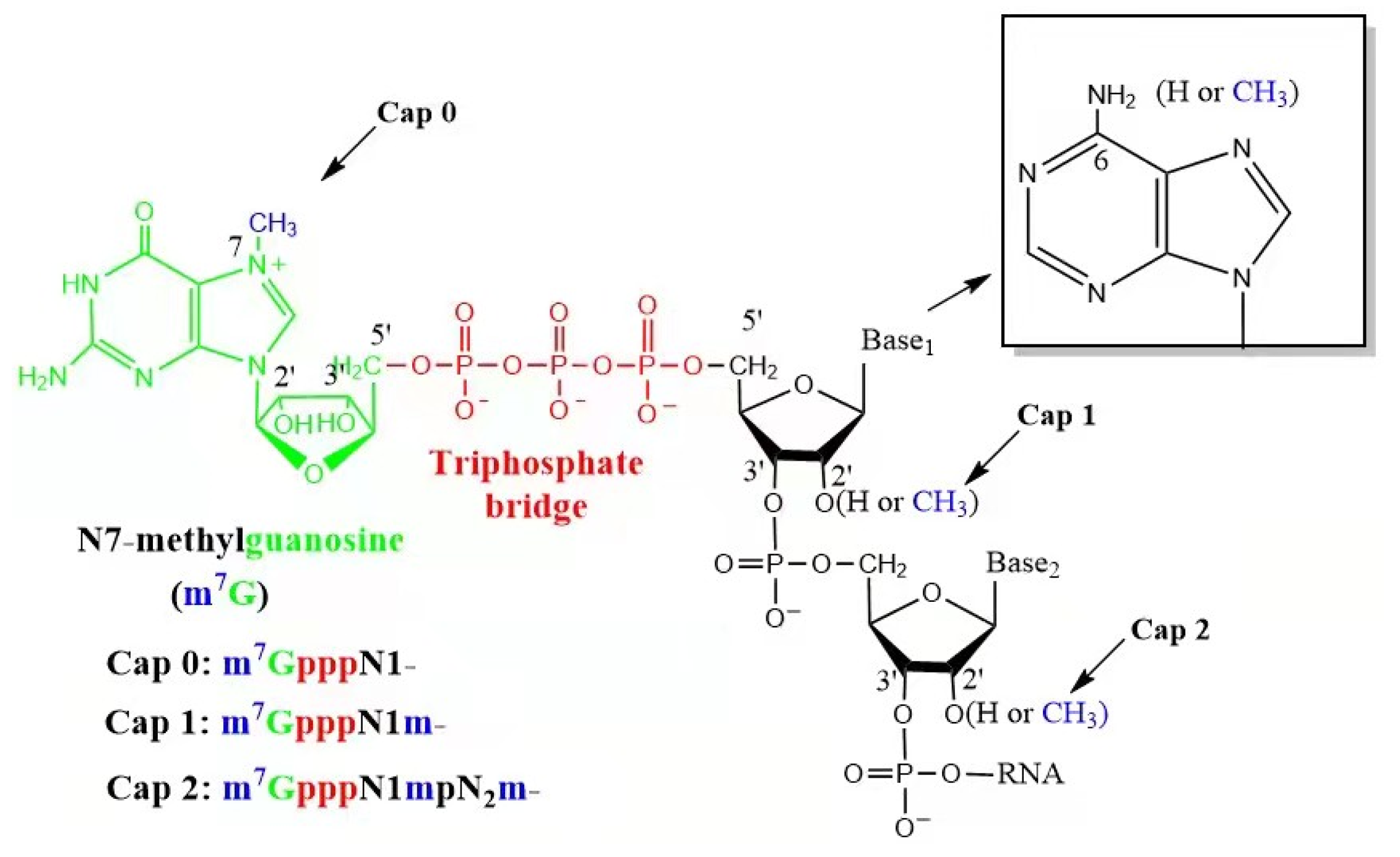
2.2. 5′ Cap
2.3. Poly(A) Tail
2.4. UTR
3. IVT mRNA Purification
3.1. The Importance of IVT mRNA Purifications
3.2. Precipitation Methods
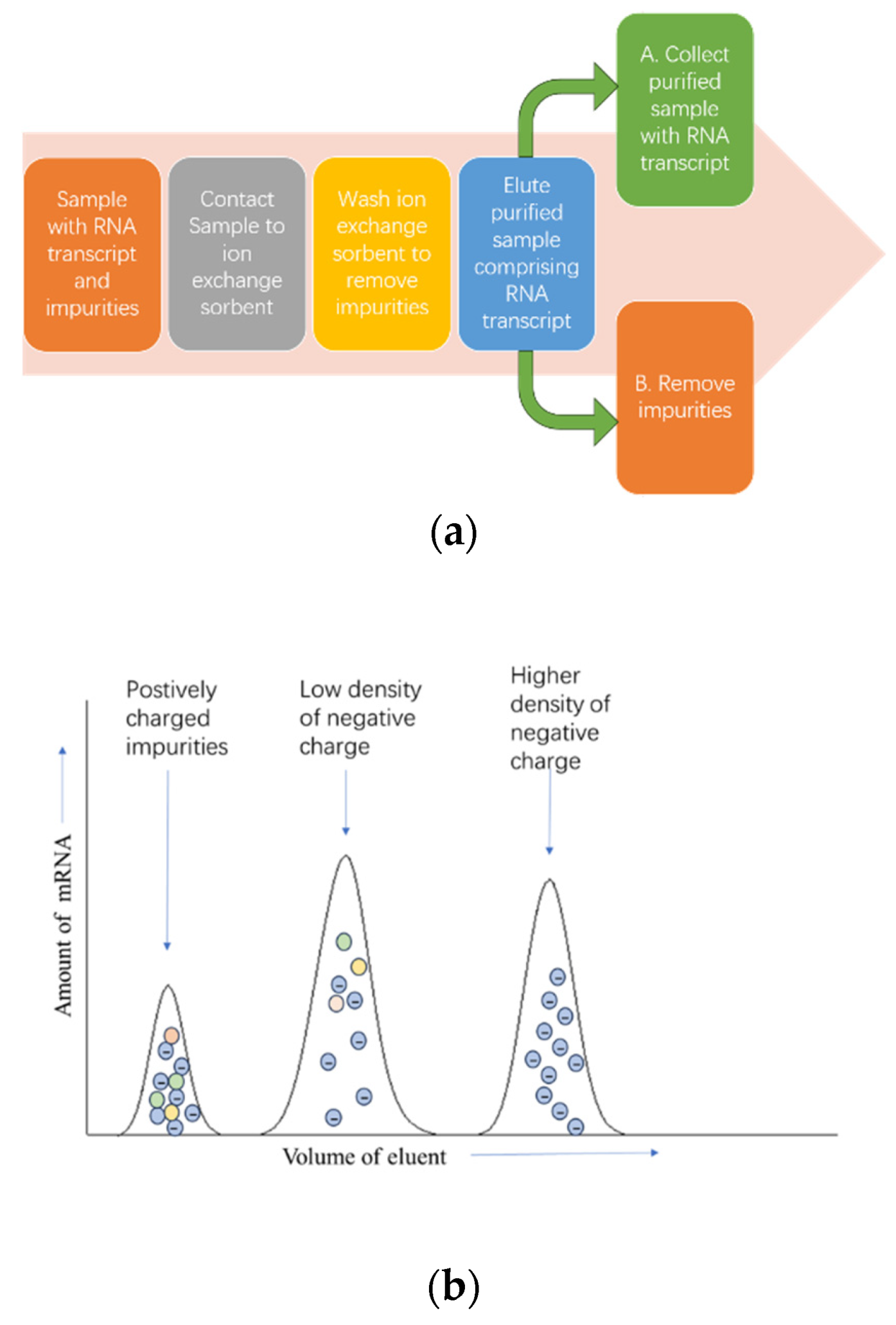
3.3. Chromatography Purification Methods
3.4. Non-Chromatography Purification Method
| Methods | Advantages | Disadvantages | |
|---|---|---|---|
| Precipitation method | Precipitation method | easy to operate | form large particles; abnormal mRNA; cationic impurities |
| Non-chromatography purification method | RNase III | effectively remove dsRNA | harm for the secondary structure of mRNA; increases the cost of purification process |
| Lower concentration of Mg2+ | reduce the dsRNA generation | affects the overall yield of the IVT process | |
| Add dispersant into the transcription system | controls the content of dsRNA | / | |
| TFF | fast and efficient | / | |
| Chromatography | SEC | simple | removes unreacted nucleotides, enzymes, short abortion transcripts, and high-molecular-weight DNA templates; time-consuming; difficult to remove impurities of similar size |
| IEC | scalable and cost-effective | / | |
| RP-HPLC | effectively removes dsRNA | toxic organic solvents; may not be conducive to maintaining the stability and biological activity of the target mRNA; loading capacity of column is limited | |
| Affinity HPLC | simple and reliable | low binding capacities and a less cost-effective process | |
| Cellulose chromatography | for large-scale production of IVT mRNA | unclear whether this method can distinguish the inherent secondary structure of dsRNA and mRNA | |
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Wolff, J.A.; Malone, R.W.; Williams, P.; Chong, W.; Acsadi, G.; Jani, A.; Felgner, P.L. Direct gene transfer into mouse muscle in vivo. Science 1990, 247, 1465–1468. [Google Scholar] [CrossRef] [PubMed]
- Polack, F.P.; Thomas, S.J.; Kitchin, N.; Absalon, J.; Gurtman, A.; Lockhart, S.; Perez, J.L.; Perez Marc, G.; Moreira, E.D.; Zerbini, C.; et al. Safety and Efficacy of the BNT162b2 mRNA COVID-19 Vaccine. N. Engl. J. Med. 2020, 383, 2603–2615. [Google Scholar] [CrossRef] [PubMed]
- Baden, L.R.; El Sahly, H.M.; Essink, B.; Kotloff, K.; Frey, S.; Novak, R.; Diemert, D.; Spector, S.A.; Rouphael, N.; Creech, C.B.; et al. Efficacy and Safety of the mRNA-1273 SARS-CoV-2 Vaccine. N. Engl. J. Med. 2021, 384, 403–416. [Google Scholar] [CrossRef] [PubMed]
- Zhu, N.; Zhang, D.; Wang, W.; Li, X.; Yang, B.; Song, J.; Zhao, X.; Huang, B.; Shi, W.; Lu, R.; et al. A Novel Coronavirus from Patients with Pneumonia in China, 2019. N. Engl. J. Med. 2020, 382, 727–733. [Google Scholar] [CrossRef] [PubMed]
- Roest, S.; Hoek, R.A.S.; Manintveld, O.C. BNT162b2 mRNA COVID-19 Vaccine in a Nationwide Mass Vaccination Setting. N. Engl. J. Med. 2021, 384, 1968–1970. [Google Scholar] [CrossRef]
- Thompson, M.G.; Burgess, J.L.; Naleway, A.L.; Tyner, H.L.; Yoon, S.K.; Meece, J.; Olsho, L.E.W.; Caban-Martinez, A.J.; Fowlkes, A.; Lutrick, K.; et al. Interim Estimates of Vaccine Effectiveness of BNT162b2 and mRNA-1273 COVID-19 Vaccines in Preventing SARS-CoV-2 Infection among Health Care Personnel, First Responders, and Other Essential and Frontline Workers—Eight U.S. Locations, December 2020–March 2021. MMWR Morb. Mortal. Wkly. Rep. 2021, 70, 495–500. [Google Scholar] [CrossRef]
- Chaudhary, N.; Weissman, D.; Whitehead, K.A. mRNA vaccines for infectious diseases: Principles, delivery and clinical translation. Nat. Rev. Drug Discov. 2021, 20, 817–838. [Google Scholar] [CrossRef]
- Jackson, L.A.; Anderson, E.J.; Rouphael, N.G.; Roberts, P.C.; Makhene, M.; Coler, R.N.; McCullough, M.P.; Chappell, J.D.; Denison, M.R.; Stevens, L.J.; et al. An mRNA Vaccine against SARS-CoV-2—Preliminary Report. N. Engl. J. Med. 2020, 383, 1920–1931. [Google Scholar] [CrossRef]
- Qin, S.; Tang, X.; Chen, Y.; Chen, K.; Fan, N.; Xiao, W.; Zheng, Q.; Li, G.; Teng, Y.; Wu, M.; et al. mRNA-based therapeutics: Powerful and versatile tools to combat diseases. Signal Transduct. Target. Ther. 2022, 7, 166. [Google Scholar] [CrossRef]
- Pardi, N.; Hogan, M.J.; Porter, F.W.; Weissman, D. mRNA vaccines—A new era in vaccinology. Nat. Rev. Drug Discov. 2018, 17, 261–279. [Google Scholar] [CrossRef]
- Arraiano, C.M.; Andrade, J.M.; Domingues, S.; Guinote, I.B.; Malecki, M.; Matos, R.G.; Moreira, R.N.; Pobre, V.; Reis, F.P.; Saramago, M.; et al. The critical role of RNA processing and degradation in the control of gene expression. FEMS Microbiol. Rev. 2010, 34, 883–923. [Google Scholar] [CrossRef] [PubMed]
- Youn, H.; Chung, J.K. Modified mRNA as an alternative to plasmid DNA (pDNA) for transcript replacement and vaccination therapy. Expert Opin. Biol. Ther. 2015, 15, 1337–1348. [Google Scholar] [CrossRef] [PubMed]
- Kwon, S.; Kwon, M.; Im, S.; Lee, K.; Lee, H. mRNA vaccines: The most recent clinical applications of synthetic mRNA. Arch. Pharm. Res. 2022, 45, 245–262. [Google Scholar] [CrossRef]
- Kis, Z.; Shattock, R.; Shah, N.; Kontoravdi, C. Emerging Technologies for Low-Cost, Rapid Vaccine Manufacture. Biotechnol. J. 2019, 14, 1800376. [Google Scholar] [CrossRef] [PubMed]
- Corbett, K.S.; Edwards, D.K.; Leist, S.R.; Abiona, O.M.; Boyoglu-Barnum, S.; Gillespie, R.A.; Himansu, S.; Schafer, A.; Ziwawo, C.T.; DiPiazza, A.T.; et al. SARS-CoV-2 mRNA vaccine design enabled by prototype pathogen preparedness. Nature 2020, 586, 567–571. [Google Scholar] [CrossRef]
- Crommelin, D.J.A.; Anchordoquy, T.J.; Volkin, D.B.; Jiskoot, W.; Mastrobattista, E. Addressing the Cold Reality of mRNA Vaccine Stability. J. Pharm. Sci. 2021, 110, 997–1001. [Google Scholar] [CrossRef]
- Zeng, C.; Zhang, C.; Walker, P.G.; Dong, Y. Formulation and Delivery Technologies for mRNA Vaccines. In mRNA Vaccines; Part of the Current Topics in Microbiology and Immunology Book Series; Springer International Publishing: Cham, Switzerland, 2020. [Google Scholar] [CrossRef]
- Hornung, V.; Barchet, W.; Schlee, M.; Hartmann, G. RNA recognition via TLR7 and TLR8. In Toll-like Receptors (TLRs) and Innate Immunity; Part of the Handbook of Experimental Pharmacology Book Series; Springer: Berlin/Heidelberg, Germany, 2008; pp. 71–86. [Google Scholar] [CrossRef]
- Kariko, K.; Muramatsu, H.; Ludwig, J.; Weissman, D. Generating the optimal mRNA for therapy: HPLC purification eliminates immune activation and improves translation of nucleoside-modified, protein-encoding mRNA. Nucleic Acids Res. 2011, 39, e142. [Google Scholar] [CrossRef]
- Akash, M.S.H.; Rehman, K. (Eds.) Column Chromatography. In Essentials of Pharmaceutical Analysis; Springer: Singapore, 2020; pp. 167–174. ISBN 978-981-15-1547-7. [Google Scholar]
- Hussain, A.; Yang, H.; Zhang, M.; Liu, Q.; Alotaibi, G.; Irfan, M.; He, H.; Chang, J.; Liang, X.J.; Weng, Y.; et al. mRNA vaccines for COVID-19 and diverse diseases. J. Control Release 2022, 345, 314–333. [Google Scholar] [CrossRef]
- Pardi, N.; Hogan, M.J.; Weissman, D. Recent advances in mRNA vaccine technology. Curr. Opin. Immunol. 2020, 65, 14–20. [Google Scholar] [CrossRef]
- Naik, R.; Peden, K. Regulatory Considerations on the Development of mRNA Vaccines. In mRNA Vaccines; Part of the Current Topics in Microbiology and Immunology Book Series; Springer International Publishing: Cham, Switzerland, 2020. [Google Scholar] [CrossRef]
- Rice, A.M.; Castillo Morales, A.; Ho, A.T.; Mordstein, C.; Muhlhausen, S.; Watson, S.; Cano, L.; Young, B.; Kudla, G.; Hurst, L.D. Evidence for Strong Mutation Bias toward, and Selection against, U Content in SARS-CoV-2: Implications for Vaccine Design. Mol. Biol. Evol. 2021, 38, 67–83. [Google Scholar] [CrossRef]
- Sullenger, B.A.; Nair, S. From the RNA world to the clinic. Science 2016, 352, 1417–1420. [Google Scholar] [CrossRef]
- Cannarozzi, G.; Schraudolph, N.N.; Faty, M.; von Rohr, P.; Friberg, M.T.; Roth, A.C.; Gonnet, P.; Gonnet, G.; Barral, Y. A role for codon order in translation dynamics. Cell 2010, 141, 355–367. [Google Scholar] [CrossRef] [PubMed]
- Shabalina, S.A.; Spiridonov, N.A.; Kashina, A. Sounds of silence: Synonymous nucleotides as a key to biological regulation and complexity. Nucleic Acids Res. 2013, 41, 2073–2094. [Google Scholar] [CrossRef] [PubMed]
- Hanson, G.; Coller, J. Codon optimality, bias and usage in translation and mRNA decay. Nat. Rev. Mol. Cell Biol. 2018, 19, 20–30. [Google Scholar] [CrossRef] [PubMed]
- Spencer, P.S.; Siller, E.; Anderson, J.F.; Barral, J.M. Silent substitutions predictably alter translation elongation rates and protein folding efficiencies. J. Mol. Biol. 2012, 422, 328–335. [Google Scholar] [CrossRef]
- Vaidyanathan, S.; Azizian, K.T.; Haque, A.; Henderson, J.M.; Hendel, A.; Shore, S.; Antony, J.S.; Hogrefe, R.I.; Kormann, M.S.D.; Porteus, M.H.; et al. Uridine Depletion and Chemical Modification Increase Cas9 mRNA Activity and Reduce Immunogenicity without HPLC Purification. Mol. Ther. Nucleic Acids 2018, 12, 530–542. [Google Scholar] [CrossRef]
- Buschmann, M.D.; Carrasco, M.J.; Alishetty, S.; Paige, M.; Alameh, M.G.; Weissman, D. Nanomaterial Delivery Systems for mRNA Vaccines. Vaccines 2021, 9, 65. [Google Scholar] [CrossRef]
- Kwon, H.; Kim, M.; Seo, Y.; Moon, Y.S.; Lee, H.J.; Lee, K.; Lee, H. Emergence of synthetic mRNA: In vitro synthesis of mRNA and its applications in regenerative medicine. Biomaterials 2018, 156, 172–193. [Google Scholar] [CrossRef]
- To, K.K.W.; Cho, W.C.S. An overview of rational design of mRNA-based therapeutics and vaccines. Expert Opin. Drug Discov. 2021, 16, 1307–1317. [Google Scholar] [CrossRef]
- Drazkowska, K.; Tomecki, R.; Warminski, M.; Baran, N.; Cysewski, D.; Depaix, A.; Kasprzyk, R.; Kowalska, J.; Jemielity, J.; Sikorski, P.J. 2’-O-Methylation of the second transcribed nucleotide within the mRNA 5’ cap impacts the protein production level in a cell-specific manner and contributes to RNA immune evasion. Nucleic Acids Res. 2022, 50, 9051–9071. [Google Scholar] [CrossRef]
- Sikorski, P.J.; Warminski, M.; Kubacka, D.; Ratajczak, T.; Nowis, D.; Kowalska, J.; Jemielity, J. The identity and methylation status of the first transcribed nucleotide in eukaryotic mRNA 5’ cap modulates protein expression in living cells. Nucleic Acids Res. 2020, 48, 1607–1626. [Google Scholar] [CrossRef] [PubMed]
- Mauer, J.; Luo, X.; Blanjoie, A.; Jiao, X.; Grozhik, A.V.; Patil, D.P.; Linder, B.; Pickering, B.F.; Vasseur, J.J.; Chen, Q.; et al. Reversible methylation of m6Am in the 5’ cap controls mRNA stability. Nature 2017, 541, 371–375. [Google Scholar] [CrossRef] [PubMed]
- Linares-Fernandez, S.; Lacroix, C.; Exposito, J.Y.; Verrier, B. Tailoring mRNA Vaccine to Balance Innate/Adaptive Immune Response. Trends Mol. Med. 2020, 26, 311–323. [Google Scholar] [CrossRef] [PubMed]
- Sahin, U.; Muik, A.; Vogler, I.; Derhovanessian, E.; Kranz, L.M.; Vormehr, M.; Quandt, J.; Bidmon, N.; Ulges, A.; Baum, A.; et al. BNT162b2 vaccine induces neutralizing antibodies and poly-specific T cells in humans. Nature 2021, 595, 572–577. [Google Scholar] [CrossRef]
- Fang, E.; Liu, X.; Li, M.; Zhang, Z.; Song, L.; Zhu, B.; Wu, X.; Liu, J.; Zhao, D.; Li, Y. Advances in COVID-19 mRNA vaccine development. Signal Transduct. Target. Ther. 2022, 7, 94. [Google Scholar] [CrossRef]
- Urbina, F.; Morales-Pison, S.; Maldonado, E. Enzymatic Protein Biopolymers as a Tool to Synthetize Eukaryotic Messenger Ribonucleic Acid (mRNA) with Uses in Vaccination, Immunotherapy and Nanotechnology. Polymers 2020, 12, 1633. [Google Scholar] [CrossRef]
- Henderson, J.M.; Ujita, A.; Hill, E.; Yousif-Rosales, S.; Smith, C.; Ko, N.; McReynolds, T.; Cabral, C.R.; Escamilla-Powers, J.R.; Houston, M.E. Cap 1 Messenger RNA Synthesis with Co-transcriptional CleanCap® Analog by In Vitro Transcription. Curr. Protoc. 2021, 1, e39. [Google Scholar] [CrossRef]
- Inagaki, M.; Abe, N.; Li, Z.; Nakashima, Y.; Acharyya, S.; Ogawa, K.; Kawaguchi, D.; Hiraoka, H.; Banno, A.; Meng, Z.; et al. Cap analogs with a hydrophobic photocleavable tag enable facile purification of fully capped mRNA with various cap structures. Nat. Commun. 2023, 14, 2657. [Google Scholar] [CrossRef]
- Kis, Z.; Kontoravdi, C.; Shattock, R.; Shah, N. Resources, Production Scales and Time Required for Producing RNA Vaccines for the Global Pandemic Demand. Vaccines 2021, 9, 3, Erratum in Vaccines 2021, 9, 205. [Google Scholar] [CrossRef]
- Pichlmair, A.; Schulz, O.; Tan, C.P.; Naslund, T.I.; Liljestrom, P.; Weber, F.; Reis e Sousa, C. RIG-I-mediated antiviral responses to single-stranded RNA bearing 5’-phosphates. Science 2006, 314, 997–1001. [Google Scholar] [CrossRef]
- Whitelaw, E.; Coates, A.; Proudfoot, N.J. Globin gene transcripts can utilize histone gene 3’ end processing signals. Nucleic Acids Res. 1986, 14, 7059–7070. [Google Scholar] [CrossRef] [PubMed]
- Holtkamp, S.; Kreiter, S.; Selmi, A.; Simon, P.; Koslowski, M.; Huber, C.; Tureci, O.; Sahin, U. Modification of antigen-encoding RNA increases stability, translational efficacy, and T-cell stimulatory capacity of dendritic cells. Blood 2006, 108, 4009–4017. [Google Scholar] [CrossRef] [PubMed]
- Goss, D.J.; Kleiman, F.E. Poly(A) binding proteins: Are they all created equal? Wiley Interdiscip. Rev. RNA 2013, 4, 167–179. [Google Scholar] [CrossRef]
- Yu, S.; Kim, V.N. A tale of non-canonical tails: Gene regulation by post-transcriptional RNA tailing. Nat. Rev. Mol. Cell Biol. 2020, 21, 542–556. [Google Scholar] [CrossRef] [PubMed]
- Tang, T.T.L.; Passmore, L.A. Recognition of Poly(A) RNA through Its Intrinsic Helical Structure. Cold Spring Harb. Symp. Quant. Biol. 2019, 84, 21–30. [Google Scholar] [CrossRef] [PubMed]
- Kormann, M.S.; Hasenpusch, G.; Aneja, M.K.; Nica, G.; Flemmer, A.W.; Herber-Jonat, S.; Huppmann, M.; Mays, L.E.; Illenyi, M.; Schams, A.; et al. Expression of therapeutic proteins after delivery of chemically modified mRNA in mice. Nat. Biotechnol. 2011, 29, 154–157. [Google Scholar] [CrossRef] [PubMed]
- Grier, A.E.; Burleigh, S.; Sahni, J.; Clough, C.A.; Cardot, V.; Choe, D.C.; Krutein, M.C.; Rawlings, D.J.; Jensen, M.C.; Scharenberg, A.M.; et al. pEVL: A Linear Plasmid for Generating mRNA IVT Templates with Extended Encoded Poly(A) Sequences. Mol. Ther. Nucleic Acids 2016, 5, e306. [Google Scholar] [CrossRef]
- Lima, S.A.; Chipman, L.B.; Nicholson, A.L.; Chen, Y.H.; Yee, B.A.; Yeo, G.W.; Coller, J.; Pasquinelli, A.E. Short poly(A) tails are a conserved feature of highly expressed genes. Nat. Struct. Mol. Biol. 2017, 24, 1057–1063. [Google Scholar] [CrossRef]
- Weissman, D. mRNA transcript therapy. Expert Rev. Vaccines 2015, 14, 265–281. [Google Scholar] [CrossRef]
- Trepotec, Z.; Geiger, J.; Plank, C.; Aneja, M.K.; Rudolph, C. Segmented poly(A) tails significantly reduce recombination of plasmid DNA without affecting mRNA translation efficiency or half-life. RNA 2019, 25, 507–518. [Google Scholar] [CrossRef]
- Preiss, T.; Muckenthaler, M.; Hentze, M.W.J.R. Poly(A)-tail-promoted translation in yeast: Implications for translational control. RNA 1998, 4, 1321–1331. [Google Scholar] [CrossRef]
- Stadler, C.R.; Bahr-Mahmud, H.; Celik, L.; Hebich, B.; Roth, A.S.; Roth, R.P.; Kariko, K.; Tureci, O.; Sahin, U. Elimination of large tumors in mice by mRNA-encoded bispecific antibodies. Nat. Med. 2017, 23, 815–817. [Google Scholar] [CrossRef] [PubMed]
- Xia, X. Detailed Dissection and Critical Evaluation of the Pfizer/BioNTech and Moderna mRNA Vaccines. Vaccines 2021, 9, 734. [Google Scholar] [CrossRef]
- Zarghampoor, F.; Azarpira, N.; Khatami, S.R.; Behzad-Behbahani, A.; Foroughmand, A.M. Improved translation efficiency of therapeutic mRNA. Gene 2019, 707, 231–238. [Google Scholar] [CrossRef] [PubMed]
- Balzer Le, S.; Onsager, I.; Lorentzen, J.A.; Lale, R. Dual UTR-A novel 5’ untranslated region design for synthetic biology applications. Synth. Biol. 2020, 5, ysaa006. [Google Scholar] [CrossRef] [PubMed]
- Leppek, K.; Das, R.; Barna, M. Functional 5’ UTR mRNA structures in eukaryotic translation regulation and how to find them. Nat. Rev. Mol. Cell Biol. 2018, 19, 158–174. [Google Scholar] [CrossRef]
- El Mouali, Y.; Balsalobre, C. 3’untranslated regions: Regulation at the end of the road. Curr. Genet. 2019, 65, 127–131. [Google Scholar] [CrossRef]
- Mayr, C. Regulation by 3’-Untranslated Regions. Annu. Rev. Genet. 2017, 51, 171–194. [Google Scholar] [CrossRef]
- Barreau, C.; Paillard, L.; Osborne, H.B. AU-rich elements and associated factors: Are there unifying principles? Nucleic Acids Res. 2005, 33, 7138–7150. [Google Scholar] [CrossRef]
- Eberhardt, W.; Doller, A.; Akool, E.S.; Pfeilschifter, J. Modulation of mRNA stability as a novel therapeutic approach. Pharmacol. Ther. 2007, 114, 56–73. [Google Scholar] [CrossRef]
- Weng, Y.; Li, C.; Yang, T.; Hu, B.; Zhang, M.; Guo, S.; Xiao, H.; Liang, X.J.; Huang, Y. The challenge and prospect of mRNA therapeutics landscape. Biotechnol. Adv. 2020, 40, 107534. [Google Scholar] [CrossRef] [PubMed]
- Asrani, K.H.; Farelli, J.D.; Stahley, M.R.; Miller, R.L.; Cheng, C.J.; Subramanian, R.R.; Brown, J.M. Optimization of mRNA untranslated regions for improved expression of therapeutic mRNA. RNA Biol. 2018, 15, 756–762. [Google Scholar] [CrossRef]
- Chen, C.Y.; Shyu, A.B. AU-rich elements: Characterization and importance in mRNA degradation. Trends Biochem. Sci. 1995, 20, 465–470. [Google Scholar] [CrossRef] [PubMed]
- Orlandini von Niessen, A.G.; Poleganov, M.A.; Rechner, C.; Plaschke, A.; Kranz, L.M.; Fesser, S.; Diken, M.; Lower, M.; Vallazza, B.; Beissert, T.; et al. Improving mRNA-Based Therapeutic Gene Delivery by Expression-Augmenting 3’ UTRs Identified by Cellular Library Screening. Mol. Ther. 2019, 27, 824–836. [Google Scholar] [CrossRef] [PubMed]
- Sample, P.J.; Wang, B.; Reid, D.W.; Presnyak, V.; McFadyen, I.J.; Morris, D.R.; Seelig, G. Human 5’ UTR design and variant effect prediction from a massively parallel translation assay. Nat. Biotechnol. 2019, 37, 803–809. [Google Scholar] [CrossRef]
- Fukuda, R.; Iwakura, Y.; Ishihama, A. Heterogeneity of RNA polymerase in Escherichia coli. I. A new holoenzyme containing a new sigma factor. J. Mol. Biol. 1974, 83, 353–367. [Google Scholar] [CrossRef]
- Weisman, G.A.; Camden, J.M.; Peterson, T.S.; Ajit, D.; Woods, L.T.; Erb, L. P2 receptors for extracellular nucleotides in the central nervous system: Role of P2X7 and P2Y(2) receptor interactions in neuroinflammation. Mol. Neurobiol. 2012, 46, 96–113. [Google Scholar] [CrossRef]
- Jahn, C.E.; Charkowski, A.O.; Willis, D.K. Evaluation of isolation methods and RNA integrity for bacterial RNA quantitation. J. Microbiol. Methods 2008, 75, 318–324. [Google Scholar] [CrossRef]
- Fleige, S.; Pfaffl, M.W. RNA integrity and the effect on the real-time qRT-PCR performance. Mol. Asp. Med. 2006, 27, 126–139. [Google Scholar] [CrossRef]
- Pereira, M.J.; Behera, V.; Walter, N.G. Nondenaturing purification of co-transcriptionally folded RNA avoids common folding heterogeneity. PLoS ONE 2010, 5, e12953. [Google Scholar] [CrossRef]
- Baiersdorfer, M.; Boros, G.; Muramatsu, H.; Mahiny, A.; Vlatkovic, I.; Sahin, U.; Kariko, K. A Facile Method for the Removal of dsRNA Contaminant from In Vitro-Transcribed mRNA. Mol. Ther. Nucleic Acids 2019, 15, 26–35. [Google Scholar] [CrossRef] [PubMed]
- Vomelova, I.; Vanickova, Z.; Sedo, A. Methods of RNA purification. All ways (should) lead to Rome. Folia Biol. 2009, 55, 243–251. [Google Scholar]
- Green, M.R.; Sambrook, J. Precipitation of RNA with Ethanol. Cold Spring Harb. Protoc. 2020, 2020, 101717. [Google Scholar] [CrossRef] [PubMed]
- Walker, S.E.; Lorsch, J. RNA purification--precipitation methods. Methods Enzymol. 2013, 530, 337–343. [Google Scholar] [CrossRef] [PubMed]
- Lukavsky, P.J.; Puglisi, J.D. Large-scale preparation and purification of polyacrylamide-free RNA oligonucleotides. RNA 2004, 10, 889–893. [Google Scholar] [CrossRef]
- Pardi, N.; Hogan, M.J.; Pelc, R.S.; Muramatsu, H.; Andersen, H.; DeMaso, C.R.; Dowd, K.A.; Sutherland, L.L.; Scearce, R.M.; Parks, R.; et al. Zika virus protection by a single low-dose nucleoside-modified mRNA vaccination. Nature 2017, 543, 248–251. [Google Scholar] [CrossRef]
- Kim, I.; McKenna, S.A.; Viani Puglisi, E.; Puglisi, J.D. Rapid purification of RNAs using fast performance liquid chromatography (FPLC). RNA 2007, 13, 289–294. [Google Scholar] [CrossRef]
- McKenna, S.A.; Kim, I.; Puglisi, E.V.; Lindhout, D.A.; Aitken, C.E.; Marshall, R.A.; Puglisi, J.D. Purification and characterization of transcribed RNAs using gel filtration chromatography. Nat. Protoc. 2007, 2, 3270–3277. [Google Scholar] [CrossRef]
- Tuttolomondo, M.; Hansen, P.L.; Mollenhauer, J.; Ditzel, H.J. One-step FPLC-size-exclusion chromatography procedure for purification of rDMBT1 6 kb with increased biological activity. Anal. Biochem. 2018, 542, 16–19. [Google Scholar] [CrossRef]
- Beck, J.D.; Reidenbach, D.; Salomon, N.; Sahin, U.; Tureci, O.; Vormehr, M.; Kranz, L.M. mRNA therapeutics in cancer immunotherapy. Mol. Cancer 2021, 20, 69. [Google Scholar] [CrossRef]
- Easton, L.E.; Shibata, Y.; Lukavsky, P.J. Rapid, nondenaturing RNA purification using weak anion-exchange fast performance liquid chromatography. RNA 2010, 16, 647–653. [Google Scholar] [CrossRef] [PubMed]
- Wu, M.Z.; Asahara, H.; Tzertzinis, G.; Roy, B. Synthesis of low immunogenicity RNA with high-temperature in vitro transcription. RNA 2020, 26, 345–360. [Google Scholar] [CrossRef] [PubMed]
- Chen, N.; Xia, P.; Li, S.; Zhang, T.; Wang, T.T.; Zhu, J. RNA sensors of the innate immune system and their detection of pathogens. IUBMB Life 2017, 69, 297–304. [Google Scholar] [CrossRef] [PubMed]
- Mu, X.; Greenwald, E.; Ahmad, S.; Hur, S. An origin of the immunogenicity of in vitro transcribed RNA. Nucleic Acids Res. 2018, 46, 5239–5249. [Google Scholar] [CrossRef]
- Nina Mencin, A.K.; Ličen, J.; Peljhan, S.; Vidič, J.; Černigoj, U.; Kostelec, T.; Štrancar, A.; Sekirnik, R. Increasing Dynamic Binding Capacity of Oligo(dT) for mRNA Purification. BioProcess Int. 2022, 20, 44–47. [Google Scholar]
- Baronti, L.; Karlsson, H.; Marusic, M.; Petzold, K. A guide to large-scale RNA sample preparation. Anal. Bioanal. Chem. 2018, 410, 3239–3252. [Google Scholar] [CrossRef]
- Azarani, A.; Hecker, K.H. RNA analysis by ion-pair reversed-phase high performance liquid chromatography. Nucleic Acids Res. 2001, 29, E7. [Google Scholar] [CrossRef]
- Aviv, H.; Leder, P. Purification of biologically active globin messenger RNA by chromatography on oligothymidylic acid-cellulose. Proc. Natl. Acad. Sci. USA 1972, 69, 1408–1412. [Google Scholar] [CrossRef]
- Korenč, M.; Mencin, N.; Puc, J.; Skok, J.; Šprinzar Nemec, K.; Martinčič Celjar, A.; Gagnon, P.; Štrancar, A.; Sekirnik, R. Chromatographic purification with CIMmultus™ Oligo dT increases mRNA stability mRNA. Cell and Gene Ther. Insights 2021, 9, 1207–1216. [Google Scholar] [CrossRef]
- Jacobsen, N.; Nielsen, P.S.; Jeffares, D.C.; Eriksen, J.; Ohlsson, H.; Arctander, P.; Kauppinen, S. Direct isolation of poly(A)+ RNA from 4 M guanidine thiocyanate-lysed cell extracts using locked nucleic acid-oligo(T) capture. Nucleic Acids Res. 2004, 32, e64. [Google Scholar] [CrossRef]
- Green, M.R.; Sambrook, J. Isolation of Poly(A)(+) Messenger RNA Using Magnetic Oligo(dT) Beads. Cold Spring Harb. Protoc. 2019, 711–714. [Google Scholar] [CrossRef]
- Zhong, Z.; McCafferty, S.; Opsomer, L.; Wang, H.; Huysmans, H.; De Temmerman, J.; Lienenklaus, S.; Portela Catani, J.P.; Combes, F.; Sanders, N.N. Corticosteroids and cellulose purification improve, respectively, the in vivo translation and vaccination efficacy of sa-mRNAs. Mol. Ther. 2021, 29, 1370–1381. [Google Scholar] [CrossRef]
- Foster, J.B.; Choudhari, N.; Perazzelli, J.; Storm, J.; Hofmann, T.J.; Jain, P.; Storm, P.B.; Pardi, N.; Weissman, D.; Waanders, A.J.; et al. Purification of mRNA Encoding Chimeric Antigen Receptor Is Critical for Generation of a Robust T-Cell Response. Human Gene Ther. 2019, 30, 168–178. [Google Scholar] [CrossRef]
- Piao, X.; Yadav, V.; Wang, E.; Chang, W.; Tau, L.; Lindenmuth, B.E.; Wang, S.X. Double-stranded RNA reduction by chaotropic agents during in vitro transcription of messenger RNA. Mol. Ther. Nucleic Acids 2022, 29, 618–624. [Google Scholar] [CrossRef] [PubMed]
- Rosa, S.S.; Prazeres, D.M.F.; Azevedo, A.M.; Marques, M.P.C. mRNA vaccines manufacturing: Challenges and bottlenecks. Vaccine 2021, 39, 2190–2200. [Google Scholar] [CrossRef] [PubMed]
- Ouranidis, A.; Davidopoulou, C.; Tashi, R.K.; Kachrimanis, K. Pharma 4.0 Continuous mRNA Drug Products Manufacturing. Pharmaceutics 2021, 13, 1371. [Google Scholar] [CrossRef]
- Zhang, N.N.; Li, X.F.; Deng, Y.Q.; Zhao, H.; Huang, Y.J.; Yang, G.; Huang, W.J.; Gao, P.; Zhou, C.; Zhang, R.R.; et al. A Thermostable mRNA Vaccine against COVID-19. Cell 2020, 182, 1271–1283.e1216. [Google Scholar] [CrossRef]
- Li, C.Y.; Liang, Z.; Hu, Y.; Zhang, H.; Setiasabda, K.D.; Li, J.; Ma, S.; Xia, X.; Kuang, Y. Cytidine-containing tails robustly enhance and prolong protein production of synthetic mRNA in cell and in vivo. Mol. Ther. Nucleic Acids 2022, 30, 300–310. [Google Scholar] [CrossRef]
- Grau, S.; Ferrandez, O.; Martin-Garcia, E.; Maldonado, R. Accidental Interruption of the Cold Chain for the Preservation of the Moderna COVID-19 Vaccine. Vaccines 2021, 9, 512. [Google Scholar] [CrossRef] [PubMed]
- Krienke, C.; Kolb, L.; Diken, E.; Streuber, M.; Kirchhoff, S.; Bukur, T.; Akilli-Ozturk, O.; Kranz, L.M.; Berger, H.; Petschenka, J.; et al. A noninflammatory mRNA vaccine for treatment of experimental autoimmune encephalomyelitis. Science 2021, 371, 145–153. [Google Scholar] [CrossRef]


| Identifier | Target | Sponsor | Name | Route of Administration | Status | Phase |
|---|---|---|---|---|---|---|
| NCT05217641 | HIV (Human Immunodeficiency Virus) | National Institute of Allergy and Infectious Diseases National Institutes of Health Department of Health and Human Services | BG505 MD39.3 BG505 MD39.3 gp151 BG505 MD39.3 gp151 CD4KO | I.M | Active, not recruiting | Ⅰ |
| NCT05398796 | Nipah Virus | National Institute of Allergy and Infectious Diseases Moderna TX, Inc. (Cambridge, MA 02139, USA). National Institutes of Health Clinical Center | mRNA-1215 | I.M | Recruiting | Ⅰ |
| NCT05430958 | Coronavirus | Inovio Pharmaceuticals | INO-4800 INO-9112 | I.M | Withdrawn | Ⅰ |
| NCT05414786 | HIV-1 | International AIDS Vaccine Initiative AURUM Tembisa Clinical Research Center for Family Health Research | mRNA-1644 | I.P | Active, not recruiting | Ⅰ |
| NCT05127434 | Respiratory Syncytial Virus | Moderna TX, Inc. | mRNA-1345 | I.M | Recruiting | Ⅱ/Ⅲ |
| NCT03713086 | Rabies | CureVac | CV7202 | I.M | Completed | Ⅰ |
| NCT05624606 | Influenza Immunization | Sanofi Pasteur | MRT5410 | I.M | Not yet recruiting | Ⅰ/Ⅱ |
| NCT05553301 | Influenza Immunization | Sanofi Pasteur | MRT5407 | I.M | Recruiting | Ⅰ/Ⅱ |
| NCT05105048 | Cytomegalovirus | Moderna TX, Inc. | mRNA-1647 | I.M | Recruiting | Ⅰ |
| NCT05085366 | Cytomegalovirus | Moderna TX, Inc. | mRNA-1647 | I.M | Recruiting | Ⅲ |
| NCT04232280 | Cytomegalovirus | Moderna TX, Inc. | mRNA-1647 | I.M | Active, not recruiting | Ⅱ |
| NCT03382405 | Cytomegalovirus | Moderna TX, Inc. | mRNA-1647/ mRNA-1443 | I.M | Completed | Ⅰ |
| NCT05164094 | Epstein–Barr Virus | Moderna TX, Inc. | mRNA-1189 | I.M | Recruiting | Ⅰ |
| NCT03392389 | Human Metapneumovirus and Human Parainfluenza | Moderna TX, Inc. | mRNA-1653 | I.M | Completed | Ⅰ |
| NCT05581641 | Malaria | BioNTech SE | BNT165b1 | I.M | Not yet recruiting | Ⅰ |
| NCT04917861 | Zika Virus | Moderna TX, Inc. | mRNA-1893 | I.M | Active, not recruiting | Ⅱ |
| NCT04064905 | Zika Virus | Moderna TX, Inc. Biomedical Advanced Research and Development Authority | mRNA-1893 | I.M | Completed | Ⅰ |
| NCT03014089 | Zika Virus | Moderna TX, Inc. Biomedical Advanced Research and Development Authority | mRNA-1325 | I.M | Completed | Ⅰ |
| NCT05566639 | Seasonal Influenza | Moderna TX, Inc. | mRNA-1010 | I.M | Recruiting | Ⅲ |
| NCT05537038 | Tuberculosis | BioNTech SE | BNT164a1/BNT164b1 | I.M | Not yet recruiting | Ⅰ |
| NCT02888756 | HIV | Rob Gruters Institut d’Investigacions Biomèdiques August Pi i Sunyer IrsiCaixa | iHIVARNA-01 Tri Mix | I.M | Terminated Has Results | Ⅱ |
| NCT05547464 | Tuberculosis | BioNTech SE | BNT164a1/BNT614b1 | I.M | Not yet recruiting | Ⅰ |
| NCT05415462 | Seasonal Influenza | Moderna TX, Inc. | mRNA-1010 | I.M | Active, not recruiting | Ⅲ |
| NCT04956575 | Seasonal Influenza | Moderna TX, Inc. | mRNA-1010 | I.M | Completed | Ⅰ/Ⅱ |
| NCT05333289 | Seasonal Influenza | Moderna TX, Inc. | mRNA-1030/mRNA-102/mRNA-1010 | I.M | Active, not recruiting | Ⅰ/Ⅱ |
| NCT02241135 | Rabies | CureVac | CV7201 | I.M | Completed | Ⅰ |
| NCT05606965 | Influenza | Moderna TX, Inc. | mRNA-1010 | I.M | Recruiting | Ⅱ |
| NCT05252338 | Seasonal Influenza | CureVac GlaxoSmithKline | CVSQIV | I.M | Recruiting | Ⅰ |
| NCT03345043 | Influenza A(H7N9) | Moderna TX, Inc. | mRNA-1851 | I.M | Completed | Ⅰ |
| NCT03076385 | Influenza A(H10N8) | Moderna TX, Inc. | mRNA-1440 | I.M | Completed | Ⅱ |
| NCT05220975 | RSV | Moderna TX, Inc. | mRNA-1345 | I.M | Recruiting | Ⅲ |
| NCT04144348 | hMPV/PIV3 | Moderna TX, Inc. | mRNA-1653 | I.M | Recruiting | Ⅲ |
| NCT04062669 | Rabies | GlaxoSmithKline | GSK3903133A | I.M | Active, not recruiting | Ⅰ |
| Enzymatic Capping | CleanCap | |
|---|---|---|
| Enzymes | RNA polymerase; Capping enzyme | RNA polymerase |
| Reaction steps | Transcription and Capping | a one-pot synthesis |
| Purification steps | 2 | 1 |
| Other materials | / | a cap analogue |
| T7 promoter | TAATACGACTCACTATAGGG | TAATACGACTCACTATAAGA |
| Same materials | DNA template; Magnesium-containing buffer; RNase inhibitor, ATP/GTP/CTP/UTP; Inorganic pyrophosphatase | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, J.; Liu, Y.; Li, C.; Xiao, Q.; Zhang, D.; Chen, Y.; Rosenecker, J.; Ding, X.; Guan, S. Recent Advances and Innovations in the Preparation and Purification of In Vitro-Transcribed-mRNA-Based Molecules. Pharmaceutics 2023, 15, 2182. https://doi.org/10.3390/pharmaceutics15092182
Zhang J, Liu Y, Li C, Xiao Q, Zhang D, Chen Y, Rosenecker J, Ding X, Guan S. Recent Advances and Innovations in the Preparation and Purification of In Vitro-Transcribed-mRNA-Based Molecules. Pharmaceutics. 2023; 15(9):2182. https://doi.org/10.3390/pharmaceutics15092182
Chicago/Turabian StyleZhang, Jingjing, Yuheng Liu, Chao Li, Qin Xiao, Dandan Zhang, Yang Chen, Joseph Rosenecker, Xiaoyan Ding, and Shan Guan. 2023. "Recent Advances and Innovations in the Preparation and Purification of In Vitro-Transcribed-mRNA-Based Molecules" Pharmaceutics 15, no. 9: 2182. https://doi.org/10.3390/pharmaceutics15092182
APA StyleZhang, J., Liu, Y., Li, C., Xiao, Q., Zhang, D., Chen, Y., Rosenecker, J., Ding, X., & Guan, S. (2023). Recent Advances and Innovations in the Preparation and Purification of In Vitro-Transcribed-mRNA-Based Molecules. Pharmaceutics, 15(9), 2182. https://doi.org/10.3390/pharmaceutics15092182








