An Innovative Bio-Vehicle for Resveratrol and Tocopherol Based on Quinoa 11S Globulin—Nanocomplex Design and Characterization
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials
2.2. 11S Purification
2.3. SDS-PAGE Electrophoresis
2.4. Molecular Docking
2.5. Preparation of 11S and TOC or RSV Mixed Systems
2.6. Circular Dichroism (CD)
2.7. Isothermal Titration Calorimetry (ITC)
2.8. Steady-State Fluorescence Measurements
2.9. Encapsulation Efficiency
2.10. Particle Size and ζ-Potential Determinations
2.11. Atomic Force Microscopy (AFM)
2.12. Antioxidant Activity of 11S-RSV or 11S-TOC Complexes
2.12.1. ABTS Assay
2.12.2. Ferric Reducing Antioxidant Power (FRAP) Assay
2.13. Statistical Analysis
3. Results
3.1. SDS-PAGE- FPLC
3.2. Molecular Docking
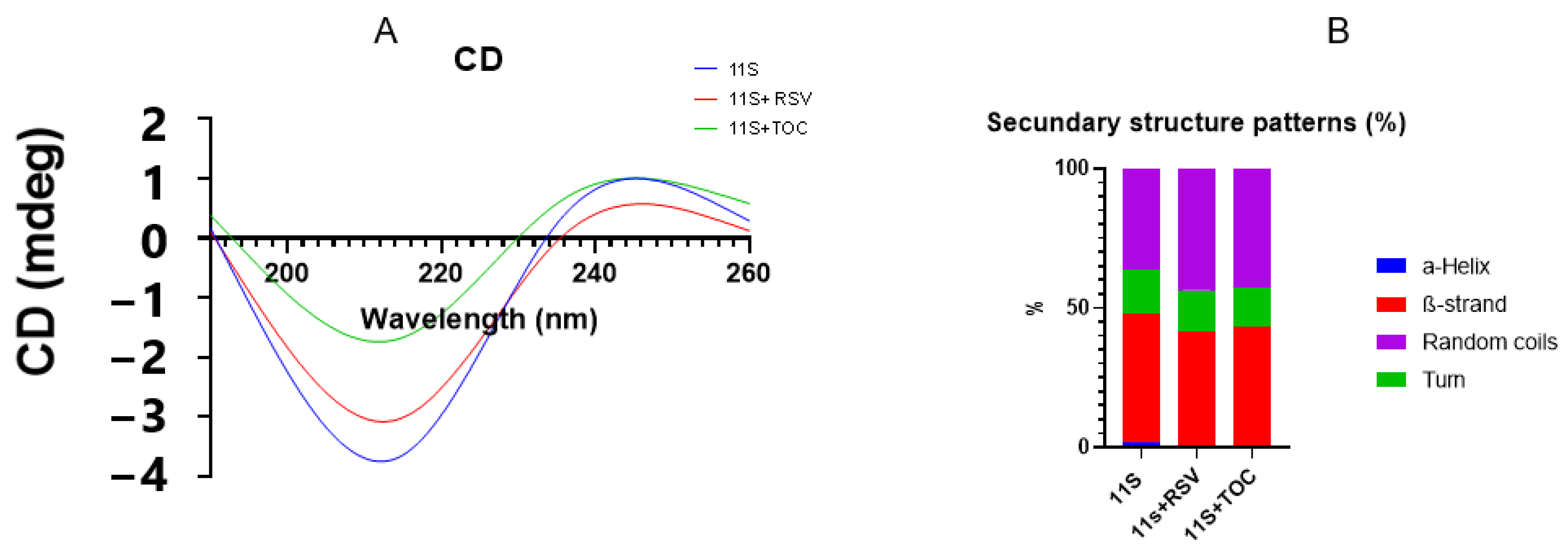
3.3. Circular Dichroism (CD)
3.4. Isothermal Titration Calorimetry (ITC)
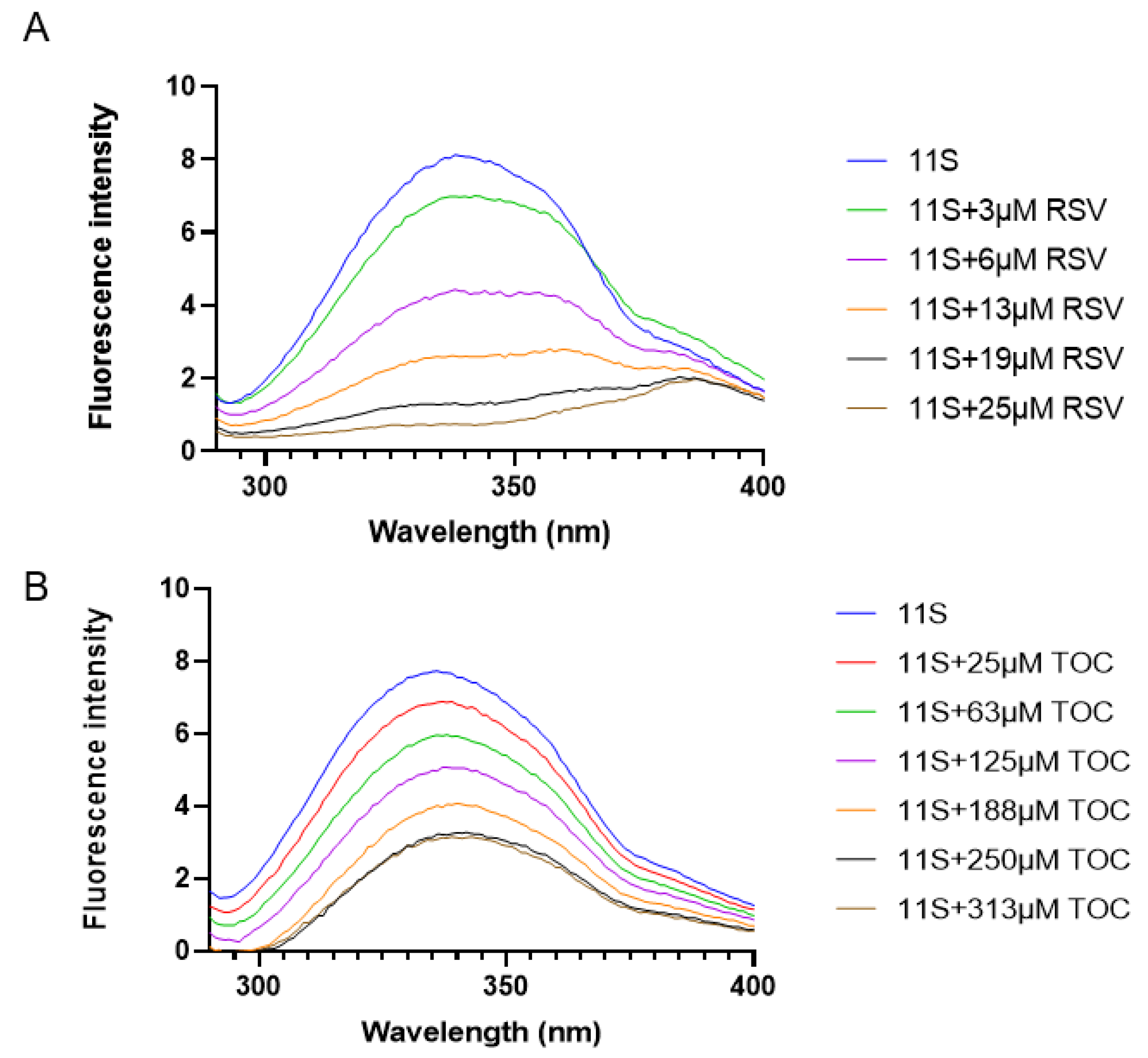
3.5. Fluorescence Spectroscopy
3.6. EE Determination
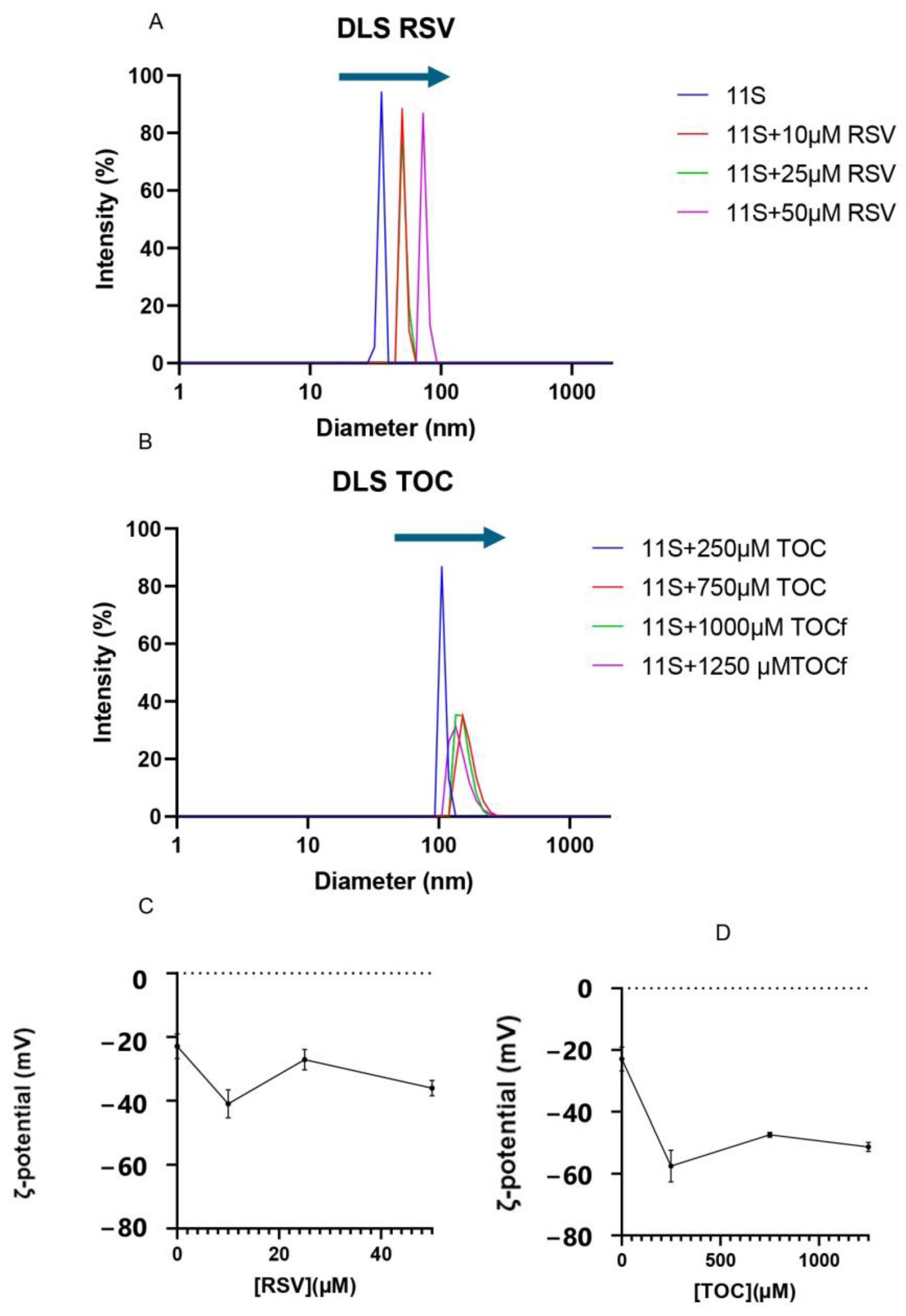
3.7. Analysis of 11S Globulin Aggregation Induced by RSV or TOC
3.8. Atomic Force Microscopy (AFM)
3.9. Antioxidant Capacity of 11S-RSV and 11S-TOC Nanocomplexes
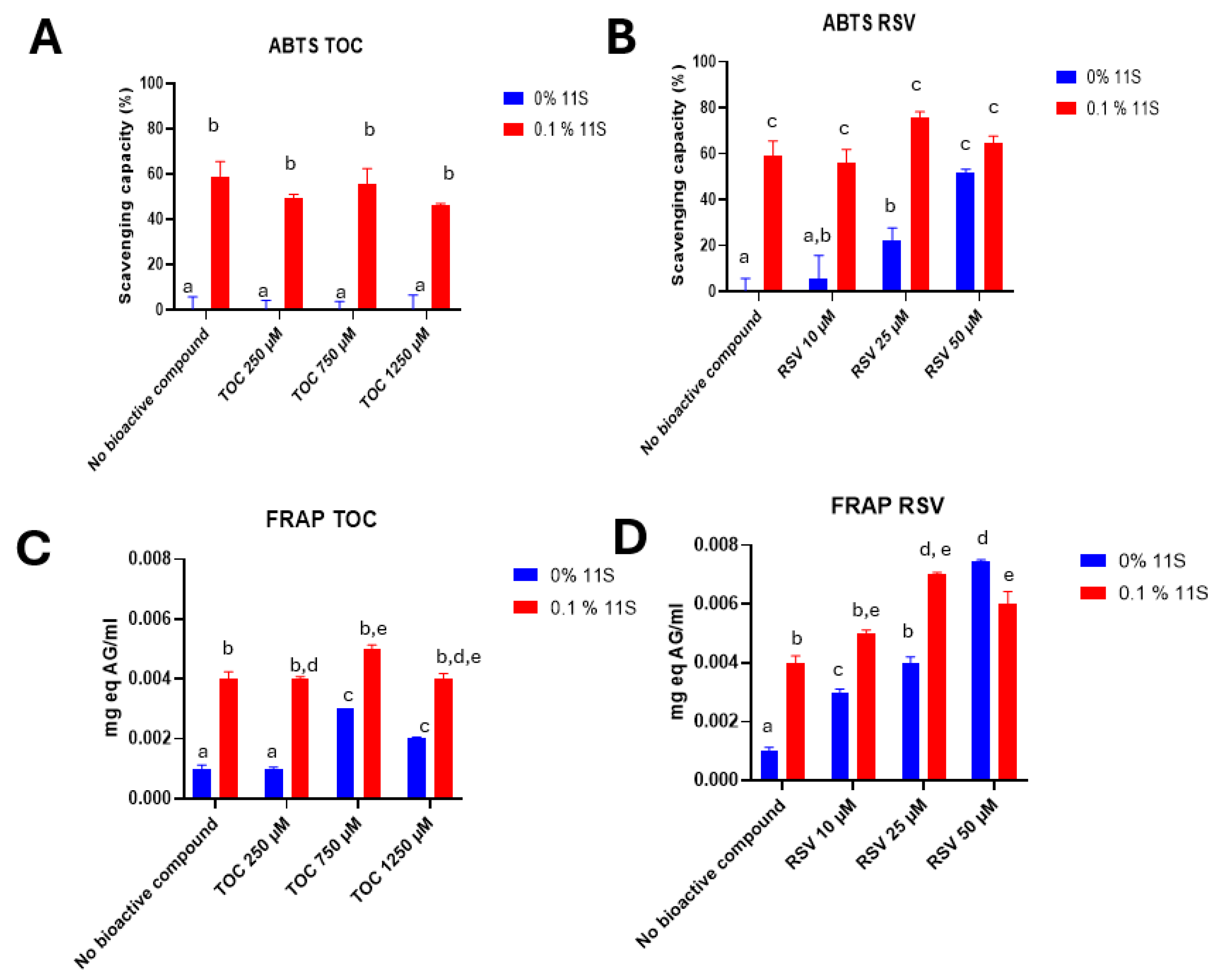
3.9.1. ABTS
3.9.2. FRAP
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- He, X.; Wang, B.; Zhao, B.; Yang, F. Ultrasonic Assisted Extraction of Quinoa (Chenopodium quinoa Willd.) Protein and Effect of Heat Treatment on Its In Vitro Digestion Characteristics. Foods 2022, 11, 771. [Google Scholar] [CrossRef]
- Lingiardi, N.; Galante, M.; de Sanctis, M.; Spelzini, D. Are Quinoa Proteins a Promising Alternative to Be Applied in Plant-Based Emulsion Gel Formulation? Food Chem. 2022, 394, 133485. [Google Scholar] [CrossRef] [PubMed]
- Kaspchak, E.; de Oliveira, M.A.S.; Simas, F.F.; Franco, C.R.C.; Silveira, J.L.M.; Mafra, M.R.; Igarashi-Mafra, L. Determination of Heat-Set Gelation Capacity of a Quinoa Protein Isolate (Chenopodium quinoa) by Dynamic Oscillatory Rheological Analysis. Food Chem. 2017, 232, 263–271. [Google Scholar] [CrossRef]
- Choi, I.; Li, N.; Zhong, Q. Enhancing Bioaccessibility of Resveratrol by Loading in Natural Porous Starch Microparticles. Int. J. Biol. Macromol. 2022, 194, 982–992. [Google Scholar] [CrossRef] [PubMed]
- Malinowska, M.A.; Sharafan, M.; Lanoue, A.; Ferrier, M.; Hano, C.; Giglioli-Guivarc’h, N.; Dziki, A.; Sikora, E.; Szopa, A. Trans-Resveratrol as a Health Beneficial Molecule: Activity, Sources, and Methods of Analysis. Sci. Radices 2023, 268–294. [Google Scholar] [CrossRef]
- Chimento, A.; De Amicis, F.; Sirianni, R.; Sinicropi, M.S.; Puoci, F.; Casaburi, I.; Saturnino, C.; Pezzi, V. Progress to Improve Oral Bioavailability and Beneficial Effects of Resveratrol. Int. J. Mol. Sci. 2019, 20, 1381. [Google Scholar] [CrossRef]
- Saini, R.K.; Keum, Y.S. Tocopherols and Tocotrienols in Plants and Their Products: A Review on Methods of Extraction, Chromatographic Separation, and Detection. Food Res. Int. 2016, 82, 59–70. [Google Scholar] [CrossRef]
- Bustos, L.F.; Judis, M.A.; Vasile, F.E.; Pérez, O.E. Molecular Interactions Involved in the Complexation Process between Buffalo Whey Proteins Concentrate and Folic Acid. Food Chem. 2022, 396, 133734. [Google Scholar] [CrossRef] [PubMed]
- Corfield, R.; Martínez, K.D.; Allievi, M.C.; Santagapita, P.; Mazzobre, F.; Schebor, C.; Pérez, O.E. Whey Proteins-Folic Acid Complexes: Formation, Isolation and Bioavailability in a Lactobacillus casei Model. Food Struct. 2020, 26, 100162. [Google Scholar] [CrossRef]
- Corfield, R.; Lalou, G.; Di Lella, S.; Martínez, K.D.; Schebor, C.; Allievi, M.C.; Pérez, O.E. Experimental and Modeling Approaches Applied to the Whey Proteins and Vitamin B9 Complexes Study. Food Hydrocoll. 2023, 142, 108834. [Google Scholar] [CrossRef]
- Liu, K.; Zha, X.Q.; Li, Q.M.; Pan, L.H.; Luo, J.P. Hydrophobic Interaction and Hydrogen Bonding Driving the Self-Assembling of Quinoa Protein and Flavonoids. Food Hydrocoll. 2021, 118, 106807. [Google Scholar] [CrossRef]
- Martínez, J.H.; Velázquez, F.; Burrieza, H.P.; Martínez, K.D.; Paula Domínguez Rubio, A.; dos Santos Ferreira, C.; del Pilar Buera, M.; Pérez, O.E. Betanin Loaded Nanocarriers Based on Quinoa Seed 11S Globulin. Impact on the Protein Structure and Antioxidant Activity. Food Hydrocoll. 2019, 87, 880–890. [Google Scholar] [CrossRef]
- Ochnio, M.E.; Martínez, J.H.; Allievi, M.C.; Palavecino, M.; Martínez, K.D.; Pérez, O.E. Proteins as Nano-Carriers for Bioactive Compounds. The Case of 7S and 11S Soy Globulins and Folic Acid Complexation. Polymers 2018, 10, 149. [Google Scholar] [CrossRef] [PubMed]
- Kiss Nanotechnology in Food Systems: A Review. Acta Aliment. 2020, 49, 460–474. [CrossRef]
- Pérez, O.E.; David-Birman, T.; Kesselman, E.; Levi-Tal, S.; Lesmes, U. Milk Protein-Vitamin Interactions: Formation of Beta-Lactoglobulin/Folic Acid Nano-Complexes and Their Impactoninvitro Gastro-Duodenal Proteolysis. Food Hydrocoll. 2014, 38, 40–47. [Google Scholar] [CrossRef]
- Laemmli Cleavage of Structural Proteins during the Assembly of the Head of bacteriophage T4. Nature 1970, 227, 680–685. [CrossRef]
- Neuhoff, V.; Arold, N.; Taube, D.; Ehrhardt, W. Improved Staining of Proteins in Polyacrylamide Gels Including Isoelectric Focusing Gels with Clear Background at Nanogram Sensitivity Using Coomassie Brilliant Blue G-250 and R-250. Electrophoresis 1988, 9, 255–262. [Google Scholar] [CrossRef] [PubMed]
- Trott, O.; Olson, A.J. AutoDock Vina: Improving the Speed and Accuracy of Docking with a New Scoring Function, Efficient Optimization, and Multithreading. J. Comput. Chem. 2010, 31, 455–461. [Google Scholar] [CrossRef]
- Bourne, P.E.; Weissig, H. Structural Bioinformatics; Wiley-Liss: Hoboken, NJ, USA, 2003; ISBN 0471201995. [Google Scholar]
- Mistry, J.; Chuguransky, S.; Williams, L.; Qureshi, M.; Salazar, G.A.; Sonnhammer, E.L.L.; Tosatto, S.C.E.; Paladin, L.; Raj, S.; Richardson, L.J.; et al. Pfam: The Protein Families Database in 2021. Nucleic Acids Res. 2021, 49, D412–D419. [Google Scholar] [CrossRef]
- Bustos, L.F.; Vasile, F.E.; Pérez, O.E. Experimental and in Silico Approaches for the Buffalo Whey Protein-Folic Acid Complexation Elucidation. Molecular Changes Impacting on Protein Structure and Functionality. Food Res. Int. 2024, 180, 114062. [Google Scholar] [CrossRef]
- Morris, G.M.; Ruth, H.; Lindstrom, W.; Sanner, M.F.; Belew, R.K.; Goodsell, D.S.; Olson, A.J. Software News and Updates AutoDock4 and AutoDockTools4: Automated Docking with Selective Receptor Flexibility. J. Comput. Chem. 2009, 30, 2785–2791. [Google Scholar] [CrossRef] [PubMed]
- Humphrey, W.; Dalke, A.; Schulten, K. VMD: Visual molecular dynamics. J. Mol. Graph. 1996, 14, 33–38. [Google Scholar] [CrossRef] [PubMed]
- Liang, L.; Subirade, M. β-Lactoglobulin/Folic Acid Complexes: Formation, Characterization, and Biological Implication. J. Phys. Chem. B 2010, 114, 6707–6712. [Google Scholar] [CrossRef] [PubMed]
- Lakowicz, J.R.; Weber, G. Quenching of Fluorescence by Oxygen. a Probe for Structural Fluctuations in Macromolecules. Biochemistry 1973, 12, 4161–4170. [Google Scholar] [CrossRef]
- Bian, Q.; Liu, J.; Tian, J.; Hu, Z. Binding of Genistein to Human Serum Albumin Demonstrated Using Tryptophan Fluorescence Quenching. Int. J. Biol. Macromol. 2004, 34, 275–279. [Google Scholar] [CrossRef] [PubMed]
- Wei, X.L.; Xiao, J.B.; Wang, Y.; Bai, Y. Which Model Based on Fluorescence Quenching Is Suitable to Study the Interaction between Trans-Resveratrol and BSA? Spectrochim. Acta A Mol. Biomol. Spectrosc. 2010, 75, 299–304. [Google Scholar] [CrossRef]
- Demirkaya, F.; Kadioglu, Y. Simple GC-FID Method Development and Validation for Determination of α-Tocopherol (Vitamin E) in Human Plasma. J. Biochem. Biophys. Methods 2007, 70, 363–368. [Google Scholar] [CrossRef]
- Carpineti, L.; Martinez, M.J.; Pilosof, A.M.R.; Pérez, O.E. β-Lactoglobulin-Carboxymethylcellulose Core-Shell Microparticles: Construction, Characterization and Isolation. J. Food Eng. 2014, 131, 65–74. [Google Scholar] [CrossRef]
- Montserrat-de la Paz, S.; Rodriguez-Martin, N.M.; Villanueva, A.; Pedroche, J.; Cruz-Chamorro, I.; Millan, F.; Millan-Linares, M.C. Evaluation of Anti-Inflammatory and Atheroprotective Properties of Wheat Gluten Protein Hydrolysates in Primary Human Monocytes. Foods 2020, 9, 854. [Google Scholar] [CrossRef]
- Rocha, G.F.; Kise, F.; Rosso, A.M.; Parisi, M.G. Potential Antioxidant Peptides Produced from Whey Hydrolysis with an Immobilized Aspartic Protease from Salpichroa Origanifolia Fruits. Food Chem. 2017, 237, 350–355. [Google Scholar] [CrossRef]
- Ruiz, G.A.; Xiao, W.; Van Boekel, M.; Minor, M.; Stieger, M. Effect of Extraction PH on Heat-Induced Aggregation, Gelation and Microstructure of Protein Isolate from Quinoa (Chenopodium quinoa Willd). Food Chem. 2016, 209, 203–210. [Google Scholar] [CrossRef] [PubMed]
- Brinegar, C.; Goundan, S. Isolation and Characterization of Chenopodin, the 11S Seed Storage Protein of Quinoa (Chenopodium quinoa). J. Agric. Food Chem 1993, 1883, 182–185. [Google Scholar] [CrossRef]
- Abugoch, L.E.; Martínez, N.E.; Añón, M.C. Influence of PH on Structure and Function of Amaranth (Amaranthus hypochondriacus) Protein Isolates. Cereal Chem. 2010, 87, 448–453. [Google Scholar] [CrossRef]
- Shahraki, S.; Heydari, A.; Saeidifar, M.; Gomroki, M. Biophysical and Computational Comparison on the Binding Affinity of Three Important Nutrients to β-Lactoglobulin: Folic Acid, Ascorbic Acid and Vitamin K3. J. Biomol. Struct. Dyn. 2018, 36, 3651–3665. [Google Scholar] [CrossRef]
- Maity, S.; Pal, S.; Sardar, S.; Sepay, N.; Parvej, H.; Chakraborty, J.; Chandra Halder, U. Multispectroscopic Analysis and Molecular Modeling to Investigate the Binding of Beta Lactoglobulin with Curcumin Derivatives. RSC Adv. 2016, 6, 112175–112183. [Google Scholar] [CrossRef]
- Vladislav Victorovich, K.; Tatyana Aleksandrovna, K.; Victor Vitoldovich, P.; Aleksander Nicolaevich, S.; Larisa Valentinovna, K.; Anastasia Aleksandrovna, A. Spectra of Tryptophan Fluorescence Are the Result of Co-Existence of Certain Most Abundant Stabilized Excited State and Certain Most Abundant Destabilized Excited State. Spectrochim. Acta A Mol. Biomol. Spectrosc. 2021, 257, 119784. [Google Scholar] [CrossRef] [PubMed]
- Mantovani, R.; Hamon, P.; Rousseau, F.; Tavares, G.; Mercadante, A.Z.; Croguennec, T.; Bouhallab, S.; Zerlotti Mer-Cadante, A.; Mantovani, R.A.; Tavares, G.M.; et al. Unraveling the Molecular Mechanisms Underlying Interactions between Caseins and Lutein. Food Res. Int. 2020, 138, 109781. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Chen, F.; Gao, C.; Feng, X.; Tang, X. Structure, Physical and Antioxidant Properties of Quinoa Protein /Hsian-Tsao Gum Composite Biodegradable Active Films. LWT 2022, 155, 112985. [Google Scholar] [CrossRef]
- Chilom, C.G.; Bacalum, M.; Stanescu, M.M.; Florescu, M. Insight into the Interaction of Human Serum Albumin with Folic Acid: A Biophysical Study. Spectrochim. Acta A Mol. Biomol. Spectrosc. 2018, 204, 648–656. [Google Scholar] [CrossRef]
- Gentile, L. Protein–Polysaccharide Interactions and Aggregates in Food Formulations. Curr. Opin. Colloid. Interface Sci. 2020, 48, 18–27. [Google Scholar] [CrossRef]
- Chandel, T.I.; Zaman, M.; Khan, M.V.; Ali, M.; Rabbani, G.; Ishtikhar, M.; Khan, R.H. A Mechanistic Insight into Protein-Ligand Interaction, Folding, Misfolding, Aggregation and Inhibition of Protein Aggregates: An Overview. Int. J. Biol. Macromol. 2018, 106, 1115–1129. [Google Scholar] [CrossRef] [PubMed]
- Shimoni, E. Using AFM to Explore Food Nanostructure. Curr. Opin. Colloid. Interface Sci. 2008, 13, 368–374. [Google Scholar] [CrossRef]
- Cao, H.; Sun, R.; Shi, J.; Li, M.; Guan, X.; Liu, J.; Huang, K.; Zhang, Y. Effect of Ultrasonic on the Structure and Quality Characteristics of Quinoa Protein Oxidation Aggregates. Ultrason. Sonochem. 2021, 77, 105685. [Google Scholar] [CrossRef] [PubMed]
- Orsini Delgado, M.C.; Nardo, A.; Pavlovic, M.; Rogniaux, H.; Añón, M.C.; Tironi, V.A. Identification and Characterization of Antioxidant Peptides Obtained by Gastrointestinal Digestion of Amaranth Proteins. Food Chem. 2016, 197, 1160–1167. [Google Scholar] [CrossRef] [PubMed]
- Gülçin, I. Antioxidant Properties of Resveratrol: A Structure-Activity Insight. Innov. Food Sci. Emerg. Technol. 2010, 11, 210–218. [Google Scholar] [CrossRef]
- Escribano, J.; Cabanes, J.; Jiménez-Atiénzar, M.; Ibañez-Tremolada, M.; Gómez-Pando, L.R.; García-Carmona, F.; Gandía-Herrero, F. Characterization of Betalains, Saponins and Antioxidant Power in Differently Colored Quinoa (Chenopodium quinoa) Varieties. Food Chem. 2017, 234, 285–294. [Google Scholar] [CrossRef]
- Zhang, X.; Wang, C.; Qi, Z.; Zhao, R.; Wang, C.; Zhang, T. Pea Protein Based Nanocarriers for Lipophilic Polyphenols: Spectroscopic Analysis, Characterization, Chemical Stability, Antioxidant and Molecular Docking. Food Res. Int. 2022, 160, 111713. [Google Scholar] [CrossRef]
- Chamizo-González, F.; Heredia, F.J.; Rodríguez-Pulido, F.J.; González-Miret, M.L.; Gordillo, B. Proteomic and Computational Characterisation of 11S Globulins from Grape Seed Flour By-Product and Its Interaction with Malvidin 3-Glucoside by Molecular Docking. Food Chem. 2022, 386, 132842. [Google Scholar] [CrossRef]
- Patnode, K.; Demchuk, Z.; Johnson, S.; Voronov, A.; Rasulev, B. Computational Protein-Ligand Docking and Experimental Study of Bioplastic Films from Soybean Protein, Zein, and Natural Modifiers. ACS Sustain. Chem. Eng. 2021, 9, 10740–10748. [Google Scholar] [CrossRef]
- Chen, S.; Han, Y.; Huang, J.; Dai, L.; Du, J.; McClements, D.J.; Mao, L.; Liu, J.; Gao, Y. Fabrication and Characterization of Layer-by-Layer Composite Nanoparticles Based on Zein and Hyaluronic Acid for Codelivery of Curcumin and Quercetagetin. ACS Appl. Mater. Interfaces 2019, 11, 16922–16933. [Google Scholar] [CrossRef]
- Ferina, J.; Daggett, V. Visualizing Protein Folding and Unfolding. J. Mol. Biol. 2019, 431, 1540–1564. [Google Scholar] [CrossRef]
- Li, X.; Ni, T. Probing the Binding Mechanisms of α-Tocopherol to Trypsin and Pepsin Using Isothermal Titration Calorimetry, Spectroscopic, and Molecular Modeling Methods. J. Biol. Phys. 2016, 42, 415–434. [Google Scholar] [CrossRef]
- Wan, Z.L.; Wang, L.Y.; Wang, J.M.; Yuan, Y.; Yang, X.Q. Synergistic Foaming and Surface Properties of a Weakly Interacting Mixture of Soy Glycinin and Biosurfactant Stevioside. J. Agric. Food Chem. 2014, 62, 6834–6843. [Google Scholar] [CrossRef] [PubMed]
- Yang, T.; Yang, H.; Fan, Y.; Li, B.; Hou, H. Interactions of Quercetin, Curcumin, Epigallocatechin Gallate and Folic Acid with Gelatin. Int. J. Biol. Macromol. 2018, 118, 124–131. [Google Scholar] [CrossRef]
- Jiang, X.Y.; Li, W.X.; Cao, H. Study of the Interaction between Trans-Resveratrol and BSA by the Multi-Spectroscopic Method. J. Solut. Chem. 2008, 37, 1609–1623. [Google Scholar] [CrossRef]
- Li, W.; Bi, D.; Yi, J.; Yao, L.; Cao, J.; Yang, P.; Li, M.; Wu, Y.; Xu, H.; Hu, Z.; et al. Soy Protein Isolate-Polyguluronate Nanoparticles Loaded with Resveratrol for Effective Treatment of Colitis. Food Chem. 2023, 410, 135418. [Google Scholar] [CrossRef]
- Khan, M.A.; Fang, Z.; Wusigale; Cheng, H.; Gao, Y.; Deng, Z.; Liang, L. Encapsulation and Protection of Resveratrol in Kafirin and Milk Protein Nanoparticles. Int. J. Food Sci. Technol. 2019, 54, 2998–3007. [Google Scholar] [CrossRef]
- Carpentier, J.; Conforto, E.; Chaigneau, C.; Vendeville, J.E.; Maugard, T. Microencapsulation and Controlled Release of α-Tocopherol by Complex Coacervation between Pea Protein and Tragacanth Gum: A Comparative Study with Arabic and Tara Gums. Innov. Food Sci. Emerg. Technol. 2022, 77, 102951. [Google Scholar] [CrossRef]
- Xu, W.; Lv, K.; Mu, W.; Zhou, S.; Yang, Y. Encapsulation of α-Tocopherol in Whey Protein Isolate/Chitosan Particles Using Oil-in-Water Emulsion with Optimal Stability and Bioaccessibility. LWT 2021, 148, 111724. [Google Scholar] [CrossRef]
- Penalva, R.; Esparza, I.; Agüeros, M.; Gonzalez-Navarro, C.J.; Gonzalez-Ferrero, C.; Irache, J.M. Casein Nanoparticles as Carriers for the Oral Delivery of Folic Acid. Food Hydrocoll. 2015, 44, 399–406. [Google Scholar] [CrossRef]
- Andreeva, Y.I.; Drozdov, A.S.; Fakhardo, A.F.; Cheplagin, N.A.; Shtil, A.A.; Vinogradov, V.V. The Controllable Destabilization Route for Synthesis of Low Cytotoxic Magnetic Nanospheres with Photonic Response. Sci. Rep. 2017, 7, 11343. [Google Scholar] [CrossRef] [PubMed]
- Fröhlich, E. The Role of Surface Charge in Cellular Uptake and Cytotoxicity of Medical Nanoparticles. Int. J. Nanomed. 2012, 7, 5577–5591. [Google Scholar] [CrossRef] [PubMed]
- Relkin, P.; Shukat, R. Food Protein Aggregates as Vitamin-Matrix Carriers: Impact of Processing Conditions. Food Chem. 2012, 134, 2141–2148. [Google Scholar] [CrossRef] [PubMed]
- Tiwari, K.; Singh, G.; Singh, S.K. Purification and Structural Characterization of N-Terminal 190 Amino Acid Deleted Essential Mammalian Protein; Transcription Termination Factor 1. ACS Omega 2022, 7, 45165–45173. [Google Scholar] [CrossRef]
- Rashidinejad, A.; Birch, E.J.; Sun-Waterhouse, D.; Everett, D.W. Addition of Milk to Tea Infusions: Helpful or Harmful? Evidence from in Vitro and in Vivo Studies on Antioxidant Properties. Crit. Rev. Food Sci. Nutr. 2017, 57, 3188–3196. [Google Scholar] [CrossRef] [PubMed]
- Pérez, O.E.; Carrera-Sánchez, C.; Rodríguez-Patino, J.M.; Pilosof, A.M.R. Adsorption Dynamics and Surface Activity at Equilibrium of Whey Proteins and Hydroxypropyl-Methyl-Cellulose Mixtures at the Air-Water Interface. Food Hydrocoll. 2007, 21, 794–803. [Google Scholar] [CrossRef]
- Wen, C.; Zhang, J.; Zhang, H.; Duan, Y.; Ma, H. Plant Protein-Derived Antioxidant Peptides: Isolation, Identification, Mechanism of Action and Application in Food Systems: A Review. Trends Food Sci. Technol. 2020, 105, 308–322. [Google Scholar] [CrossRef]
- Elias, R.J.; Kellerby, S.S.; Decker, E.A. Antioxidant Activity of Proteins and Peptides. Crit. Rev. Food Sci. Nutr. 2008, 48, 430–441. [Google Scholar] [CrossRef]
- Biskup, I.; Golonka, I.; Gamian, A.; Sroka, Z. Antioxidant activity of selected phenols estimated by ABTS and FRAP methods. Adv. Hyg. Exp. Med. 2013, 67, 958–963. [Google Scholar] [CrossRef]
- Schlesier, K.; Harwat, M.; Böhm, V.; Bitsch, R. Assessment of Antioxidant Activity by Using Different in Vitro Methods. Free Radic. Res. 2002, 36, 177–187. [Google Scholar] [CrossRef]





| Complex | Binding Energy (Kcal/Mol) | Hydrogen Bonds | Hydrophobic Interactions | Saline Bridge | Total |
|---|---|---|---|---|---|
| 11S-RSV | −5.6 | 4 | 4 | 0 | 8 |
| 11S-TOC | −6.2 | 1 | 10 | 1 | 12 |
| Bioactive Compound | n | Kd (µM) | Ka (M−1) | ΔH (kcal/mol) | ΔS (cal/K × mol) | ΔG (kcal/mol) |
|---|---|---|---|---|---|---|
| TOC | 0.85 ± 0.03 | 340 ± 40 | (2.9 ± 0.8) × 103 | 11 ± 1 | 25 ± 3 | 4.7 ± 0.2 |
| RSV | 0.91 ± 0.04 | 48.2 ± 1 | (2.1 ± 0.3) × 104 | 10 ± 2 | 15 ± 1 | 5.9 ± 0.1 |
| TOC | RSV | |
|---|---|---|
| Stern–Volmer model | ||
| Ksv (M−1) | 4800 | 385,700 |
| Kq (M−1 s−1) | 1.66 × 1012 | 1.13 × 1014 |
| n | 1.04 | 2.35 |
| R2 | 0.9931 | 0.9406 |
| Scatchard model | ||
| Ks (M−1) | 151,478 | 3,852,674 |
| n | 8.87 | 0.90 |
| R2 | 0.9422 | 0.9707 |
| Förster resonance energy transfer (FRET) | ||
| J (λ) (cm3/M) | 1.14 × 1013 | 2.67 × 1013 |
| R0 (nm) | 5.39 | 6.22 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rubinstein, A.J.; Garcia Liñares, G.; Boeris, V.; Pérez, O.E. An Innovative Bio-Vehicle for Resveratrol and Tocopherol Based on Quinoa 11S Globulin—Nanocomplex Design and Characterization. Pharmaceutics 2024, 16, 1118. https://doi.org/10.3390/pharmaceutics16091118
Rubinstein AJ, Garcia Liñares G, Boeris V, Pérez OE. An Innovative Bio-Vehicle for Resveratrol and Tocopherol Based on Quinoa 11S Globulin—Nanocomplex Design and Characterization. Pharmaceutics. 2024; 16(9):1118. https://doi.org/10.3390/pharmaceutics16091118
Chicago/Turabian StyleRubinstein, Alejandra J., Guadalupe Garcia Liñares, Valeria Boeris, and Oscar E. Pérez. 2024. "An Innovative Bio-Vehicle for Resveratrol and Tocopherol Based on Quinoa 11S Globulin—Nanocomplex Design and Characterization" Pharmaceutics 16, no. 9: 1118. https://doi.org/10.3390/pharmaceutics16091118





