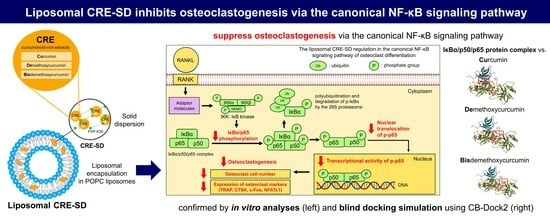
Modified Curcuminoid-Rich Extract Liposomal CRE-SDInhibits Osteoclastogenesis via the Canonical NF-κB Signaling Pathway
Abstract
:1. Introduction
2. Materials and Methods
2.1. In Vitro Experiments
2.1.1. Cells and Reagents
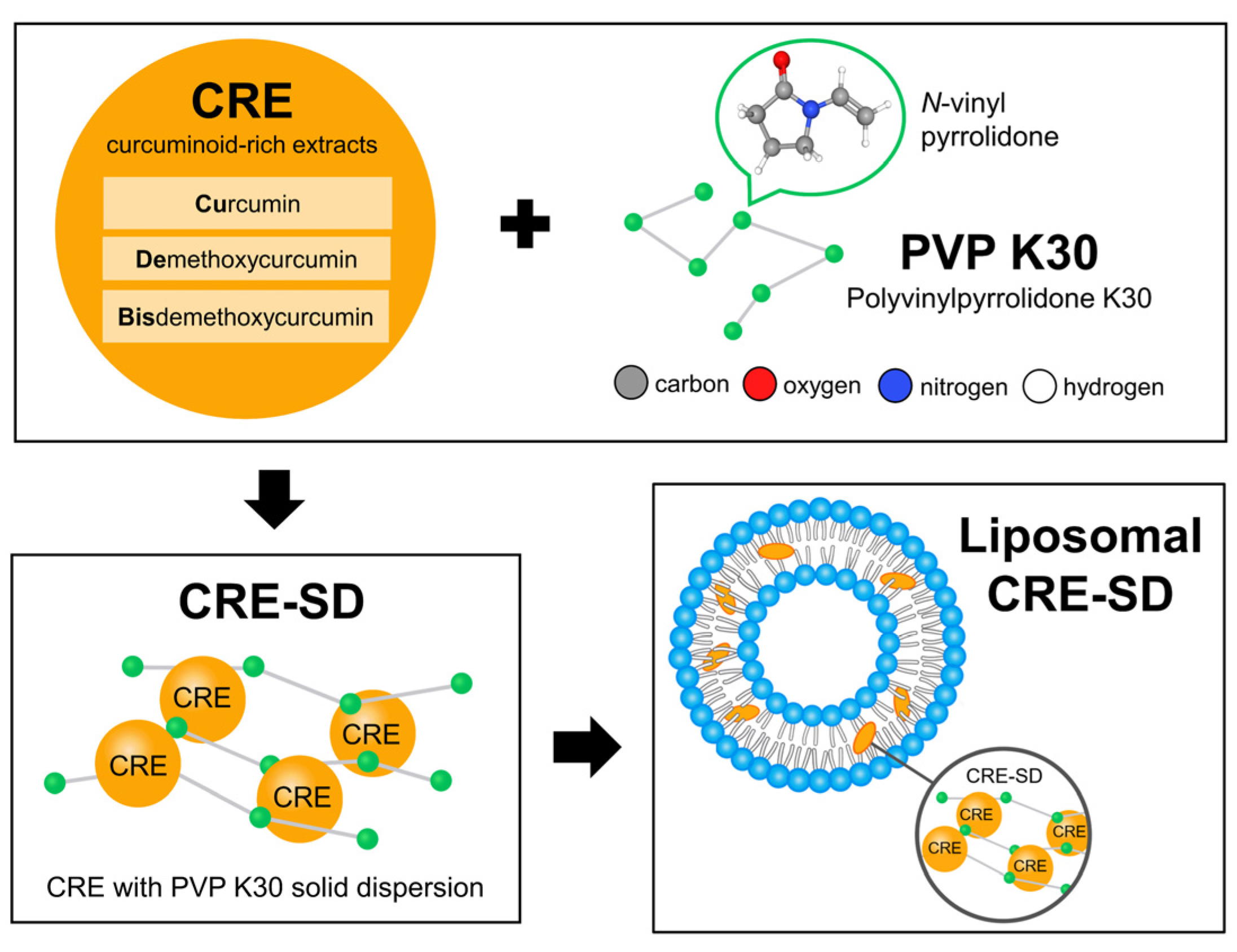
2.1.2. Preparation, Characterization, and In Vitro Release of Liposomal CRE-SD
2.1.3. Culture of RAW 264.7 Cells
2.1.4. Evaluation of Optimal RANKL Incubation Time in RAW 264.7 Cells Treated with CRE-SD-Free POPC Liposomes (CRE-SD-FREE-LIP)
2.1.5. Viability Examination of Liposomal CRE-SD-Treated RAW 264.7 Cells
2.1.6. Confirmation of Osteoclastogenic Inhibition in Liposomal CRE-SD-Treated RANKL-Stimulated RAW 264.7 Cells
2.1.7. Primer Sequences for RT–PCR and qRT–PCR Amplification
| CTSK | Forward: ATGTGGGGGCTCAAGGTTCTG | |
| Reverse: CATATGGGAAAGCATCTTCAGAGTC | ||
| c-Fos | Forward: CCAGTCAAGAGCATCAGCAA | |
| Reverse: AAGTAGTGCAGCCCGGAGTA | ||
| NFATc1 | Forward: CCGTTGCTTCCAGAAAATAACA | |
| Reverse: TGTGGGATGTGAACTCGGAA | ||
| GAPDH (internal control) | ||
| Forward: AAATGGTGAAGGTCGGTGTG | ||
| Reverse: GAATTTGCCGTGAGTGGAGT | ||
2.1.8. Detection of Phosphorylation of p65 and Iκbα Proteins in Liposomal CRE-SD-Treated RANKL-Stimulated RAW 264.7 Cells
2.1.9. Confirmation of the Inhibitory Effect of Liposomal CRE-SD on Nuclear Translocation and Transcriptional Activity of Phosphorylated p65 (p-p65)
2.1.10. Detecting Reactive Oxygen Species (ROS) Production in Liposomal CRE-SD-Treated RANKL-Stimulated RAW 264.7 Cells
2.1.11. Statistical Analysis
2.2. In Silico Analyses
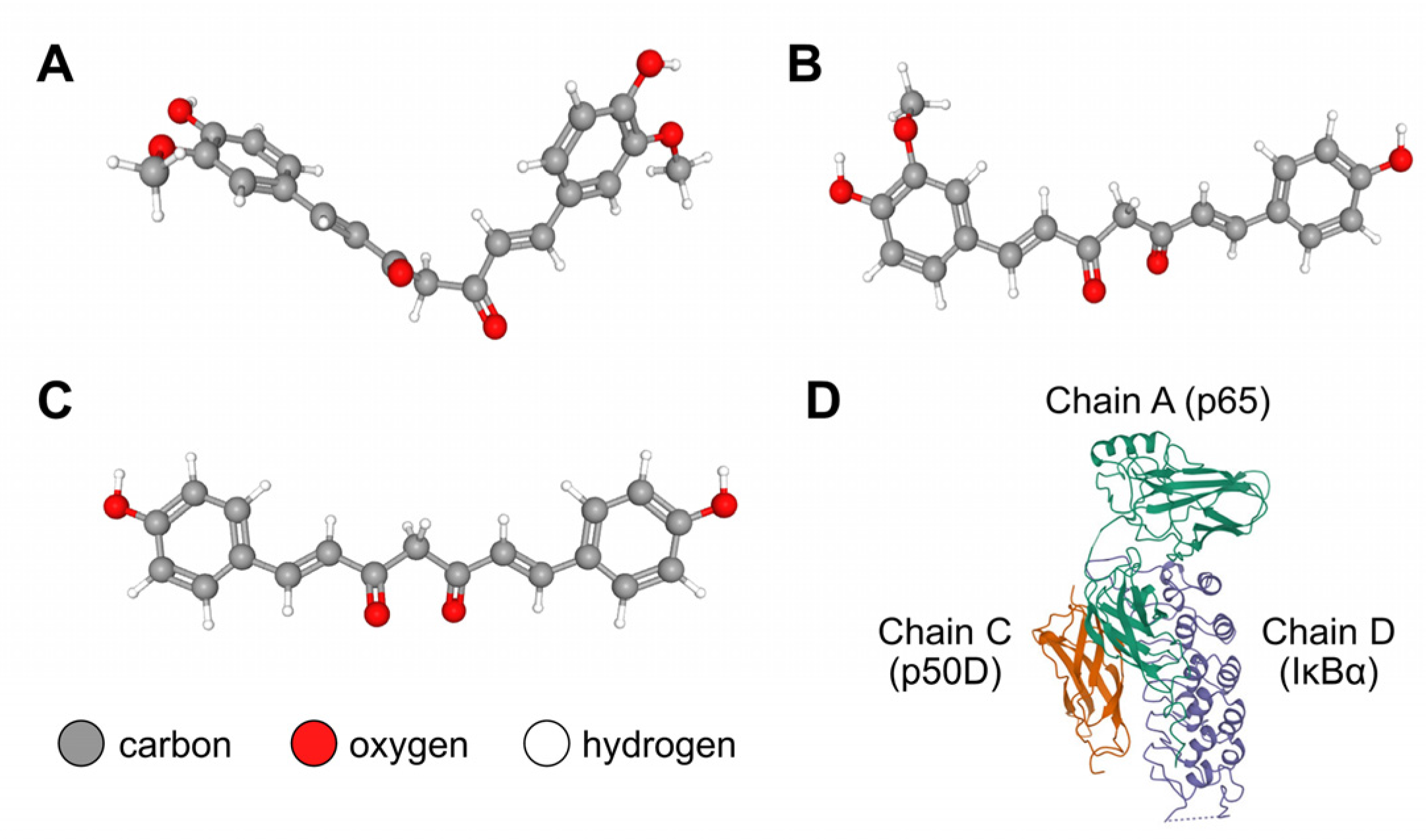
2.2.1. Retrieval of the Structures of the IκBα/p50/p65 Complex and Curcuminoids
2.2.2. Blind Docking Simulation between the IκBα/p50/p65 Complex and Cu/De/Bis
3. Results
3.1. Liposomal CRE-SD Characterization
3.2. In Vitro Assessment of CRE-SD Release
3.3. Optimization of RANKL Incubation Time
3.4. Optimization of Liposomal CRE-SD Concentration
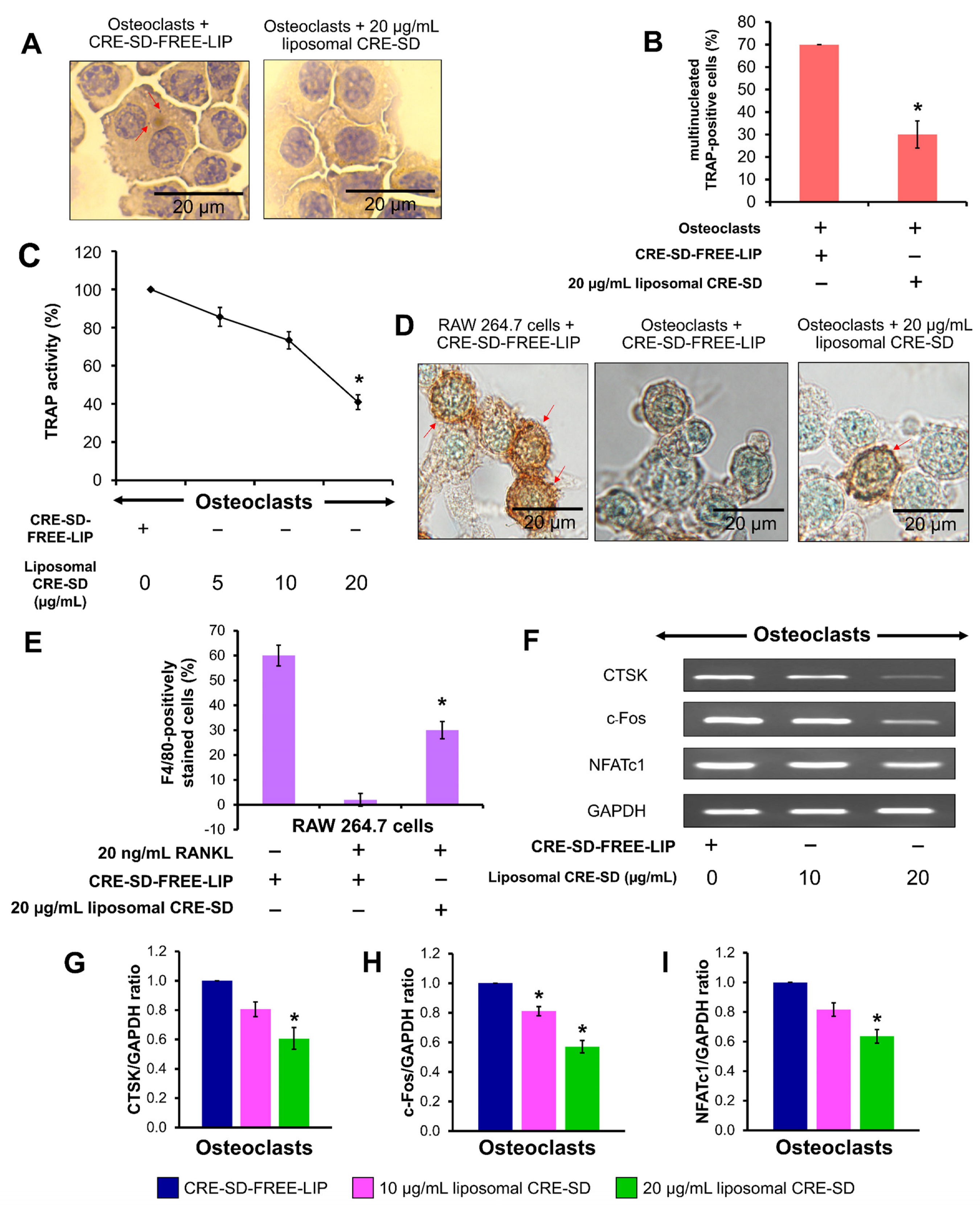
3.5. Inhibitory Effect of Liposomal CRE-SD on Osteoclastogenesis
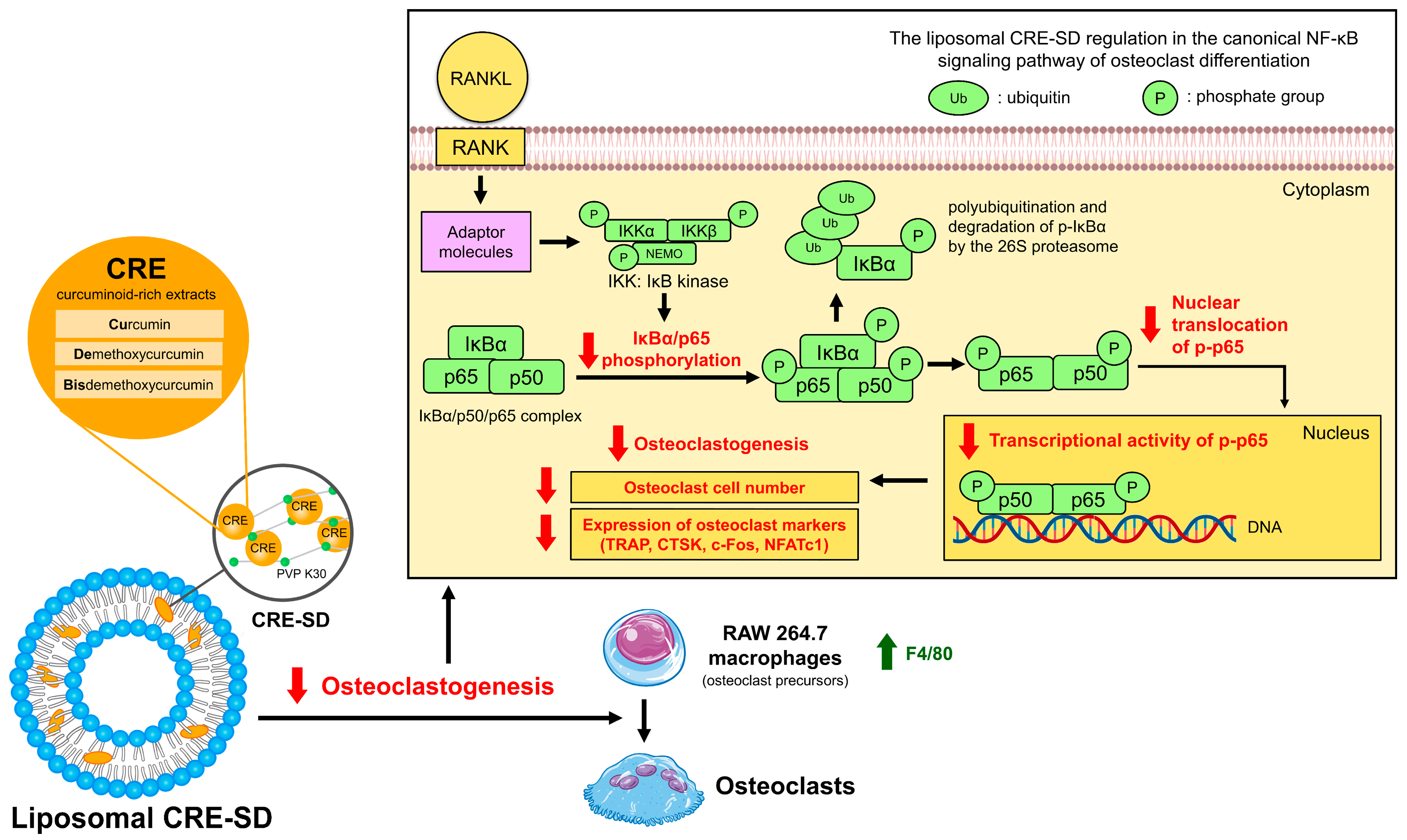
3.6. Suppressive Effect of Liposomal CRE-SD on IκBα/p65 Phosphorylation in Osteoclastogenesis
3.7. Inhibitory Effect of Liposomal CRE-SD on Nuclear Translocation and Transcriptional Activity of p-p65 in Osteoclastogenesis
3.8. Inhibition of Liposomal CRE-SD on Intracellular ROS Production in Osteoclastogenesis
3.9. Blind Docking Simulation between IκBα/p50/p65 Protein Complex and Cu/De/Bis
3.10. The Inhibitory Effect of Liposomal CRE-SD on Osteoclastogenesis via the Canonical NF-κB Signaling Pathway
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Holliday, L.S.; Patel, S.S.; Rody, W.J., Jr. RANKL and RANK in extracellular vesicles: Surprising new players in bone remodeling. Extracell. Vesicles Circ. Nucl. Acids 2021, 2, 18–28. [Google Scholar] [CrossRef] [PubMed]
- Salhotra, A.; Shah, H.N.; Levi, B.; Longaker, M.T. Mechanisms of bone development and repair. Nat. Rev. Mol. Cell Biol. 2020, 21, 696–711. [Google Scholar] [CrossRef] [PubMed]
- Zhao, W.; Huang, Z.; Lin, Y.; Lan, J.; Gao, X. Inhibition effect of zoledronate on the osteoclast differentiation of RAW264.7 induced by titanium particles. BioMed Res. Int. 2021, 2021, 5578088. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Y.; Liu, H.; Li, J.; Ma, Y.; Song, C.; Wang, Y.; Li, P.; Chen, Y.; Zhang, Z. Evaluation of culture conditions for osteoclastogenesis in RAW264.7 cells. PLoS ONE 2022, 17, e0277871. [Google Scholar] [CrossRef]
- Yao, Y.; Cai, X.; Ren, F.; Ye, Y.; Wang, F.; Zheng, C.; Qian, Y.; Zhang, M. The macrophage-osteoclast axis in osteoimmunity and osteo-related diseases. Front. Immunol. 2021, 12, 664871. [Google Scholar] [CrossRef]
- Al-Bari, A.A.; Al Mamun, A. Current advances in regulation of bone homeostasis. FASEB Bioadv. 2020, 2, 668–679. [Google Scholar] [CrossRef] [PubMed]
- Jiménez, J.A.; Lawlor, E.R.; Lyssiotis, C.A. Amino acid metabolism in primary bone sarcomas. Front. Oncol. 2022, 12, 1001318. [Google Scholar] [CrossRef] [PubMed]
- Hauser, B.; Raterman, H.; Ralston, S.H.; Lems, W.F. The effect of anti-rheumatic drugs on the skeleton. Calcif. Tissue Int. 2022, 111, 445–456. [Google Scholar] [CrossRef]
- Lu, J.; Hu, D.; Ma, C.; Shuai, B. Advances in our understanding of the mechanism of action of drugs (including traditional Chinese medicines) for the intervention and treatment of osteoporosis. Front. Pharmacol. 2022, 13, 938447. [Google Scholar] [CrossRef]
- Oh, K.-K.; Adnan, M.; Cho, D.-H. Drug investigation to dampen the comorbidity of rheumatoid arthritis and osteoporosis via molecular docking test. Curr. Issues Mol. Biol. 2022, 44, 1046–1061. [Google Scholar] [CrossRef]
- Xue, S.-T.; Wang, Y.-L.; Han, X.-W.; Yi, H.; Jiang, W.; Si, S.-Y.; Guo, H.-F.; Li, Z.-R. Novel cathepsin K inhibitors block osteoclasts in vitro and increase spinal bone density in zebrafish. RSC Adv. 2019, 9, 8600–8607. [Google Scholar] [CrossRef]
- LeBoff, M.S.; Greenspan, S.L.; Insogna, K.L.; Lewiecki, E.M.; Saag, K.G.; Singer, A.J.; Siris, E.S. The clinician’s guide to prevention and treatment of osteoporosis. Osteoporos. Int. 2022, 33, 2049–2102. [Google Scholar] [CrossRef] [PubMed]
- Amalraj, A.; Pius, A.; Gopi, S.; Gopi, S. Biological activities of curcuminoids, other biomolecules from turmeric and their derivatives—A review. J. Tradit. Complement. Med. 2017, 7, 205–233. [Google Scholar] [CrossRef]
- Hewlings, S.J.; Kalman, D.S. Curcumin: A review of its effects on human health. Foods 2017, 6, 92. [Google Scholar] [CrossRef]
- Fuloria, S.; Mehta, J.; Chandel, A.; Sekar, M.; Rani, N.N.I.M.; Begum, M.Y.; Subramaniyan, V.; Chidambaram, K.; Thangavelu, L.; Nordin, R.; et al. A comprehensive review on the therapeutic potential of Curcuma longa Linn. in relation to its major active constituent curcumin. Front. Pharmacol. 2022, 13, 820806. [Google Scholar] [CrossRef] [PubMed]
- Bresciani, L.; Favari, C.; Calani, L.; Francinelli, V.; Riva, A.; Petrangolini, G.; Allegrini, P.; Mena, P.; Del Rio, D. The effect of formulation of curcuminoids on their metabolism by human colonic microbiota. Molecules 2020, 25, 940. [Google Scholar] [CrossRef] [PubMed]
- Shen, X.; Sun, X.; Chen, H.; Lu, B.; Qin, Y.; Zhang, C.; Liang, G.; Wang, J.; Yu, P.; Su, L.; et al. Demethoxycucumin protects MDA-MB-231 cells induced bone destruction through JNK and ERK pathways inhibition. Cancer Chemother. Pharmacol. 2021, 87, 487–499. [Google Scholar] [CrossRef] [PubMed]
- Wei, J.-X.; Luo, Y.; Xu, Y.; Xiao, J.-H. Osteoinductive activity of bisdemethoxycurcumin and its synergistic protective effect with human amniotic mesenchymal stem cells against ovariectomy-induced osteoporosis mouse model. Biomed. Pharmacother. 2022, 146, 112605. [Google Scholar] [CrossRef] [PubMed]
- Sivani, B.M.; Azzeh, M.; Patnaik, R.; Pantea Stoian, A.; Rizzo, M.; Banerjee, Y. Reconnoitering the therapeutic role of curcumin in disease prevention and treatment: Lessons learnt and future directions. Metabolites 2022, 12, 639. [Google Scholar] [CrossRef]
- Huang, C.; Lu, H.-F.; Chen, Y.-H.; Chen, J.-C.; Chou, W.-H.; Huang, H.-C. Curcumin, demethoxycurcumin, and bisdemethoxycurcumin induced caspase-dependent and –independent apoptosis via Smad or Akt signaling pathways in HOS cells. BMC Complement. Med. Ther. 2020, 20, 68. [Google Scholar] [CrossRef]
- Urošević, M.; Nikolić, L.; Gajić, I.; Nikolić, V.; Dinić, A.; Miljković, V. Curcumin: Biological activities and modern pharmaceutical forms. Antibiotics 2022, 11, 135. [Google Scholar] [CrossRef]
- Racz, L.Z.; Racz, C.P.; Pop, L.-C.; Tomoaia, G.; Mocanu, A.; Barbu, I.; Sárközi, M.; Roman, I.; Avram, A.; Tomoaia-Cotisel, M.; et al. Strategies for improving bioavailability, bioactivity, and physical-chemical behavior of curcumin. Molecules 2022, 27, 6854. [Google Scholar] [CrossRef]
- Song, J.G.; Noh, H.-M.; Lee, S.H.; Han, H.-K. Lipid/clay-based solid dispersion formulation for improving the oral bioavailability of curcumin. Pharmaceutics 2022, 14, 2269. [Google Scholar] [CrossRef] [PubMed]
- Song, J.-W.; Liu, Y.-S.; Guo, Y.-R.; Zhong, W.-X.; Guo, Y.-P.; Guo, L. Nano-liposomes double loaded with curcumin and tetrandrine: Preparation, characterization, hepatotoxicity and anti-tumor effects. Int. J. Mol. Sci. 2022, 23, 6858. [Google Scholar] [CrossRef] [PubMed]
- Capece, D.; Verzella, D.; Flati, I.; Arboretto, P.; Cornice, J.; Franzoso, G. NF-κB: Blending metabolism, immunity, and inflammation. Trends Immunol. 2022, 43, 757–775. [Google Scholar] [CrossRef] [PubMed]
- Liu, P.; Li, Y.; Wang, W.; Bai, Y.; Jia, H.; Yuan, Z.; Yang, Z. Role and mechanisms of the NF-ĸB signaling pathway in various developmental processes. Biomed. Pharmacother. 2022, 153, 113513. [Google Scholar] [CrossRef]
- Florio, T.J.; Lokareddy, R.K.; Yeggoni, D.P.; Sankhala, R.S.; Ott, C.A.; Gillilan, R.E.; Cingolani, G. Differential recognition of canonical NF-κB dimers by Importin α3. Nat. Commun. 2022, 13, 1207. [Google Scholar] [CrossRef]
- Jimi, E.; Katagiri, T. Critical roles of NF-κB signaling molecules in bone metabolism revealed by genetic mutations in osteopetrosis. Int. J. Mol. Sci. 2022, 23, 7995. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Lenardo, M.J.; Baltimore, D. 30 Years of NF-κB: A blossoming of relevance to human pathobiology. Cell 2017, 168, 37–57. [Google Scholar] [CrossRef]
- Yu, H.; Lin, L.; Zhang, Z.; Zhang, H.; Hu, H. Targeting NF-κB pathway for the therapy of diseases: Mechanism and clinical study. Signal Transduct. Target. Ther. 2020, 5, 209. [Google Scholar] [CrossRef] [PubMed]
- Chauhan, A.; Islam, A.U.; Prakash, H.; Singh, S. Phytochemicals targeting NF-κB signaling: Potential anti-cancer interventions. J. Pharm. Anal. 2022, 12, 394–405. [Google Scholar] [CrossRef] [PubMed]
- Bharti, A.C.; Takada, Y.; Aggarwal, B.B. Curcumin (diferuloylmethane) inhibits receptor activator of NF-κB ligand-induced NF-κB activation in osteoclast precursors and suppresses osteoclastogenesis. J. Immunol. 2004, 172, 5940–5947. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.; Zhu, K.; Yuan, X.; Zhang, X.; Qian, Y.; Cheng, T. Curcumin has immunomodulatory effects on RANKL-stimulated osteoclastogenesis in vitro and titanium nanoparticle-induced bone loss in vivo. J. Cell. Mol. Med. 2020, 24, 1553–1567. [Google Scholar] [CrossRef]
- Cheemanapalli, S.; Chinthakunta, N.; Shaikh, N.M.; Shivaranjani, V.; Pamuru, R.R.; Chitta, S.K. Comparative binding studies of curcumin and tangeretin on up-stream elements of NF-kB cascade: A combined molecular docking approach. Netw. Model. Anal. Health Inform. Bioinform. 2019, 8, 15. [Google Scholar] [CrossRef]
- Saeed, M.E.M.; Yücer, R.; Dawood, M.; Hegazy, M.-E.F.; Drif, A.; Ooko, E.; Kadioglu, O.; Seo, E.-J.; Kamounah, F.S.; Titinchi, S.J.; et al. In silico and in vitro screening of 50 curcumin compounds as EGFR and NF-κB inhibitors. Int. J. Mol. Sci. 2022, 23, 3966. [Google Scholar] [CrossRef] [PubMed]
- Lateh, L.; Yuenyongsawad, S.; Chen, H.; Panichayupakaranant, P. A green method for preparation of curcuminoid-rich Curcuma longa extract and evaluation of its anticancer activity. Pharmacogn. Mag. 2019, 15, 730–735. [Google Scholar] [CrossRef]
- Lateh, L.; Kaewnopparat, N.; Panichayupakaranant, P. A method for increasing the water-solubility of curcuminoids and its products. Petty Patent 2018, 14639. [Google Scholar]
- Sinjari, B.; Pizzicannella, J.; D’Aurora, M.; Zappacosta, R.; Gatta, V.; Fontana, A.; Trubiani, O.; Diomede, F. Curcumin/liposome nanotechnology as delivery platform for anti-inflammatory activities via NFkB/ERK/pERK pathway in human dental pulp treated with 2-hydroxyethyl methacrylate (HEMA). Front. Physiol. 2019, 10, 633. [Google Scholar] [CrossRef]
- Pengjam, Y.; Panichayupakaranant, P.; Tanrattanakul, V. Curcuminoid (CRE-Ter)/liposome as delivery platform for anti-osteoclastogenesis via NF-κB/ERK pathways in RANKL-induced RAW 264.7 cells through PLA foams. Heliyon 2021, 7, e07823. [Google Scholar] [CrossRef]
- Waddell, L.A.; Lefevre, L.; Bush, S.J.; Raper, A.; Young, R.; Lisowski, Z.M.; McCulloch, M.E.B.; Muriuki, C.; Sauter, K.A.; Clark, E.L.; et al. ADGRE1 (EMR1, F4/80) is a rapidly-evolving gene expressed in mammalian monocyte-macrophages. Front. Immunol. 2018, 9, 2246. [Google Scholar] [CrossRef]
- Pengjam, Y.; Prajantasen, T.; Tonwong, N.; Panichayupakaranant, P. Downregulation of miR-21 gene expression by CRE-Ter to modulate osteoclastogenesis: De novo mechanism. Biochem. Biophys. Rep. 2021, 26, 101002. [Google Scholar] [CrossRef]
- Berman, H.M.; Westbrook, J.; Feng, Z.; Gilliland, G.; Bhat, T.N.; Weissig, H.; Shindyalov, I.N.; Bourne, P.E. The Protein Data Bank. Nucleic Acids Res. 2000, 28, 235–242. [Google Scholar] [CrossRef]
- Kim, S.; Chen, J.; Cheng, T.; Gindulyte, A.; He, J.; He, S.; Li, Q.; Shoemaker, B.A.; Thiessen, P.A.; Yu, B.; et al. PubChem 2023 update. Nucleic Acids Res. 2023, 51, D1373–D1380. [Google Scholar] [CrossRef]
- Liu, Y.; Yang, X.; Gan, J.; Chen, S.; Xiao, Z.-X.; Cao, Y. CB-Dock2: Improved protein–ligand blind docking by integrating cavity detection, docking and homologous template fitting. Nucleic Acids Res. 2022, 50, W159–W164. [Google Scholar] [CrossRef]
- Hegde, M.; Girisa, S.; BharathwajChetty, B.; Vishwa, R.; Kunnumakkara, A.B. Curcumin formulations for better bioavailability: What we learned from clinical trials thus far? ACS Omega 2023, 8, 10713–10746. [Google Scholar] [CrossRef] [PubMed]
- Kunnumakkara, A.B.; Hegde, M.; Parama, D.; Girisa, S.; Kumar, A.; Daimary, U.D.; Garodia, P.; Yenisetti, S.C.; Oommen, O.V.; Aggarwal, B.B. Role of turmeric and curcumin in prevention and treatment of chronic diseases: Lessons learned from clinical trials. ACS Pharmacol. Transl. Sci. 2023, 6, 447–518. [Google Scholar] [CrossRef]
- Khanizadeh, F.; Rahmani, A.; Asadollahi, K.; Ahmadi, M.R.H. Combination therapy of curcumin and alendronate modulates bone turnover markers and enhances bone mineral density in postmenopausal women with osteoporosis. Arch. Endocrinol. Metab. 2018, 62, 438–445. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Wang, M.; Wang, Y.; Pan, J.; Sun, C.; Zeng, Z.; Ren, S.; Cui, H.; Zhao, X. Construction and characterization of novel hydrophilic nanospheres loaded with lambda-cyhalothrin via ultrasonic emulsification–solvent evaporation. Int. J. Mol. Sci. 2022, 23, 14063. [Google Scholar] [CrossRef]
- Danaei, M.; Dehghankhold, M.; Ataei, S.; Hasanzadeh Davarani, F.; Javanmard, R.; Dokhani, A.; Khorasani, S.; Mozafari, M.R. Impact of particle size and polydispersity index on the clinical applications of lipidic nanocarrier systems. Pharmaceutics 2018, 10, 57. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Gong, J.; Wei, Z. Strategies for liposome drug delivery systems to improve tumor treatment efficacy. AAPS PharmSciTech 2021, 23, 27. [Google Scholar] [CrossRef] [PubMed]
- Peram, M.R.; Jalalpure, S.S.; Palkar, M.B.; Diwan, P.V. Stability studies of pure and mixture form of curcuminoids by reverse phase-HPLC method under various experimental stress conditions. Food Sci. Biotechnol. 2017, 26, 591–602. [Google Scholar] [CrossRef] [PubMed]
- Takechi-Haraya, Y.; Ohgita, T.; Demizu, Y.; Saito, H.; Izutsu, K.I.; Sakai-Kato, K. Current status and challenges of analytical methods for evaluation of size and surface modification of nanoparticle-based drug formulations. AAPS PharmSciTech 2022, 23, 150. [Google Scholar] [CrossRef] [PubMed]
- Wahyudiono; He, J.; Hu, X.; Machmudah, S.; Yasuda, K.; Takami, S.; Kanda, H.; Goto, M. Curcumin-loaded liposome preparation in ultrasound environment under pressurized carbon dioxide. Foods 2022, 11, 1469. [Google Scholar] [CrossRef]
- Csicsák, D.; Szolláth, R.; Kádár, S.; Ambrus, R.; Bartos, C.; Balogh, E.; Antal, I.; Köteles, I.; Tőzsér, P.; Bárdos, V.; et al. The effect of the particle size reduction on the biorelevant solubility and dissolution of poorly soluble drugs with different acid-base character. Pharmaceutics 2023, 15, 278. [Google Scholar] [CrossRef]
- Lee, M.-K. Liposomes for enhanced bioavailability of water-insoluble drugs: In vivo evidence and recent approaches. Pharmaceutics 2020, 12, 264. [Google Scholar] [CrossRef] [PubMed]
- Paradkar, A.; Ambike, A.A.; Jadhav, B.K.; Mahadik, K.R. Characterization of curcumin–PVP solid dispersion obtained by spray drying. Int. J. Pharm. 2004, 271, 281–286. [Google Scholar] [CrossRef]
- Chhouk, K.; Diono, W.; Kanda, H.; Goto, M. Micronization for enhancement of curcumin dissolution via electrospraying technique. ChemEngineering 2018, 2, 60. [Google Scholar] [CrossRef]
- Machmudah, S.; Winardi, S.; Wahyudiono; Kanda, H.; Goto, M. Formation of fine particles from curcumin/PVP by the supercritical antisolvent process with a coaxial nozzle. ACS Omega 2020, 5, 6705–6714. [Google Scholar] [CrossRef]
- Laouini, A.; Charcosset, C.; Fessi, H.; Holdich, R.G.; Vladisavljević, G.T. Preparation of liposomes: A novel application of microengineered membranes—Investigation of the process parameters and application to the encapsulation of vitamin E. RSC Adv. 2013, 3, 4985–4994. [Google Scholar] [CrossRef]
- Sarkar, N.; Bose, S. Liposome-encapsulated curcumin-loaded 3D printed scaffold for bone tissue engineering. ACS Appl. Mater. Interfaces 2019, 11, 17184–17192. [Google Scholar] [CrossRef]
- Ileri Ercan, N. Understanding interactions of curcumin with lipid bilayers: A coarse-grained molecular dynamics study. J. Chem. Inf. Model. 2019, 59, 4413–4426. [Google Scholar] [CrossRef]
- Yeh, C.-C.; Su, Y.-H.; Lin, Y.-J.; Chen, P.-J.; Shi, C.-S.; Chen, C.-N.; Chang, H.-I. Evaluation of the protective effects of curcuminoid (curcumin and bisdemethoxycurcumin)-loaded liposomes against bone turnover in a cell-based model of osteoarthritis. Drug Des. Dev. Ther. 2015, 9, 2285–2300. [Google Scholar] [CrossRef]
- Gallego-Selles, A.; Galvan-Alvarez, V.; Martinez-Canton, M.; Garcia-Gonzalez, E.; Morales-Alamo, D.; Santana, A.; Gonzalez-Henriquez, J.J.; Dorado, C.; Calbet, J.A.L.; Martin-Rincon, M. Fast regulation of the NF-κB signalling pathway in human skeletal muscle revealed by high-intensity exercise and ischaemia at exhaustion: Role of oxygenation and metabolite accumulation. Redox Biol. 2022, 55, 102398. [Google Scholar] [CrossRef]
- Reis, J.; Ramos, A. In sickness and in health: The oxygen reactive species and the bone. Front. Bioeng. Biotechnol. 2021, 9, 745911. [Google Scholar] [CrossRef] [PubMed]
- Quirós-Fallas, M.I.; Vargas-Huertas, F.; Quesada-Mora, S.; Azofeifa-Cordero, G.; Wilhelm-Romero, K.; Vásquez-Castro, F.; Alvarado-Corella, D.; Sánchez-Kopper, A.; Navarro-Hoyos, M. Polyphenolic HRMS characterization, contents and antioxidant activity of Curcuma longa rhizomes from Costa Rica. Antioxidants 2022, 11, 620. [Google Scholar] [CrossRef] [PubMed]
- Trott, O.; Olson, A.J. AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010, 31, 455–461. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Cao, Y.; Zhang, L. Exploring the computational methods for protein-ligand binding site prediction. Comput. Struct. Biotechnol. J. 2020, 18, 417–426. [Google Scholar] [CrossRef] [PubMed]
- Tian, W.; Chen, C.; Lei, X.; Zhao, J.; Liang, J. CASTp 3.0: Computed atlas of surface topography of proteins. Nucleic Acids Res. 2018, 46, W363–W367. [Google Scholar] [CrossRef]
- Riedlinger, T.; Liefke, R.; Meier-Soelch, J.; Jurida, L.; Nist, A.; Stiewe, T.; Kracht, M.; Schmitz, M.L. NF-κB p65 dimerization and DNA-binding is important for inflammatory gene expression. FASEB J. 2019, 33, 4188–4202. [Google Scholar] [CrossRef]






| Liposome Suspension | Liposome Diameter (nm) | Polydispersity Index (PDI) | Zeta Potential at pH 7.4 (mV) |
|---|---|---|---|
| CRE-SD-FREE-LIP | 328.0 ± 14.5 | 0.45 ± 0.11 | −22.22 ± 0.51 |
| Liposomal CRE-SD | 380.1 ± 20.5 | 0.43 ± 0.13 | −31.00 ± 0.48 |
| Binding Pocket | Cavity Volume (Å3) | Center (x, y, z) | Cavity Size (x, y, z) |
|---|---|---|---|
| 1 | 5316 | 21.2, 32.5, 4.8 | 30, 30, 30 |
| 2 | 2633 | 49.8, 32.5, 31.0 | 21, 22, 29 |
| 3 | 1341 | 28.0, 30.0, 26.4 | 15, 20, 17 |
| 4 | 1291 | 44.2, 16.7, 44.0 | 21, 15, 24 |
| 5 | 654 | 42.0, 25.7, 49.6 | 18, 16, 14 |
| Ligand | Binding Pocket | Vina Score (kcal/mol) | Cavity Volume (Å3) | Docking Center (x, y, z) | Docking Size (x, y, z) |
|---|---|---|---|---|---|
| Curcumin | 1 | −8.0 | 5316 | 21, 33, 5 | 35, 35, 35 |
| Demethoxycurcumin | 2 | −9.2 | 2633 | 50, 33, 31 | 27, 27, 34 |
| Bisdemethoxycurcumin | 2 | −8.8 | 2633 | 50, 33, 31 | 26, 26, 34 |
| Ligand | Contact Residues |
|---|---|
| Curcumin | Chain C: THR256 ALA257 PRO324 PRO344 PHE345 LEU346 Chain D: GLU138 ARG140 GLY144 HIS173 LYS177 ALA178 THR179 ASN180 TYR181 ASN182 GLY183 THR185 GLN212 PRO214 |
| Demethoxycurcumin | Chain A: GLN29 ARG30 LYS79 HIS181 PRO182 PHE184 VAL219 GLN220 LYS221 GLU222 GLN241 VAL244 HIS245 ARG246 GLN247 Chain D: TYR251 TRP258 MET279 LEU280 PRO281 GLU282 SER283 GLU287 SER288 |
| Bisdemethoxycurcumin | Chain A: LYS221 GLU222 VAL244 HIS245 ARG246 GLN247 Chain C: ASN247 LYS249 VAL251 ASP271 Chain D: TYR248 GLN249 GLY250 TYR251 PRO281 GLU282 SER283 GLU284 GLU287 SER288 TYR289 ASP290 THR291 GLU292 SER293 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jantarawong, S.; Swangphon, P.; Lauterbach, N.; Panichayupakaranant, P.; Pengjam, Y. Modified Curcuminoid-Rich Extract Liposomal CRE-SDInhibits Osteoclastogenesis via the Canonical NF-κB Signaling Pathway. Pharmaceutics 2023, 15, 2248. https://doi.org/10.3390/pharmaceutics15092248
Jantarawong S, Swangphon P, Lauterbach N, Panichayupakaranant P, Pengjam Y. Modified Curcuminoid-Rich Extract Liposomal CRE-SDInhibits Osteoclastogenesis via the Canonical NF-κB Signaling Pathway. Pharmaceutics. 2023; 15(9):2248. https://doi.org/10.3390/pharmaceutics15092248
Chicago/Turabian StyleJantarawong, Sompot, Piyawut Swangphon, Natda Lauterbach, Pharkphoom Panichayupakaranant, and Yutthana Pengjam. 2023. "Modified Curcuminoid-Rich Extract Liposomal CRE-SDInhibits Osteoclastogenesis via the Canonical NF-κB Signaling Pathway" Pharmaceutics 15, no. 9: 2248. https://doi.org/10.3390/pharmaceutics15092248
APA StyleJantarawong, S., Swangphon, P., Lauterbach, N., Panichayupakaranant, P., & Pengjam, Y. (2023). Modified Curcuminoid-Rich Extract Liposomal CRE-SDInhibits Osteoclastogenesis via the Canonical NF-κB Signaling Pathway. Pharmaceutics, 15(9), 2248. https://doi.org/10.3390/pharmaceutics15092248