A Nucleus-Targeting WT1 Antagonistic Peptide Encapsulated in Polymeric Nanomicelles Combats Refractory Chronic Myeloid Leukemia
Abstract
:1. Introduction
2. Materials and Methods
2.1. Chemicals and Reagents
2.2. Chemically Synthesized Peptides
2.3. Cell Lines and Animals
2.4. Immunoblot Assay
2.5. Intracellular Localization of WT1 and WIP2W
2.6. Pull-Down Assay
2.7. Cytotoxicity Assay
2.8. Cell Cycle Analysis
2.9. RNA Sequencing and Differentially Expressed Gene Analysis
2.10. Preparation of M—WIP2W and Empty PEG-PE Micelles
2.11. Characterization of M—WIP2W and PEG-PE Micelles
2.12. Encapsulation Efficiency of Micelles to WIP2W
2.13. Cellular Uptake of WIP2W
2.14. In Vivo Biodistribution of WIP2W and M—WIP2W
2.15. In Vivo Antitumor Efficiency of WIP2W and M—WIP2W
2.16. Statistical Analysis
3. Results
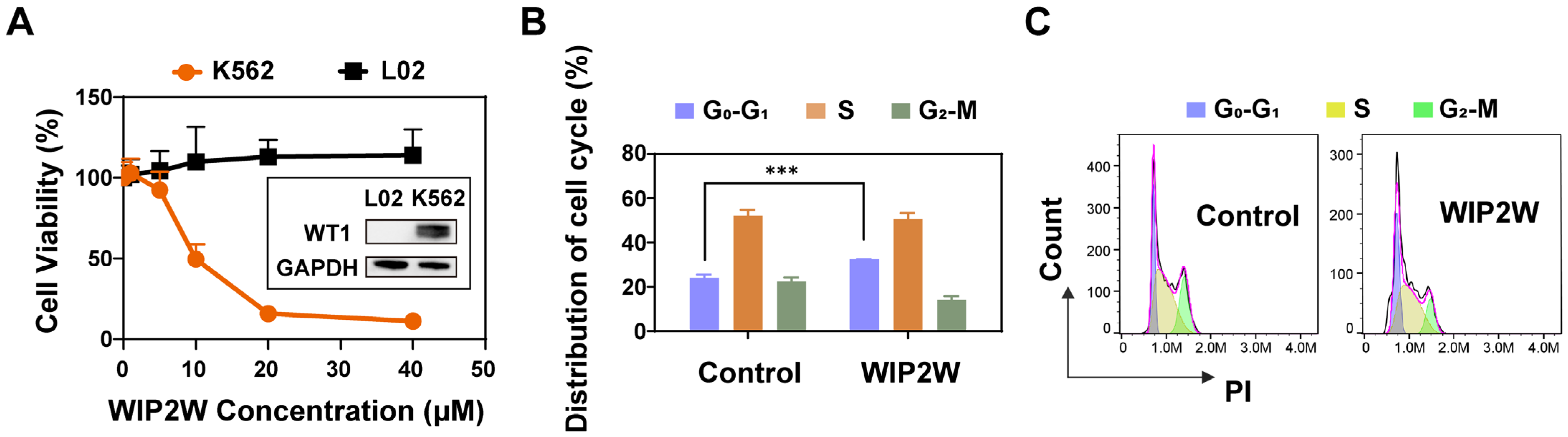
3.1. Synthetic WIP2W Specifically Binds to the WT1 Protein
3.2. Anti-BP-CML Effect of WIP2W In Vitro
3.3. Preparation, Characterization, and Cytotoxicity of M—WIP2W
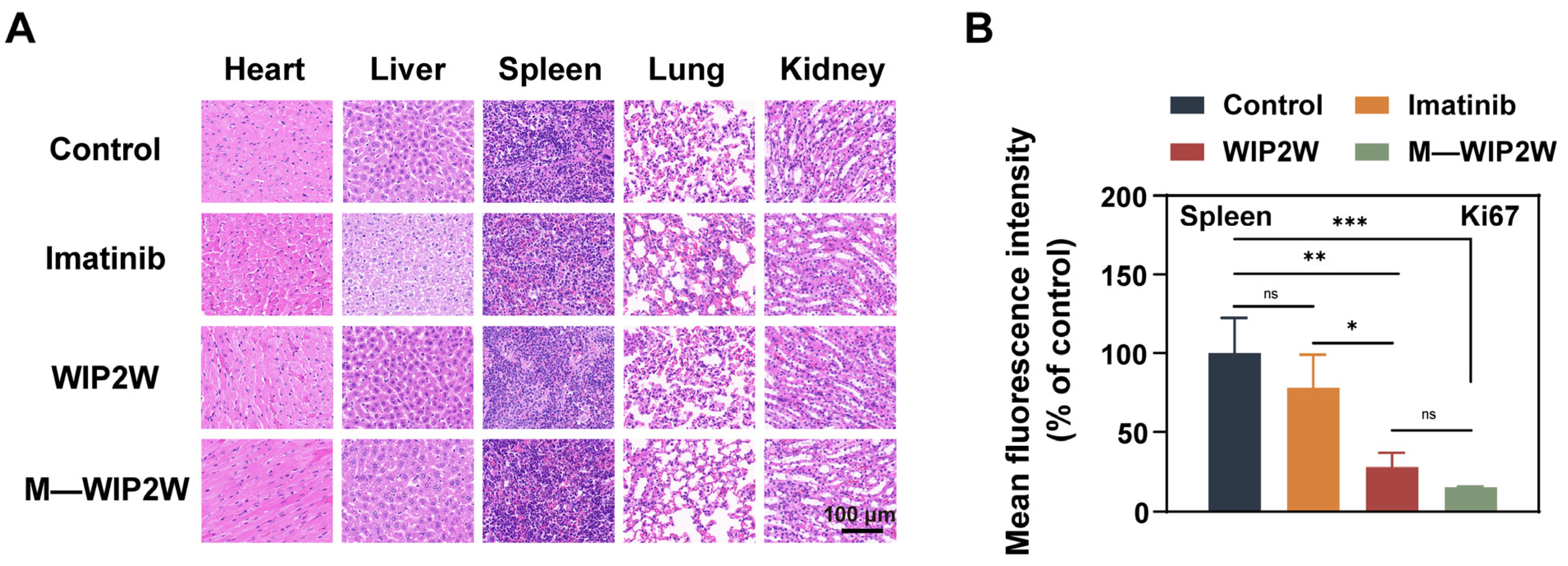
3.4. Anti-BP-CML Effect of WIP2W and M—WIP2W In Vivo
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Jabbour, E.; Kantarjian, H. Chronic myeloid leukemia: 2022 update on diagnosis, therapy, and monitoring. Am. J. Hematol. 2022, 97, 1236–1256. [Google Scholar] [CrossRef] [PubMed]
- Rowley, J.A. A New Consistent Chromosomal Abnormality in Chronic Myelogenous Leukaemia identified by Quinacrine Fluorescence and Giemsa Staining. Nature 1973, 243, 290–293. [Google Scholar] [CrossRef]
- Mandanas, R.A.; Leibowitz, D.S.; Gharehbaghi, K.; Tauchi, T.; Burgess, G.S.; Miyazawa, K.; Jayaram, H.N.; Boswel, H.S. Role of p21 RAS in p210 bcr-abl transformation of murine myeloid cells. Blood 1993, 82, 1838–1847. [Google Scholar] [CrossRef] [PubMed]
- Raitano, A.B.; Halpern, J.R.; Hambuch, T.M.; Sawyers, C.L. The Bcr-Abl leukemia oncogene activates Jun kinase and requires Jun for transformation. Proc. Natl. Acad. Sci. USA 1995, 92, 11746–11750. [Google Scholar] [CrossRef]
- Sawyers, C.L.; Callahan, W.; Witte, O.N. Dominant negative MYC blocks transformation by ABL oncogenes. Cell 1992, 70, 901–910. [Google Scholar] [CrossRef] [PubMed]
- Osman, A.E.G.; Deininger, M.W. Chronic Myeloid Leukemia: Modern therapies, current challenges and future directions. Blood Rev. 2021, 49, 100825. [Google Scholar] [CrossRef] [PubMed]
- Cohen, P. Protein kinases—The major drug targets of the twenty-first century? Nat. Rev. Drug Discov. 2002, 1, 309–315. [Google Scholar] [CrossRef]
- Copland, M.; Hamilton, A.; Elrick, L.J.; Baird, J.W.; Allan, E.K.; Jordanides, N.; Barow, M.; Mountford, J.C.; Holyoake, T.L. Dasatinib (BMS-354825) targets an earlier progenitor population than imatinib in primary CML but does not eliminate the quiescent fraction. Blood 2006, 107, 4532–4539. [Google Scholar] [CrossRef]
- Saglio, G.D.; Kim, D.W.; Issaragrisil, S.; le Coutre, P.; Etienne, G.; Lobo, C.; Pasquini, R.; Clark, R.E.; Hochhaus, A.; Hughes, T.P.; et al. Nilotinib versus imatinib for newly diagnosed chronic myeloid leukemia. N. Engl. J. Med. 2010, 362, 2251–2259. [Google Scholar] [CrossRef]
- Hoy, S.M. Ponatinib: A review of its use in adults with chronic myeloid leukaemia or Philadelphia chromosome-positive acute lymphoblastic leukaemia. Drugs 2014, 74, 793–806. [Google Scholar] [CrossRef]
- Wang, L.; Li, L.; Chen, R.; Huang, X.; Ye, X. Understanding and Monitoring Chronic Myeloid Leukemia Blast Crisis: How to Better Manage Patients. Cancer Manag. Res. 2021, 13, 4987–5000. [Google Scholar] [CrossRef] [PubMed]
- Yamaguchi, H.; Takezako, N.; Ohashi, K.; Oba, K.; Kumagai, T.; Kozai, Y.; Wakita, H.; Yamamoto, K.; Fujita, A.; Igarashi, T.; et al. Treatment-free remission after first-line dasatinib treatment in patients with chronic myeloid leukemia in the chronic phase: The D-NewS Study of the Kanto CML Study Group. Int. J. Hematol. 2020, 111, 401–408. [Google Scholar] [CrossRef] [PubMed]
- Zheng, C.; Li, L.; Haak, M.; Brors, B.; Frank, O.; Giehl, M.; Fabarius, A.; Schatz, M.; Weisser, A.; Lorentz, C.; et al. Gene expression profiling of CD34+ cells identifies a molecular signature of chronic myeloid leukemia blast crisis. Leukemia 2006, 20, 1028–1034. [Google Scholar] [CrossRef]
- Morris, E.L.; Dutcher, J.P. Blastic phase of chronic myelogenous leukemia. Clin. Adv. Hematol. Oncol. 2005, 3, 547–552. [Google Scholar] [PubMed]
- Copland, M. Treatment of blast phase chronic myeloid leukaemia: A rare and challenging entity. Br. J. Haematol. 2022, 199, 665–678. [Google Scholar] [CrossRef] [PubMed]
- Cheever, M.A.; Allison, J.P.; Ferris, A.S.; Finn, O.J.; Hastings, B.M.; Hecht, T.T.; Mellman, I.; Prindiville, S.A.; Viner, J.L.; Weiner, L.M.; et al. The Prioritization of Cancer Antigens: A National Cancer Institute Pilot Project for the Acceleration of Translational Research. Clin. Cancer Res. 2009, 15, 5323–5337. [Google Scholar] [CrossRef] [PubMed]
- Kerst, G.; Bergold, N.; Gieseke, F.; Coustan-Smith, E.; Lang, P.; Kalinova, M.; Handgretinger, R.; Trka, J.; Müller, I. WT1 protein expression in childhood acute leukemia. Am. J. Hematol. 2008, 83, 382–386. [Google Scholar] [CrossRef]
- Lopotová, T.; Polák, J.; Schwarz, J.; Klamová, H.; Moravcová, J. Expression of four major WT1 splicing variants in acute and chronic myeloid leukemia patients analyzed by newly developed four real-time RT PCRs. Blood Cells Mol. Dis. 2012, 49, 41–47. [Google Scholar] [CrossRef]
- Brett, A.; Pandey, S.; Fraizer, G. The Wilms’ tumor gene (WT1) regulates E-cadherin expression and migration of prostate cancer cells. Mol. Cancer 2013, 12, 3. [Google Scholar] [CrossRef]
- Qi, X.W.; Zhang, F.; Wu, H.; Liu, J.L.; Zong, B.G.; Xu, C.; Jiang, J. Wilms’ tumor 1 (WT1) expression and prognosis in solid cancer patients: A systematic review and meta-analysis. Sci. Rep. 2015, 5, 8924. [Google Scholar] [CrossRef]
- McCarty, G.; Awad, O.; Loeb, D.M. WT1 protein directly regulates expression of vascular endothelial growth factor and is a mediator of tumor response to hypoxia. J. Biol. Chem. 2011, 286, 43634–43643. [Google Scholar] [CrossRef] [PubMed]
- Rosenfeld, C.; Cheever, M.A.; Gaiger, A. WT1 in acute leukemia, chronic myelogenous leukemia and myelodysplastic syndrome: Therapeutic potential of WT1 targeted therapies. Leukemia 2003, 17, 1301–1312. [Google Scholar] [CrossRef] [PubMed]
- Inoue, K.; Sugiyama, H.; Ogawa, H.; Nakagawa, M.; Yamagami, T.; Miwa, H.; Kita, K.; Hiraoka, A.; Masaoka, T.; Nasu, K.; et al. WT1 as a New Prognostic Factor and a New Marker for the Detection of Minimal Residual Disease in Acute Leukemia. Blood 1994, 84, 3071–3079. [Google Scholar] [CrossRef] [PubMed]
- Na, I.K.; Kreuzer, K.A.; Lupberger, J.; Dörken, B.; Coutre, P.l. Quantitative RT-PCR of Wilms tumor gene transcripts (WT1) for the molecular monitoring of patients with accelerated phase bcr/abl + CML. Leuk. Res. 2005, 29, 343–345. [Google Scholar] [CrossRef]
- Pei, H.; Guo, W.; Peng, Y.; Xiong, H.; Chen, Y. Targeting key proteins involved intranscriptional regulation for cancer therapy: Current strategies and future prospective. Med. Res. Rev. 2022, 42, 1607–1660. [Google Scholar] [CrossRef]
- Shi, M.; McHugh, K.J. Strategies for overcoming protein and peptide instability in biodegradable drug delivery systems. Adv. Drug Deliv. Rev. 2023, 199, 114904. [Google Scholar] [CrossRef]
- Mehrotra, N.; Kharbanda, S.; Singh, H. Peptide-based combination nanoformulations for cancer therapy. Nanomedicine 2020, 15, 2201–2217. [Google Scholar] [CrossRef]
- Norouzi, P.; Mirmohammadi, M.; Tehrani, M.H.H. Anticancer peptides mechanisms, simple and complex. Chem. Biol. Interact. 2022, 368, 110194. [Google Scholar] [CrossRef]
- Massaoka, M.H.; Matsuo, A.L.; Figueiredo, C.R.; Girola, N.; Faria, C.F.; Azevedo, R.A.; Travassos, L.R. A novel cell-penetrating peptide derived from WT1 enhances p53 activity, induces cell senescence and displays antimelanoma activity in xeno- and syngeneic systems. FEBS Open Bio 2014, 4, 153–161. [Google Scholar] [CrossRef]
- Li, X.; Guo, H.; Duan, H.; Yang, Y.; Meng, J.; Liu, J.; Wang, C.; Xu, H. Improving chemotherapeutic efficiency in acute myeloid leukemia treatments by chemically synthesized peptide interfering with CXCR4/CXCL12 axis. Sci. Rep. 2015, 5, 16228. [Google Scholar] [CrossRef]
- Xu, S.; Zhang, M.; Fang, X.; Meng, J.; Xing, H.; Yan, D.; Liu, J.; Yang, Y.; Wen, T.; Zhang, W.; et al. A novel CD123-targeted therapeutic peptide loaded by micellar delivery system combats refractory acute myeloid leukemia. J. Hematol. Oncol. 2021, 14, 193. [Google Scholar] [CrossRef] [PubMed]
- Meng, J.; Ge, Y.; Xing, H.; Wei, H.; Xu, S.; Liu, J.; Yan, D.; Wen, T.; Wang, M.; Fang, X.; et al. Synthetic CXCR4 Antagonistic Peptide Assembling with Nanoscaled Micelles Combat Acute Myeloid Leukemia. Small 2020, 16, e2001890. [Google Scholar] [CrossRef] [PubMed]
- Jiang, M.; Fang, X.; Ma, L.; Liu, M.; Chen, M.; Liu, J.; Yang, Y.; Wang, C. A nucleus-targeting peptide antagonist towards EZH2 displays therapeutic efficacy for lung cancer. Int. J. Pharm. 2022, 622, 121894. [Google Scholar] [CrossRef]
- Lee, M.F.; Poh, C.L. Strategies to improve the physicochemical properties of peptide-based drugs. Pharm. Res. 2023, 40, 617–632. [Google Scholar] [CrossRef] [PubMed]
- Patra, J.K.; Das, G.; Fraceto, L.F.; Campos, E.V.R.; Rodriguez-Torres, L.S.; Acosta-Torres, L.S.; Diaz-Torres, R.G.; Grillo, R.; Swamy, M.K.; Sharma, S.; et al. Nano based drug delivery systems: Recent developments and future prospects. J. Nanobiotechnol. 2018, 16, 71. [Google Scholar] [CrossRef] [PubMed]
- Peer, D.; Karp, J.; Hong, S.; Farokhzad, O.C.; Margalit, R.; Langer, R. Nanocarriers as an emerging platform for cancer therapy. Nature Nanotechnol. 2007, 2, 751–760. [Google Scholar] [CrossRef]
- Shi, J.; Kantoff, P.W.; Wooster, R.; Farokhzad, O.C. Cancer nanomedicine: Progress, challenges and opportunities. Nat. Rev. Cancer 2017, 17, 20–37. [Google Scholar] [CrossRef] [PubMed]
- Hanafy, N.A.N.; El-Kemary, M.; Leporatti, S. Micelles Structure Development as a Strategy to Improve Smart Cancer Therapy. Cancers 2018, 10, 238. [Google Scholar] [CrossRef]
- Allen, T.M.; Cullis, P.R. Liposomal drug delivery systems: From concept to clinical applications. Adv. Drug Deliv. Rev. 2013, 65, 36–48. [Google Scholar] [CrossRef]
- Wang, S.; Chen, Y.; Guo, J.; Huang, Q. Liposomes for Tumor Targeted Therapy: A Review. Int. J. Mol. Sci. 2023, 24, 2643. [Google Scholar] [CrossRef]
- Torchilin, V.P. Micellar Nanocarriers: Pharmaceutical Perspectives. Pharm. Res. 2007, 24, 1–16. [Google Scholar] [CrossRef]
- Mohammed, H.; Khan, R.; Singh, V.; Yusuf, M.; Akhtar, N.; Sulaiman, G.; Albukhaty, S.; Abdellatif, A.; Khan, M.; Mohammed, S.; et al. Solid lipid nanoparticles for targeted natural and synthetic drugs delivery in high-incidence cancers, and other diseases: Roles of preparation methods, lipid composition, transitional stability, and release profiles in nanocarriers’ development. Nanotechnol. Rev. 2023, 12, 20220517. [Google Scholar] [CrossRef]
- Fang, X.; Zhang, K.; Jiang, M.; Ma, L.; Liu, J.; Xu, H.; Yang, Y.; Wang, C. Enhanced lymphatic delivery of nanomicelles encapsulating CXCR4-recognizing peptide and doxorubicin for the treatment of breast cancer. Int. J. Pharm. 2021, 594, 120183. [Google Scholar] [CrossRef] [PubMed]
- Cerrato, C.P.; Langel, Ü. An update on cell-penetrating peptides with intracellular organelle targeting. Expert Opin. Drug Deliv. 2022, 19, 133–146. [Google Scholar] [CrossRef] [PubMed]
- Bottens, R.A.; Yamada, T. Cell-Penetrating Peptides (CPPs) as Therapeutic and Diagnostic Agents for Cancer. Cancers 2022, 14, 5546. [Google Scholar] [CrossRef]
- Lin, J.T.; Chen, H.; Wang, D.; Xiong, L.; Li, J.Z.; Chen, G.; Chen, G.B. Nuclear-targeted p53 and DOX co-delivery of chitosan derivatives for cancer therapy in vitro and in vivo. Colloids Surf. B 2019, 183, 110440. [Google Scholar] [CrossRef]
- Xu, J.; Wang, F.; Ye, L.; Wang, R.; Zhao, L.; Yang, X.; Ji, J.; Liu, A.; Zhai, G. Penetrating peptides: Applications in drug delivery. J. Drug Deliv. Sci. Technol. 2023, 84, 104475. [Google Scholar] [CrossRef]
- Yang, S.; Meng, J.; Yang, Y.; Liu, H.; Wang, C.; Liu, J.; Zhang, Y.; Wang, C.; Xu, H. A HSP60-targeting peptide for cell apoptosis imaging. Oncogenesis 2016, 5, e201. [Google Scholar] [CrossRef]
- Wang, Y.; Wang, R.; Lu, X.; Lu, W.; Zhang, C.; Liang, W. Pegylated phospholipids-based self-assembly with water-soluble drugs. Pharm. Res. 2010, 27, 361–370. [Google Scholar] [CrossRef]
- Klein, E.; Ben-Bassat, H.; Neumann, H.; Ralph, P.; Zeuthen, J.; Polliack, A.; Vánky, F. Properties of the K562 cell line, derived from a patient with chronic myeloid leukemia. Int. J. Cancer 1976, 18, 421–431. [Google Scholar] [CrossRef]
- Li, Y.; Wang, J.; Li, X.; Jia, Y.; Huai, L.; He, K.; Yu, P.; Wang, M.; Xing, H.; Rao, Q.; et al. Role of the Wilms’ tumor 1 gene in the aberrant biological behavior of leukemic cells and the related mechanisms. Oncol. Rep. 2014, 32, 2680–2686. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.; Wang, J.; Xu, D. Cell-penetrating peptides as noninvasive transmembrane vectors for the development of novel multifunctional drug-delivery systems. J. Control. Release 2016, 229, 130–139. [Google Scholar] [CrossRef] [PubMed]
- Frankel, A.D.; Pabo, C.O. Cellular uptake of the tat protein from human immunodeficiency virus. Cell 1988, 55, 1189–1193. [Google Scholar] [CrossRef]
- Wang, J.; Hu, X.; Xiang, D. Nanoparticle drug delivery systems: An excellent carrier for tumor peptide vaccines. Drug Deliv. 2018, 25, 1319–1327. [Google Scholar] [CrossRef]
- Jain, A.; Jain, A.; Gulbake, A.; Shilpi, S.; Hurkat, P.; Jain, S.K. Peptide and protein delivery using new drug delivery systems. Crit. Rev. Ther. Drug Carr. Syst. 2013, 30, 293–329. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Wang, Y.; Liang, W. Delivery of drugs to cell membranes by encapsulation in PEG-PE micelles. J. Control. Release 2012, 160, 637–651. [Google Scholar] [CrossRef]
- Ding, Y.; Xu, Y.; Yang, W.; Niu, P.; Li, X.; Chen, Y.; Li, Z.; Liu, Y.; An, Y.; Liu, Y.; et al. Investigating the EPR effect of nanomedicines in human renal tumors via ex vivo perfusion strategy. Nano Today 2020, 35, 100970. [Google Scholar] [CrossRef]
- Zhang, Y.N.; Poon, W.; Tavares, A.J.; McGilvray, I.D.; Chan, W.C.W. Nanoparticle-liver interactions: Cellular uptake and hepatobiliary elimination. J. Control. Release 2016, 240, 332–348. [Google Scholar] [CrossRef] [PubMed]
- Yu, M.; Zheng, J. Clearance Pathways and Tumor Targeting of Imaging Nanoparticles. ACS Nano 2015, 9, 6655–6674. [Google Scholar] [CrossRef]
- Sun, Z.; Huang, J.; Fishelson, Z.; Wang, C.; Zhang, S. Cell-Penetrating Peptide-Based Delivery of Macromolecular Drugs: Development, Strategies, and Progress. Biomedicines 2023, 11, 1971. [Google Scholar] [CrossRef]
- Baccarani, M.; Zaccaria, A.; Bagnara, G.P.; Santucci, M.A.; Brunelli, M.A.; Tura, S. The relevance of extramedullary hemopoiesis to the staging of chronic myeloid leukemia. Boll. Ist. Sieroter. Milan. 1978, 57, 257–270. [Google Scholar] [PubMed]
- Wang, F.; Wang, Y.; Zhang, X.; Zhang, W.; Guo, S.; Jin, F. Recent progress of cell-penetrating peptides as new carriers for intracellular cargo delivery. J. Control. Release 2014, 174, 126–136. [Google Scholar] [CrossRef]
- Kolhar, P.; Anselmo, A.C.; Gupta, V.; Pant, K.; Prabhakarpandian, B.; Ruoslahti, E.; Mitragotri, S. Using shape effects to target antibody-coated nanoparticles to lung and brain endothelium. Proc. Natl. Acad. Sci. USA 2013, 110, 10753–10758. [Google Scholar] [CrossRef]
- Jiang, Y.; Huo, S.; Mizuhara, T.; Das, R.; Lee, Y.W.; Hou, S.; Moyano, D.F.; Duncan, B.; Liang, X.J.; Rotello, V.M. The Interplay of Size and Surface Functionality on the Cellular Uptake of Sub-10 nm Gold Nanoparticles. ACS Nano 2015, 9, 9986–9993. [Google Scholar] [CrossRef] [PubMed]
- Braun, T.P.; Eide, C.A.; Druker, B.J. Response and Resistance to BCR-ABL1-Targeted Therapies. Cancer Cell 2020, 37, 530–542. [Google Scholar] [CrossRef] [PubMed]
- Lyczek, A.; Berger, B.T.; Rangwala, A.M.; Paung, Y.; Tom, J.; Philipose, H.; Guo, J.; Albanese, S.K.; Robers, M.B.; Knapp, S.; et al. Mutation in Abl kinase with altered drug-binding kinetics indicates a novel mechanism of imatinib resistance. Proc. Natl. Acad. Sci. USA 2021, 118, e2111451118. [Google Scholar] [CrossRef]
- Svensson, E.; Vidovic, K.; Lassen, C.; Richter, J.; Olofsson, T.; Fioretos, T.; Gullberg, U. Deregulation of the Wilms’ tumour gene 1 protein (WT1) by BCR/ABL1 mediates resistance to imatinib in human leukaemia cells. Leukemia 2007, 21, 2485–2494. [Google Scholar] [CrossRef]
- Kienle, D.L. The spleen in hematologic malignancies. Ther. Umsch. 2013, 70, 163–169. [Google Scholar] [CrossRef]
- Bührer, E.D.; Amrein, M.A.; Forster, S.; Isringhausen, S.; Schürch, C.M.; Bhate, S.S.; Brodie, T.; Zindel, J.; Stroka, D.; Sayed, M.A.; et al. Splenic red pulp macrophages provide a niche for CML stem cells and induce therapy resistance. Leukemia 2022, 36, 2634–2646. [Google Scholar] [CrossRef]


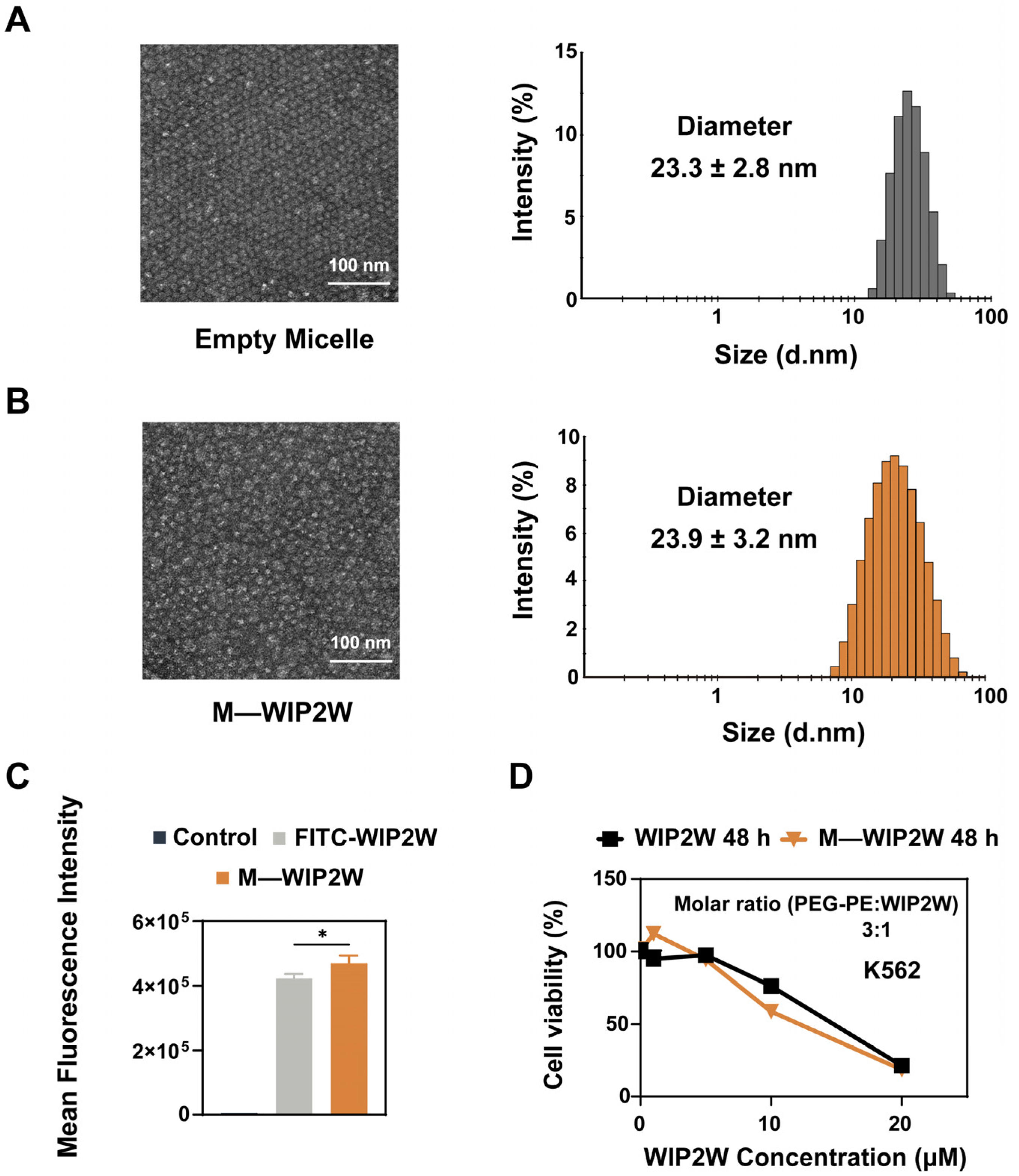

| Name | Diameter (nm) | Zeta Potential (mV) | PDI |
|---|---|---|---|
| Empty Micelle | 23.3 ± 2.8 | −4.0 ± 0.6 | 0.188 |
| M—WIP2W | 23.9 ± 3.2 | 5.3 ± 0.3 | 0.253 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, M.; Fang, X.; Du, R.; Meng, J.; Liu, J.; Liu, M.; Yang, Y.; Wang, C. A Nucleus-Targeting WT1 Antagonistic Peptide Encapsulated in Polymeric Nanomicelles Combats Refractory Chronic Myeloid Leukemia. Pharmaceutics 2023, 15, 2305. https://doi.org/10.3390/pharmaceutics15092305
Chen M, Fang X, Du R, Meng J, Liu J, Liu M, Yang Y, Wang C. A Nucleus-Targeting WT1 Antagonistic Peptide Encapsulated in Polymeric Nanomicelles Combats Refractory Chronic Myeloid Leukemia. Pharmaceutics. 2023; 15(9):2305. https://doi.org/10.3390/pharmaceutics15092305
Chicago/Turabian StyleChen, Mengting, Xiaocui Fang, Rong Du, Jie Meng, Jingyi Liu, Mingpeng Liu, Yanlian Yang, and Chen Wang. 2023. "A Nucleus-Targeting WT1 Antagonistic Peptide Encapsulated in Polymeric Nanomicelles Combats Refractory Chronic Myeloid Leukemia" Pharmaceutics 15, no. 9: 2305. https://doi.org/10.3390/pharmaceutics15092305
APA StyleChen, M., Fang, X., Du, R., Meng, J., Liu, J., Liu, M., Yang, Y., & Wang, C. (2023). A Nucleus-Targeting WT1 Antagonistic Peptide Encapsulated in Polymeric Nanomicelles Combats Refractory Chronic Myeloid Leukemia. Pharmaceutics, 15(9), 2305. https://doi.org/10.3390/pharmaceutics15092305






