Lipid-Based Nanoparticles Fused with Natural Killer Cell Plasma Membrane Proteins for Triple-Negative Breast Cancer Therapy
Abstract
1. Introduction
2. Experimental Methods
2.1. Plasmid
2.2. Expression and Purification of Recombinant Human Protamine Protein (PA)
2.3. NK-92 Culture and Plasma Membrane Protein Isolation
2.4. Cell Culture
2.5. Synthesis and Characterization of NK-LNP with pDNA
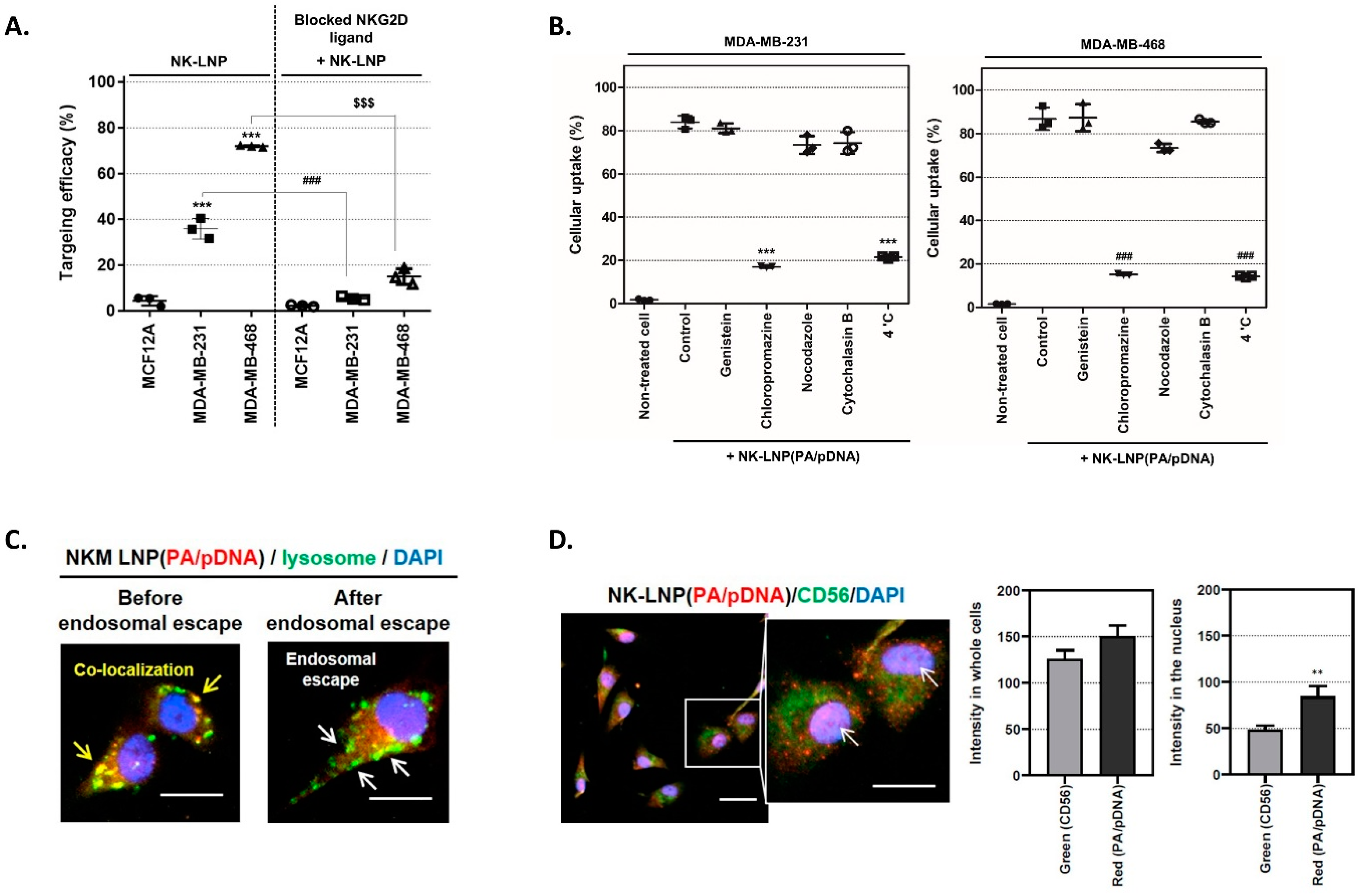
2.6. Targeting and Transfection Efficiency of NK-LNP
2.7. Fluorescence Image Analysis
2.8. Western Blot Analysis
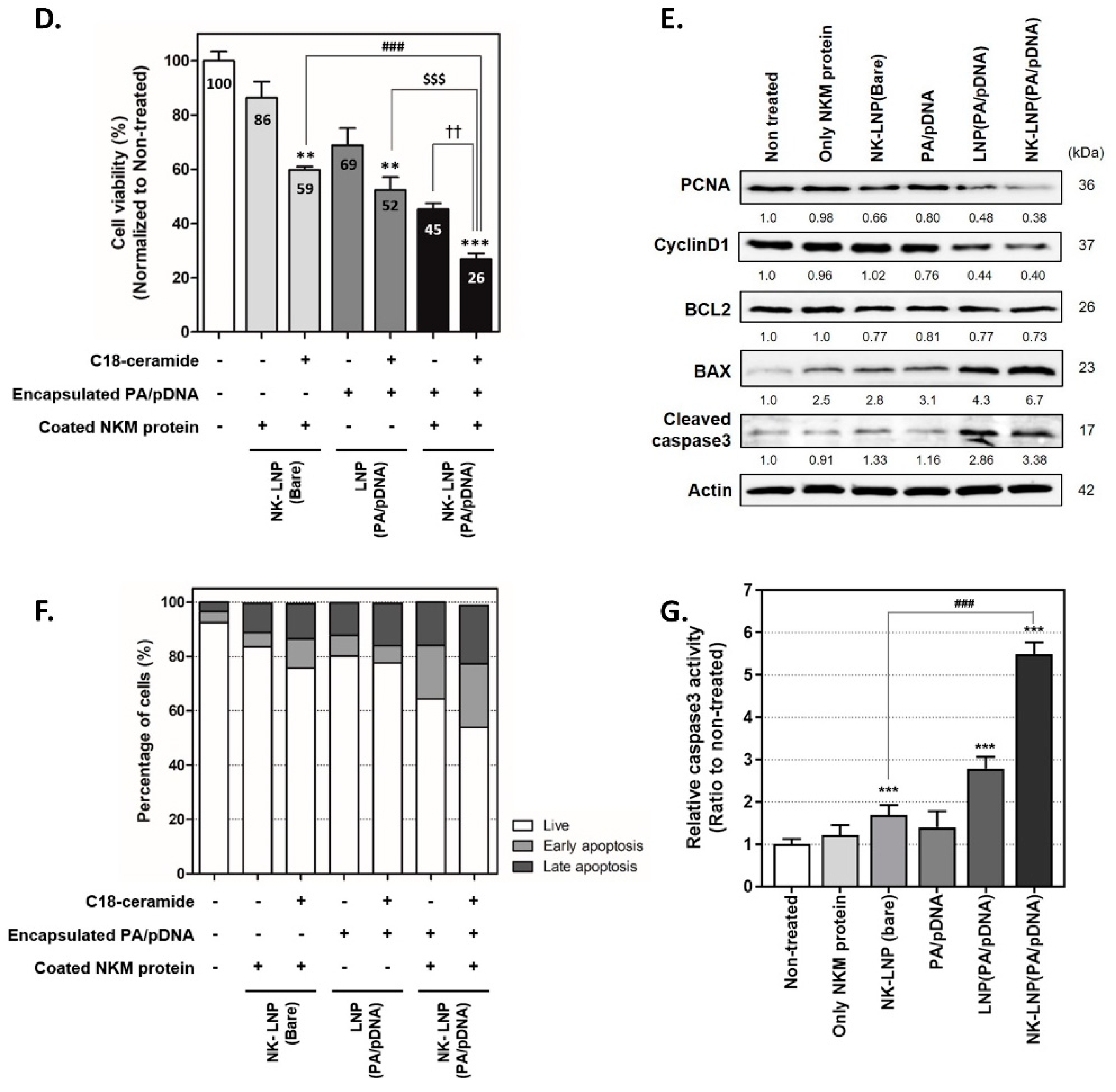
2.9. Cell Viability, Migration, Invasion, and Apoptosis Assay
2.10. THP1 Immune Cell Real-Time PCR Analysis and Cytokine Detection
2.11. Statistical Analysis
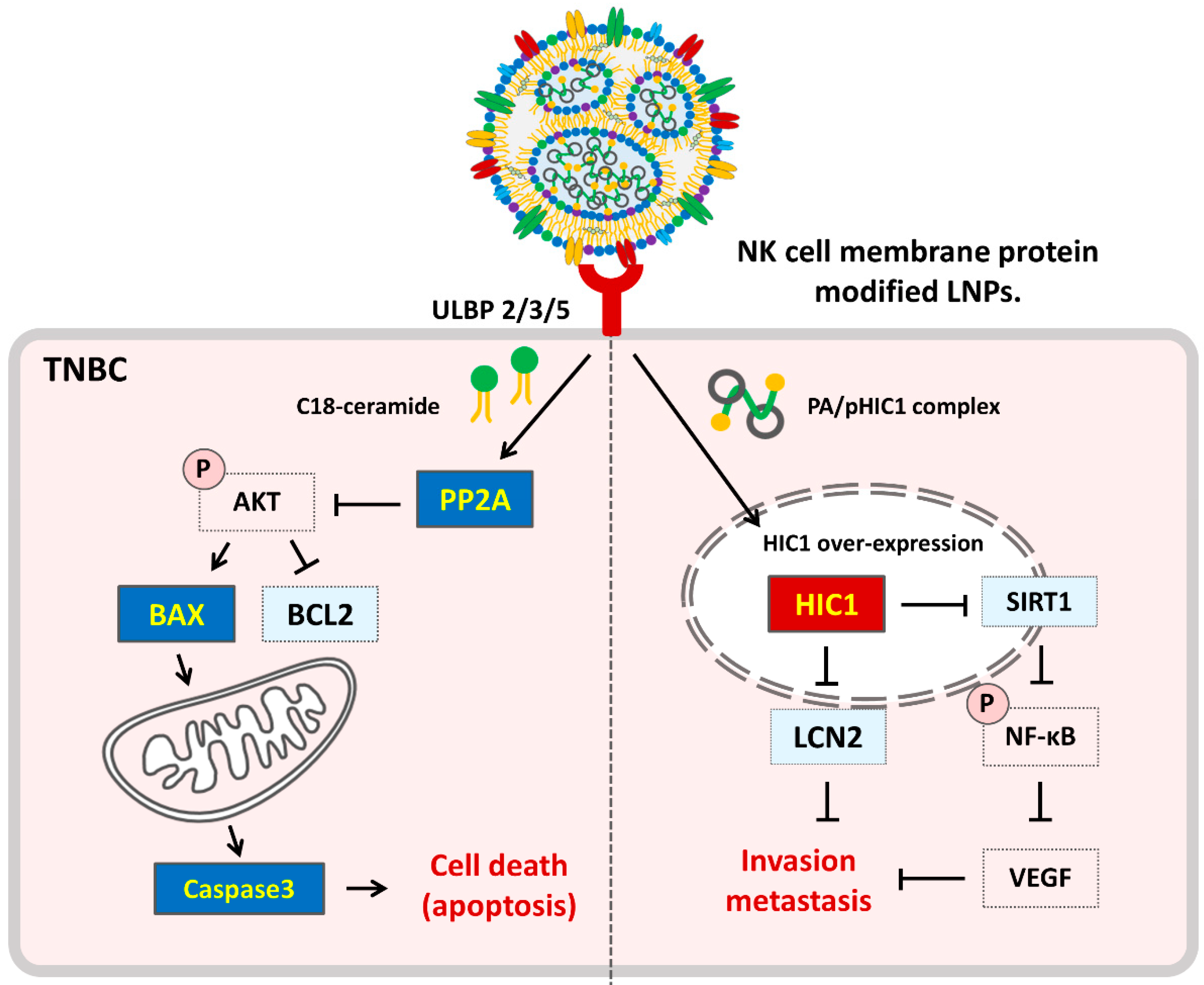
3. Results
3.1. Isolation of NK Cell Plasma Membrane Proteins and Complexes with Protamine
3.2. Synthesis and Characterization of NK-LNP(PA/pDNA)
3.3. Specific Targeting Ability and Cellular Uptake Mechanism
3.4. Therapeutic Effect of NK-LNP(PA/pDNA) in TNBC
3.5. Immune Cell Regulation Effect of NK-LNP(PA/pDNA)
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Loibl, S.; Poortmans, P.; Morrow, M.; Denkert, C.; Curigliano, G. Breast cancer. Lancet 2021, 397, 1750–1769. [Google Scholar] [CrossRef] [PubMed]
- Manjunath, M.; Choudhary, B. Triple-negative breast cancer: A run-through of features, classification and current therapies. Oncol. Lett. 2021, 22, 512–532. [Google Scholar] [CrossRef] [PubMed]
- Yin, L.; Duan, J.J.; Bian, X.W.; Yu, S.C. Triple-negative breast cancer molecular subtyping and treatment progress. Breast Cancer Res. 2020, 22, 61–73. [Google Scholar] [CrossRef] [PubMed]
- Hsu, J.Y.; Chang, C.J.; Cheng, J.S. Survival, treatment regimens and medical costs of women newly diagnosed with metastatic triple-negative breast cancer. Sci. Rep. 2022, 12, 729–736. [Google Scholar] [CrossRef]
- Mittendorf, E.A.; Philips, A.V.; Meric-Bernstam, F.; Qiao, N.; Wu, Y.; Harrington, S.; Su, X.; Wang, Y.; Gonzalez-Angulo, A.M.; Akcakanat, A.; et al. PD-L1 expression in triple-negative breast cancer. Cancer Immunol. Res. 2014, 2, 361–370. [Google Scholar] [CrossRef]
- Kagihara, J.A.; Andress, M.; Diamond, J.R. Nab-paclitaxel and atezolizumab for the treatment of PD-L1-positive, metastatic triple-negative breast cancer: Review and future directions. Expert Rev. Precis. Med. Drug Dev. 2020, 5, 59–65. [Google Scholar] [CrossRef]
- Fleuriel, C.; Touka, M.; Boulay, G.; Guérardel, C.; Rood, B.R.; Leprince, D. HIC1 (Hypermethylated in Cancer 1) epigenetic silencing in tumors. Int. J. Biochem. Cell Biol. 2009, 41, 26–33. [Google Scholar] [CrossRef]
- Li, P.; Liu, X.; Dong, Z.; Ling, Z. Epigenetic silencing of HIC1 promotes epithelial-mesenchymal transition and drives progression in esophageal squamous cell carcinoma. Oncotarget 2015, 6, 38151–38165. [Google Scholar] [CrossRef]
- Chen, W.Y.; Wang, D.H.; Yen, R.C.; Luo, J.; Gu, W.; Baylin, S.B. Tumor suppressor HIC1 directly regulates SIRT1 to modulate p53-dependent DNA-damage responses. Cell 2005, 123, 437–448. [Google Scholar] [CrossRef]
- Dehennaut, V.; Loison, I.; Boulay, G.; Van Rechem, C.; Leprince, D. Identification of p21 (CIP1/WAF1) as a direct target gene of HIC1 (Hypermethylated in Cancer 1). Biochem. Biophys. Res. Commun. 2013, 430, 49–53. [Google Scholar] [CrossRef]
- Hu, C.; Yang, K.; Li, M.; Huang, W.; Zhang, F.; Wang, H. Lipocalin 2: A potential therapeutic target for breast cancer metastasis. Onco Targets Ther. 2020, 11, 8099–8106. [Google Scholar] [CrossRef]
- Cheng, G.; Sun, X.; Wang, J.; Xiao, G.; Wang, X.; Fan, X.; Zu, L.; Hao, M.; Qu, Q.; Mao, Y.; et al. HIC1 silencing in triple-negative breast cancer drives progression through misregulation of LCN2. Cancer Res. 2014, 74, 862–872. [Google Scholar] [CrossRef] [PubMed]
- Grösch, S.; Schiffmann, S.; Geisslinger, G. Chain length-specific properties of ceramides. Prog. Lipid Res. 2012, 51, 50–62. [Google Scholar] [CrossRef] [PubMed]
- Stith, J.L.; Velazquez, F.N.; Obeid, L.M. Advances in determining signaling mechanisms of ceramide and role in disease. J. Lipid Res. 2019, 60, 913–918. [Google Scholar] [CrossRef] [PubMed]
- Koybasi, S.; Senkal, C.E.; Sundararaj, K.; Spassieva, S.; Bielawski, J.; Osta, W.; Day, T.A.; Jiang, J.C.; Jazwinski, S.M.; Hannun, Y.A.; et al. Defects in cell growth regulation by C18:0-ceramide and longevity assurance gene 1 in human head and neck squamous cell carcinomas. J. Biol. Chem. 2004, 79, 44311–44319. [Google Scholar] [CrossRef] [PubMed]
- Park, W.J.; Park, J.W. The effect of altered sphingolipid acyl chain length on various disease models. Biol. Chem. 2015, 396, 693–705. [Google Scholar] [CrossRef]
- Wang, Z.; Wen, L.; Zhu, F.; Wang, Y.; Xie, Q.; Chen, Z.; Li, Y. Overexpression of ceramide synthase 1 increases C18-ceramide and leads to lethal autophagy in human glioma. Oncotarget 2017, 8, 104022–104036. [Google Scholar] [CrossRef]
- Smyth, M.J.; Hayakawa, Y.; Takeda, K.; Yagita, H. New aspects of natural-killer-cell surveillance and therapy of cancer. Nat. Rev. Cancer 2022, 2, 850–861. [Google Scholar] [CrossRef]
- Wang, W.; Jiang, J.; Wu, C. CAR-NK for tumor immunotherapy: Clinical transformation and future prospects. Cancer Lett. 2020, 472, 175–180. [Google Scholar] [CrossRef]
- Liu, S.; Galat, V.; Galat, Y.; Lee, Y.K.A.; Wainwright, D.; Wu, J. NK cell-based cancer immunotherapy: From basic biology to clinical development. J. Hematol. Oncol. 2021, 14, 7–23. [Google Scholar] [CrossRef]
- Pitchaimani, A.; Nguyen, T.D.T.; Aryal, S. Natural killer cell membrane infused biomimetic liposomes for targeted tumor therapy. Biomaterials 2018, 160, 124–137. [Google Scholar] [CrossRef] [PubMed]
- Deng, G.; Sun, Z.; Li, S.; Peng, X.; Li, W.; Zhou, L.; Ma, Y.; Gong, P.; Cai, L. Cell-membrane immunotherapy based on natural killer cell membrane coated nanoparticles for the effective inhibition of primary and abscopal tumor growth. ACS Nano 2018, 26, 12096–12108. [Google Scholar] [CrossRef] [PubMed]
- Pitchaimani, A.; Nguyen, T.D.T.; Marasini, R.; Eliyapura, A.; Azizi, T.; Jaberi-Douraki, M.; Aryal, S. Biomimetic natural killer membrane camouflaged polymeric nanoparticle for targeted bioimaging. Adv. Funct. Mater. 2019, 29, 1806817. [Google Scholar] [CrossRef]
- Gong, P.; Wang, Y.; Zhang, P.; Yang, Z.; Deng, W.; Sun, Z.; Yang, M.; Li, X.; Ma, G.; Deng, G.; et al. Immunocyte membrane-coated nanoparticles for cancer immunotherapy. Cancers 2020, 13, 77–93. [Google Scholar] [CrossRef]
- Smith, M.; Young, H.; Hurlstone, A.; Wellbrock, C. Differentiation of THP1 cells into macrophages for transwell co-culture assay with melanoma cells. BioProtocol 2015, 5, e1638–e1644. [Google Scholar] [CrossRef]
- Zhang, Y.; Shi, L.; Mei, H.; Zhang, J.; Zhu, Y.; Han, X.; Zhu, D. Inflamed macrophage microvesicles induce insulin resistance in human adipocytes. Nutr. Metab. 2015, 12, 21–34. [Google Scholar] [CrossRef] [PubMed]
- Berges, C.; Naujokat, C.; Tinapp, S.; Wieczore, H.; Höh, A.; Sadeghi, M.; Opelz, G.; Daniel, V. A cell line model for the differentiation of human dendritic cells. Biochem. Biophys. Res. Commun. 2005, 333, 896–907. [Google Scholar] [CrossRef]
- Han, Y.; Xie, W.; Song, D.G.; Powell, D.J., Jr. Control of triple-negative breast cancer using ex vivo self-enriched, costimulated NKG2D CAR T cells. J. Hematol. Oncol. 2018, 11, 92–104. [Google Scholar] [CrossRef]
- Glover, D.J.; Ng, S.M.; Mechler, A.; Martin, L.L.; Jans, D.A. Multifunctional protein nanocarriers for targeted nuclear gene delivery in nondividing cells. FASEB J. 2009, 23, 2996–3006. [Google Scholar] [CrossRef]
- Zhang, L.; Wang, P.; Feng, Q.; Wang, N.; Chen, Z.; Huang, Y.; Zheng, W.; Jiang, X. Lipid nanoparticle-mediated efficient delivery of CRISPR/Cas9 for tumor therapy. NPG Asia Mater. 2017, 9, e441–e448. [Google Scholar] [CrossRef]
- Won, E.J.; Park, H.; Chang, S.H.; Kim, J.H.; Kwon, H.; Cho, Y.S.; Yoon, T.J. One-shot dual gene editing for drug-resistant pancreatic cancer therapy. Biomaterials 2021, 279, 121252–121261. [Google Scholar] [CrossRef] [PubMed]
- Alizadeh, J.; da Silva Rosa, S.C.; Weng, X.; Jacobs, J.; Lorzadeh, S.; Ravandi, A.; Vitorino, R.; Pecic, S.; Zivkovic, A.; Stark, H.; et al. Ceramides and ceramide synthases in cancer: Focus on apoptosis and autophagy. Eur. J. Cell Biol. 2023, 102, 151337–151354. [Google Scholar] [CrossRef]
- Zhu, Q.Y.; Wang, Z.; Ji, C.; Cheng, L.; Yang, Y.L.; Ren, J.; Jin, Y.H.; Wang, Q.J.; Gu, X.J.; Bi, Z.G.; et al. C6-ceramide synergistically potentiates the anti-tumor effects of histone deacetylase inhibitors via AKT dephosphorylation and α-tubulin hyperacetylation both in vitro and in vivo. Cell Death Dis. 2011, 2, e117–e128. [Google Scholar] [CrossRef] [PubMed]
- Kadomoto, S.; Izumi, K.; Mizokami, A. Macrophage polarity and disease control. Int. J. Mol. Sci. 2021, 23, 144. [Google Scholar] [CrossRef]
- Xie, D.; Wang, Q.; Wu, G. Research progress in inducing immunogenic cell death of tumor cells. Front Immunol. 2022, 13, 1017400–1017420. [Google Scholar]
- Seong, J.; Kim, K. Activation of cellular players in adaptive immunity via exogenous delivery of tumor cell lysates. Pharmaceutics 2022, 14, 1358. [Google Scholar] [CrossRef] [PubMed]
- Kroemer, G.; Galluzzi, L.; Keep, O.; Zitvogel, L. Immunogenic cell death in cancer therapy. Annu. Rev. Immunol. 2013, 31, 51–72. [Google Scholar] [CrossRef]
- Hernandez, C.; Huebener, P.; Schwabe, R.F. Damage-associated molecular patterns in cancer: A double-edged sword. Oncogene 2016, 17, 5931–5941. [Google Scholar] [CrossRef]
- Aleksandra, M.D.; Shaun, M.; Abhishek, D.G.; Patrizia, A. Immature, semi-mature, and fully mature dendritic cells: Toward a DC-Cancer cells interface that augments anticancer immunity. Front Immunol. 2013, 4, 438–451. [Google Scholar]
- Garufi, G.; Palazzo, A.; Paris, I.; Orlandi, A.; Cassano, A.; Tortora, G.; Scambia, G.; Bria, E.; Carbognin, L. Neoadjuvant therapy for triple-negative breast cancer: Potential predictive biomarkers of activity and efficacy of platinum chemotherapy, PARP- and immune-checkpoint-inhibitors. Expert Opin. Pharmacother. 2020, 21, 687–699. [Google Scholar] [CrossRef]




Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Won, E.-J.; Lee, M.; Lee, E.-K.; Baek, S.-H.; Yoon, T.-J. Lipid-Based Nanoparticles Fused with Natural Killer Cell Plasma Membrane Proteins for Triple-Negative Breast Cancer Therapy. Pharmaceutics 2024, 16, 1142. https://doi.org/10.3390/pharmaceutics16091142
Won E-J, Lee M, Lee E-K, Baek S-H, Yoon T-J. Lipid-Based Nanoparticles Fused with Natural Killer Cell Plasma Membrane Proteins for Triple-Negative Breast Cancer Therapy. Pharmaceutics. 2024; 16(9):1142. https://doi.org/10.3390/pharmaceutics16091142
Chicago/Turabian StyleWon, Eun-Jeong, Myungchul Lee, Eui-Kyung Lee, Seung-Hoon Baek, and Tae-Jong Yoon. 2024. "Lipid-Based Nanoparticles Fused with Natural Killer Cell Plasma Membrane Proteins for Triple-Negative Breast Cancer Therapy" Pharmaceutics 16, no. 9: 1142. https://doi.org/10.3390/pharmaceutics16091142
APA StyleWon, E.-J., Lee, M., Lee, E.-K., Baek, S.-H., & Yoon, T.-J. (2024). Lipid-Based Nanoparticles Fused with Natural Killer Cell Plasma Membrane Proteins for Triple-Negative Breast Cancer Therapy. Pharmaceutics, 16(9), 1142. https://doi.org/10.3390/pharmaceutics16091142









