Dual Effect by Chemical Electron Transfer Enhanced siRNA Lipid Nanoparticles: Reactive Oxygen Species-Triggered Tumor Cell Killing Aggravated by Nrf2 Gene Silencing
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials and Chemicals
2.2. Preparation of LNPs
2.3. DPA Degradation Assay
2.4. Gene Silencing Efficiency Assay
2.5. Cellular Uptake Study
2.6. Endosomal Escape Assay
2.7. Cell Apoptosis Assay
2.8. RT-qPCR Assay
2.9. Statistical Analysis
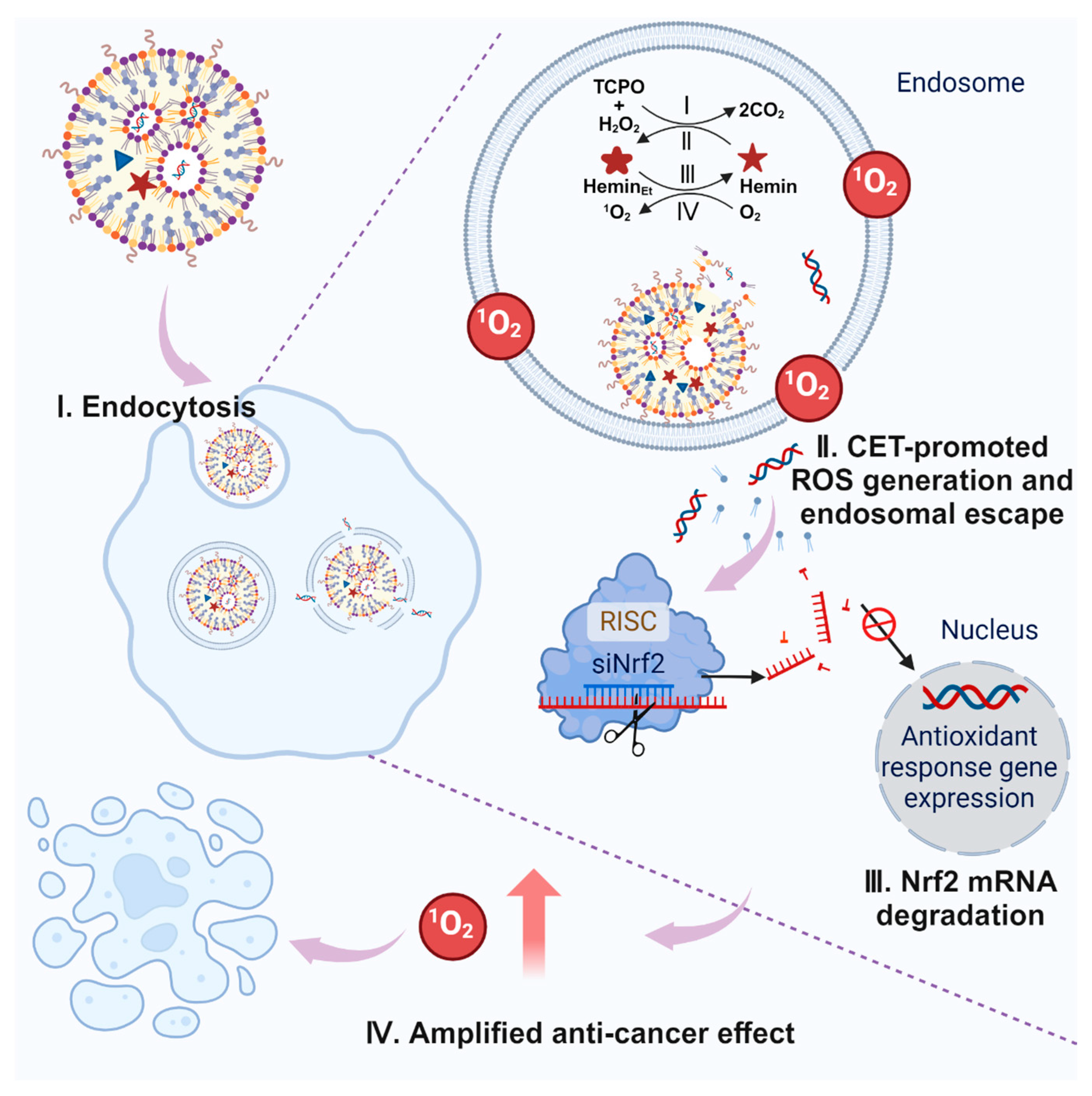
3. Results and Discussion
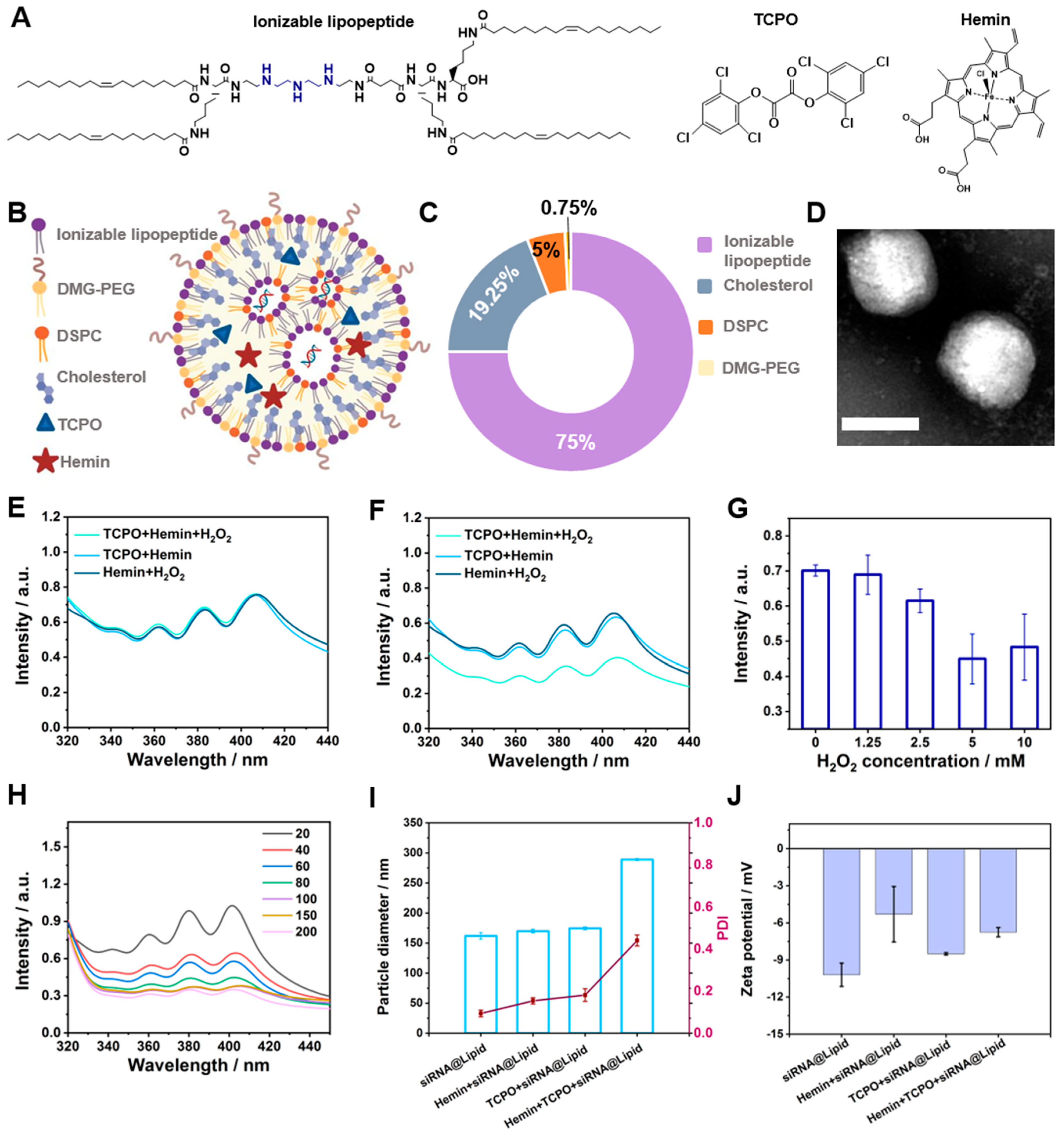
3.1. Design and Preparation of LNPs
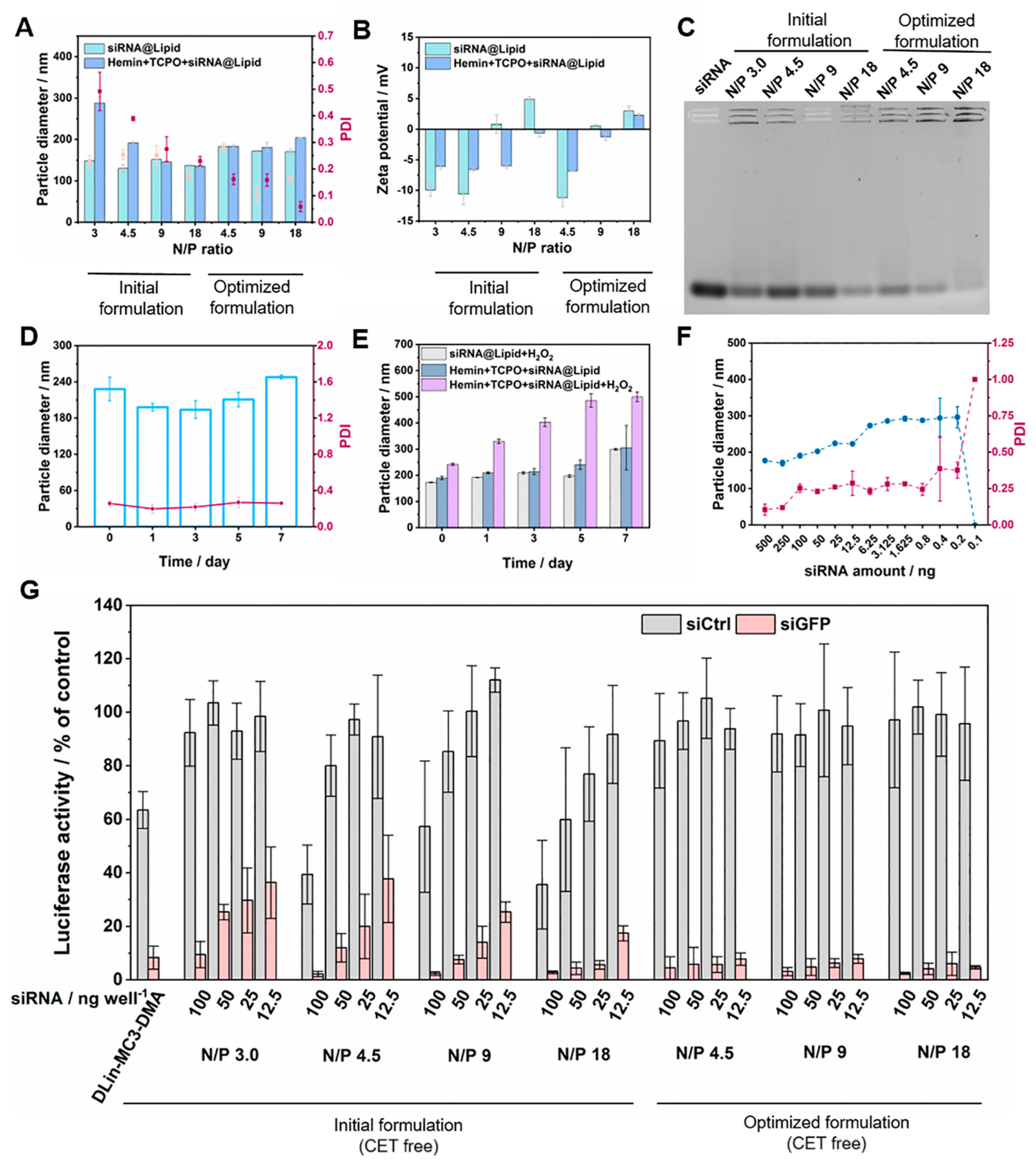
3.2. Optimization of LNPs
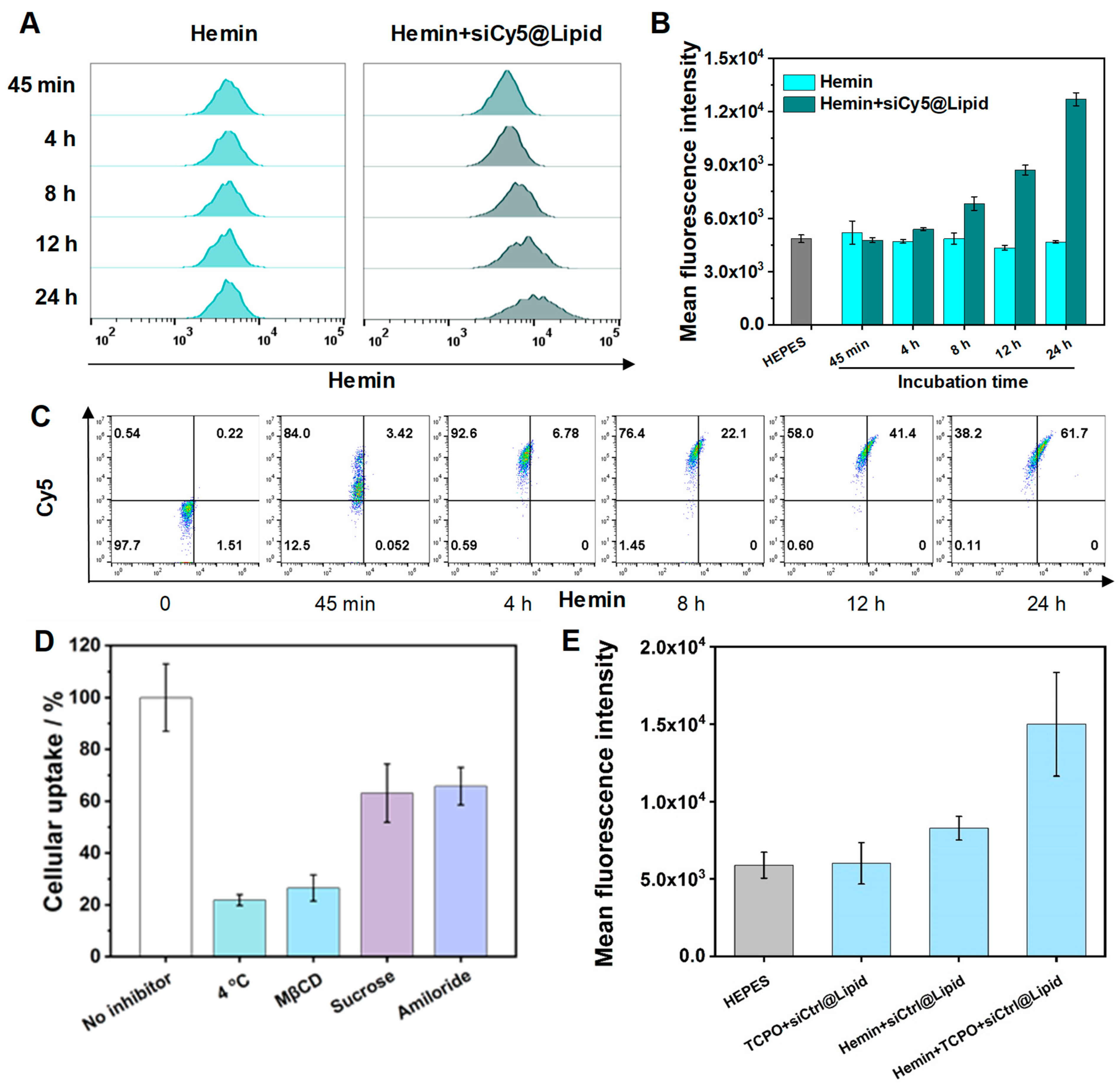
3.3. Evaluation of Cellular Uptake and Cellular ROS Production
3.4. CET-Related Gene Silence Enhancement, Endosomal Escape, and Cancer Cell Killing
3.5. CET-Enhanced Cancer Cell Killing
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ni, H.; Hatit, M.Z.C.; Zhao, K.; Loughrey, D.; Lokugamage, M.P.; Peck, H.E.; Del Cid, A.; Muralidharan, A.; Kim, Y.; Santangelo, P.J.; et al. Piperazine-Derived Lipid Nanoparticles Deliver mRNA to Immune Cells In Vivo. Nat. Commun. 2022, 13, 4766. [Google Scholar] [CrossRef]
- Liu, S.; Cheng, Q.; Wei, T.; Yu, X.; Johnson, L.T.; Farbiak, L.; Siegwart, D.J. Membrane-Destabilizing Ionizable Phospholipids for Organ-Selective mRNA Delivery and CRISPR–Cas Gene Editing. Nat. Mater. 2021, 20, 701–710. [Google Scholar] [CrossRef] [PubMed]
- Dammes, N.; Goldsmith, M.; Ramishetti, S.; Dearling, J.L.J.; Veiga, N.; Packard, A.B.; Peer, D. Conformation-Sensitive Targeting of Lipid Nanoparticles for RNA Therapeutics. Nat. Nanotechnol. 2021, 16, 1030–1038. [Google Scholar] [CrossRef]
- Cullis, P.R.; Hope, M.J. Lipid Nanoparticle Systems for Enabling Gene Therapies. Mol. Ther. 2017, 25, 1467–1475. [Google Scholar] [CrossRef]
- Miao, L.; Zhang, Y.; Huang, L. mRNA Vaccine for Cancer Immunotherapy. Mol. Cancer 2021, 20, 41. [Google Scholar] [CrossRef]
- Melamed, J.R.; Yerneni, S.S.; Arral, M.L.; LoPresti, S.T.; Chaudhary, N.; Sehrawat, A.; Muramatsu, H.; Alameh, M.-G.; Pardi, N.; Weissman, D.; et al. Ionizable Lipid Nanoparticles Deliver mRNA to Pancreatic β Cells via Macrophage-Mediated Gene Transfer. Sci. Adv. 2023, 9, eade1444. [Google Scholar] [CrossRef] [PubMed]
- Palanki, R.; Bose, S.K.; Dave, A.; White, B.M.; Berkowitz, C.; Luks, V.; Yaqoob, F.; Han, E.; Swingle, K.L.; Menon, P.; et al. Ionizable Lipid Nanoparticles for Therapeutic Base Editing of Congenital Brain Disease. ACS Nano 2023, 17, 13594–13610. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Jiang, A.Y.; Raji, I.; Atyeo, C.; Raimondo, T.M.; Gordon, A.G.R.; Rhym, L.H.; Samad, T.; MacIsaac, C.; Witten, J.; et al. Enhancing the Immunogenicity of Lipid-Nanoparticle mRNA Vaccines by Adjuvanting the Ionizable Lipid and the mRNA. Nat. Biomed. Eng. 2023. online ahead of print. [Google Scholar] [CrossRef]
- Riley, R.S.; Kashyap, M.V.; Billingsley, M.M.; White, B.; Alameh, M.-G.; Bose, S.K.; Zoltick, P.W.; Li, H.; Zhang, R.; Cheng, A.Y.; et al. Ionizable Lipid Nanoparticles for in Utero mRNA Delivery. Sci. Adv. 2021, 7, eaba1028. [Google Scholar] [CrossRef]
- Meulewaeter, S.; Nuytten, G.; Cheng, M.H.Y.; De Smedt, S.C.; Cullis, P.R.; De Beer, T.; Lentacker, I.; Verbeke, R. Continuous Freeze-Drying of Messenger RNA Lipid Nanoparticles Enables Storage at Higher Temperatures. J. Control. Release 2023, 357, 149–160. [Google Scholar] [CrossRef]
- Hammel, M.; Fan, Y.; Sarode, A.; Byrnes, A.E.; Zang, N.; Kou, P.; Nagapudi, K.; Leung, D.; Hoogenraad, C.C.; Chen, T.; et al. Correlating the Structure and Gene Silencing Activity of Oligonucleotide-Loaded Lipid Nanoparticles Using Small-Angle X-Ray Scattering. ACS Nano 2023, 17, 11454–11465. [Google Scholar] [CrossRef]
- Schoenmaker, L.; Witzigmann, D.; Kulkarni, J.A.; Verbeke, R.; Kersten, G.; Jiskoot, W.; Crommelin, D.J.A. mRNA-Lipid Nanoparticle COVID-19 Vaccines: Structure and Stability. Int. J. Pharm. 2021, 601, 120586. [Google Scholar] [CrossRef]
- Maugeri, M.; Nawaz, M.; Papadimitriou, A.; Angerfors, A.; Camponeschi, A.; Na, M.; Hölttä, M.; Skantze, P.; Johansson, S.; Sundqvist, M.; et al. Linkage between Endosomal Escape of LNP-mRNA and Loading into EVs for Transport to Other Cells. Nat. Commun. 2019, 10, 4333. [Google Scholar] [CrossRef] [PubMed]
- Blersch, J.; Francisco, V.; Rebelo, C.; Jiménez-Balsa, A.; Antunes, H.; Gonzato, C.; Pinto, S.; Simões, S.; Liedl, K.; Haupt, K.; et al. A Light-Triggerable Nanoparticle Library for the Controlled Release of Non-Coding RNAs. Angew. Chem. Int. Ed. 2020, 59, 1985–1991. [Google Scholar] [CrossRef]
- Dowdy, S.F. Endosomal Escape of RNA Therapeutics: How Do We Solve This Rate-Limiting Problem? RNA 2023, 29, 396–401. [Google Scholar] [CrossRef]
- Martens, T.F.; Remaut, K.; Demeester, J.; De Smedt, S.C.; Braeckmans, K. Intracellular Delivery of Nanomaterials: How to Catch Endosomal Escape in the Act. Nano Today 2014, 9, 344–364. [Google Scholar] [CrossRef]
- Vermeulen, L.M.P.; De Smedt, S.C.; Remaut, K.; Braeckmans, K. The Proton Sponge Hypothesis: Fable or Fact? Eur. J. Pharm. Biopharm. 2018, 129, 184–190. [Google Scholar] [CrossRef] [PubMed]
- Herrera-Barrera, M.; Ryals, R.C.; Gautam, M.; Jozic, A.; Landry, M.; Korzun, T.; Gupta, M.; Acosta, C.; Stoddard, J.; Reynaga, R.; et al. Peptide-Guided Lipid Nanoparticles Deliver mRNA to the Neural Retina of Rodents and Nonhuman Primates. Sci. Adv. 2023, 9, eadd4623. [Google Scholar] [CrossRef]
- Nakamura, T.; Nakade, T.; Yamada, K.; Sato, Y.; Harashima, H. The Hydrophobic Tail of a pH-Sensitive Cationic Lipid Influences siRNA Transfection Activity and Toxicity in Human NK Cell Lines. Int. J. Pharm. 2021, 609, 121140. [Google Scholar] [CrossRef] [PubMed]
- Winkeljann, B.; Keul, D.C.; Merkel, O.M. Engineering Poly- and Micelleplexes for Nucleic Acid Delivery–A Reflection on Their Endosomal Escape. J. Control. Release 2023, 353, 518–534. [Google Scholar] [CrossRef]
- Liu, S.; Wang, X.; Yu, X.; Cheng, Q.; Johnson, L.T.; Chatterjee, S.; Zhang, D.; Lee, S.M.; Sun, Y.; Lin, T.-C.; et al. Zwitterionic Phospholipidation of Cationic Polymers Facilitates Systemic mRNA Delivery to Spleen and Lymph Nodes. J. Am. Chem. Soc. 2021, 143, 21321–21330. [Google Scholar] [CrossRef]
- Klipp, A.; Burger, M.; Leroux, J.-C. Get out or Die Trying: Peptide- and Protein-Based Endosomal Escape of RNA Therapeutics. Adv. Drug Deliv. Rev. 2023, 200, 115047. [Google Scholar] [CrossRef]
- Zhang, X.; Zhao, W.; Nguyen, G.N.; Zhang, C.; Zeng, C.; Yan, J.; Du, S.; Hou, X.; Li, W.; Jiang, J.; et al. Functionalized Lipid-like Nanoparticles for In Vivo mRNA Delivery and Base Editing. Sci. Adv. 2020, 6, eabc2315. [Google Scholar] [CrossRef]
- Carrasco, M.J.; Alishetty, S.; Alameh, M.-G.; Said, H.; Wright, L.; Paige, M.; Soliman, O.; Weissman, D.; Cleveland, T.E.; Grishaev, A.; et al. Ionization and Structural Properties of mRNA Lipid Nanoparticles Influence Expression in Intramuscular and Intravascular Administration. Commun. Biol. 2021, 4, 956. [Google Scholar] [CrossRef]
- Teplensky, M.H.; Fantham, M.; Poudel, C.; Hockings, C.; Lu, M.; Guna, A.; Aragones-Anglada, M.; Moghadam, P.Z.; Li, P.; Farha, O.K.; et al. A Highly Porous Metal-Organic Framework System to Deliver Payloads for Gene Knockdown. Chem 2019, 5, 2926–2941. [Google Scholar] [CrossRef]
- Gilleron, J.; Querbes, W.; Zeigerer, A.; Borodovsky, A.; Marsico, G.; Schubert, U.; Manygoats, K.; Seifert, S.; Andree, C.; Stöter, M.; et al. Image-Based Analysis of Lipid Nanoparticle–Mediated siRNA Delivery, Intracellular Trafficking and Endosomal Escape. Nat. Biotechnol. 2013, 31, 638–646. [Google Scholar] [CrossRef]
- Ndeupen, S.; Qin, Z.; Jacobsen, S.; Bouteau, A.; Estanbouli, H.; Igyártó, B.Z. The mRNA-LNP Platform’s Lipid Nanoparticle Component Used in Preclinical Vaccine Studies Is Highly Inflammatory. iScience 2021, 24, 103479. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Q.; Wei, T.; Farbiak, L.; Johnson, L.T.; Dilliard, S.A.; Siegwart, D.J. Selective Organ Targeting (SORT) Nanoparticles for Tissue-Specific mRNA Delivery and CRISPR–Cas Gene Editing. Nat. Nanotechnol. 2020, 15, 313–320. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Su, Z.; Zhao, W.; Zhang, X.; Momin, N.; Zhang, C.; Wittrup, K.D.; Dong, Y.; Irvine, D.J.; Weiss, R. Multifunctional Oncolytic Nanoparticles Deliver Self-Replicating IL-12 RNA to Eliminate Established Tumors and Prime Systemic Immunity. Nat. Cancer 2020, 1, 882–893. [Google Scholar] [CrossRef]
- Li, B.; Zhao, M.; Lai, W.; Zhang, X.; Yang, B.; Chen, X.; Ni, Q. Activatable NIR-II Photothermal Lipid Nanoparticles for Improved Messenger RNA Delivery. Angew. Chem. Int. Ed. 2023, 62, 3–9. [Google Scholar]
- Zhang, Q.; Kuang, G.; He, S.; Lu, H.; Cheng, Y.; Zhou, D.; Huang, Y. Photoactivatable Prodrug-Backboned Polymeric Nanoparticles for Efficient Light-Controlled Gene Delivery and Synergistic Treatment of Platinum-Resistant Ovarian Cancer. Nano Lett. 2020, 20, 3039–3049. [Google Scholar] [CrossRef] [PubMed]
- Rebelo, C.; Reis, T.; Guedes, J.; Saraiva, C.; Rodrigues, A.F.; Simões, S.; Bernardino, L.; Peça, J.; Pinho, S.L.C.; Ferreira, L. Efficient Spatially Targeted Gene Editing Using a Near-Infrared Activatable Protein-Conjugated Nanoparticle for Brain Applications. Nat. Commun. 2022, 13, 4135. [Google Scholar] [CrossRef] [PubMed]
- Li, G.; Zhang, Y.; Li, J. A Hybrid Nanoassembly for Ultrasound-Inducible Cytosolic siRNA Delivery and Cancer Sono-Gene Therapy. Ultrason. Sonochem. 2023, 92, 106262. [Google Scholar] [CrossRef] [PubMed]
- Wan, G.; Chen, X.; Wang, H.; Hou, S.; Wang, Q.; Cheng, Y.; Chen, Q.; Lv, Y.; Chen, H.; Zhang, Q. Gene Augmented Nuclear-Targeting Sonodynamic Therapy via Nrf2 Pathway-Based Redox Balance Adjustment Boosts Peptide-Based Anti-PD-L1 Therapy on Colorectal Cancer. J. Nanobiotechnol. 2021, 19, 347. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Wang, S.; Chen, Y.; Xin, H.; Zhang, S.; Wu, D.; Xue, Y.; Zha, M.; Li, H.; Li, K.; et al. Non-Invasive Activation of Intratumoural Gene Editing for Improved Adoptive T-Cell Therapy in Solid Tumours. Nat. Nanotechnol. 2023, 18, 933–944. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Chen, Y.; Xin, H.; Wan, T.; Ping, Y. Near-Infrared Optogenetic Engineering of Photothermal NanoCRISPR for Programmable Genome Editing. Proc. Natl. Acad. Sci. USA 2020, 117, 2395–2405. [Google Scholar] [CrossRef] [PubMed]
- Berg, K.; Folini, M.; Prasmickaite, L.; Selbo, P.; Bonsted, A.; Engesaeter, B.; Zaffaroni, N.; Weyergang, A.; Dietzea, A.; Maelandsmo, G.; et al. Photochemical Internalization: A New Tool for Drug Delivery. Curr. Pharm. Biotechnol. 2007, 8, 362–372. [Google Scholar] [CrossRef] [PubMed]
- Mo, Y.; Cheng, M.H.Y.; D’Elia, A.; Doran, K.; Ding, L.; Chen, J.; Cullis, P.R.; Zheng, G. Light-Activated siRNA Endosomal Release (LASER) by Porphyrin Lipid Nanoparticles. ACS Nano 2023, 17, 4688–4703. [Google Scholar] [CrossRef] [PubMed]
- Zhang, F.; Lin, Y.; Höhn, M.; Wagner, E. Chemical-Electron-Transfer-Based Lipopolyplexes for Enhanced siRNA Delivery. Cell Reports Phys. Sci. 2023, 4, 101444. [Google Scholar] [CrossRef]
- Fröhlich, T.; Edinger, D.; Kläger, R.; Troiber, C.; Salcher, E.; Badgujar, N.; Martin, I.; Schaffert, D.; Cengizeroglu, A.; Hadwiger, P.; et al. Structure-Activity Relationships of siRNA Carriers Based on Sequence-Defined Oligo (Ethane Amino) Amides. J. Control. Release 2012, 160, 532–541. [Google Scholar] [CrossRef]
- Schaffert, D.; Troiber, C.; Salcher, E.E.; Fröhlich, T.; Martin, I.; Badgujar, N.; Dohmen, C.; Edinger, D.; Kläger, R.; Maiwald, G.; et al. Solid-Phase Synthesis of Sequence-Defined T-, i-, and U-Shape Polymers for pDNA and siRNA Delivery. Angew. Chem. Int. Ed. 2011, 50, 8986–8989. [Google Scholar] [CrossRef] [PubMed]
- Schaffert, D.; Badgujar, N.; Wagner, E. Novel Fmoc-Polyamino Acids for Solid-Phase Synthesis of Defined Polyamidoamines. Org. Lett. 2011, 13, 1586–1589. [Google Scholar] [CrossRef] [PubMed]
- Dohmen, C.; Edinger, D.; Fröhlich, T.; Schreiner, L.; Lächelt, U.; Troiber, C.; Rädler, J.; Hadwiger, P.; Vornlocher, H.P.; Wagner, E. Nanosized Multifunctional Polyplexes for Receptor-Mediated siRNA Delivery. ACS Nano 2012, 6, 5198–5208. [Google Scholar] [CrossRef] [PubMed]
- Rui, Y.; Wilson, D.R.; Tzeng, S.Y.; Yamagata, H.M.; Sudhakar, D.; Conge, M.; Berlinicke, C.A.; Zack, D.J.; Tuesca, A.; Green, J.J. High-Throughput and High-Content Bioassay Enables Tuning of Polyester Nanoparticles for Cellular Uptake, Endosomal Escape, and Systemic In Vivo Delivery of mRNA. Sci. Adv. 2022, 8, eabk2855. [Google Scholar] [CrossRef] [PubMed]
- Lin, Y.; Wilk, U.; Pöhmerer, J.; Hörterer, E.; Höhn, M.; Luo, X.; Mai, H.; Wagner, E.; Lächelt, U. Folate Receptor-Mediated Delivery of Cas9 RNP for Enhanced Immune Checkpoint Disruption in Cancer Cells. Small 2023, 19, 2205318. [Google Scholar] [CrossRef] [PubMed]
- El-Beltagi, H.S.; Mohamed, H.I. Reactive Oxygen Species, Lipid Peroxidation and Antioxidative Defense Mechanism. Not. Bot. Horti Agrobot. Cluj-Napoca 2013, 41, 44. [Google Scholar] [CrossRef]
- Stamenkovic, A.; Pierce, G.N.; Ravandi, A. Phospholipid Oxidation Products in Ferroptotic Myocardial Cell Death. Am. J. Physiol. Circ. Physiol. 2019, 317, H156–H163. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Zhang, X.; Zhang, C.; Yan, J.; Hou, X.; Du, S.; Zeng, C.; Zhao, W.; Deng, B.; McComb, D.W.; et al. Biomimetic Nanoparticles Deliver mRNAs Encoding Costimulatory Receptors and Enhance T Cell Mediated Cancer Immunotherapy. Nat. Commun. 2021, 12, 7264. [Google Scholar] [CrossRef]
- Lee, K.; Conboy, M.; Park, H.M.; Jiang, F.; Kim, H.J.; Dewitt, M.A.; Mackley, V.A.; Chang, K.; Rao, A.; Skinner, C.; et al. Nanoparticle Delivery of Cas9 Ribonucleoprotein and Donor DNA In Vivo Induces Homology-Directed DNA Repair. Nat. Biomed. Eng. 2017, 1, 889–901. [Google Scholar] [CrossRef]
- Smith, R.; Wafa, E.I.; Geary, S.M.; Ebeid, K.; Alhaj-Suliman, S.O.; Salem, A.K. Cationic Nanoparticles Enhance T Cell Tumor Infiltration and Antitumor Immune Responses to a Melanoma Vaccine. Sci. Adv. 2022, 8, eabk3150. [Google Scholar] [CrossRef]
- Sies, H.; Jones, D.P. Reactive Oxygen Species (ROS) as Pleiotropic Physiological Signalling Agents. Nat. Rev. Mol. Cell Biol. 2020, 21, 363–383. [Google Scholar] [CrossRef] [PubMed]
- Deng, S.; Li, X.; Liu, S.; Chen, J.; Li, M.; Chew, S.Y.; Leong, K.W.; Cheng, D. Codelivery of CRISPR-Cas9 and Chlorin E6 for Spatially Controlled Tumor-Specific Gene Editing with Synergistic Drug Effects. Sci. Adv. 2020, 6, eabb4005. [Google Scholar] [CrossRef] [PubMed]
- Shi, S.; Wang, Y.; Wang, B.; Chen, Q.; Wan, G.; Yang, X.; Zhang, J.; Zhang, L.; Li, C.; Wang, Y. Homologous-Targeting Biomimetic Nanoparticles for Photothermal Therapy and Nrf2-siRNA Amplified Photodynamic Therapy against Oral Tongue Squamous Cell Carcinoma. Chem. Eng. J. 2020, 388, 124268. [Google Scholar] [CrossRef]
- Ulasov, A.V.; Rosenkranz, A.A.; Georgiev, G.P.; Sobolev, A.S. Nrf2/Keap1/ARE Signaling: Towards Specific Regulation. Life Sci. 2022, 291, 120111. [Google Scholar] [CrossRef] [PubMed]
- Vomund, S.; Schäfer, A.; Parnham, M.J.; Brüne, B.; Von Knethen, A. Nrf2, the Master Regulator of Anti-Oxidative Responses. Int. J. Mol. Sci. 2017, 18, 2772. [Google Scholar] [CrossRef] [PubMed]
- Li, R.; Jia, Z.; Zhu, H. Regulation of Nrf2 Signaling. React. Oxyg. Species Apex N.C. 2019, 8, 312–322. [Google Scholar] [CrossRef]
- Valentine, M.T.; Fordyce, P.M.; Block, S.M. Eg5 Steps It Up! Cell Div. 2006, 1, 31. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, F.; Burghardt, T.; Höhn, M.; Wagner, E. Dual Effect by Chemical Electron Transfer Enhanced siRNA Lipid Nanoparticles: Reactive Oxygen Species-Triggered Tumor Cell Killing Aggravated by Nrf2 Gene Silencing. Pharmaceutics 2024, 16, 779. https://doi.org/10.3390/pharmaceutics16060779
Zhang F, Burghardt T, Höhn M, Wagner E. Dual Effect by Chemical Electron Transfer Enhanced siRNA Lipid Nanoparticles: Reactive Oxygen Species-Triggered Tumor Cell Killing Aggravated by Nrf2 Gene Silencing. Pharmaceutics. 2024; 16(6):779. https://doi.org/10.3390/pharmaceutics16060779
Chicago/Turabian StyleZhang, Fengrong, Tobias Burghardt, Miriam Höhn, and Ernst Wagner. 2024. "Dual Effect by Chemical Electron Transfer Enhanced siRNA Lipid Nanoparticles: Reactive Oxygen Species-Triggered Tumor Cell Killing Aggravated by Nrf2 Gene Silencing" Pharmaceutics 16, no. 6: 779. https://doi.org/10.3390/pharmaceutics16060779




