Advances in Nanoparticles as Non-Viral Vectors for Efficient Delivery of CRISPR/Cas9
Abstract
:1. Introduction
2. Mechanism of CRISPR/Cas9 Genome Editing System
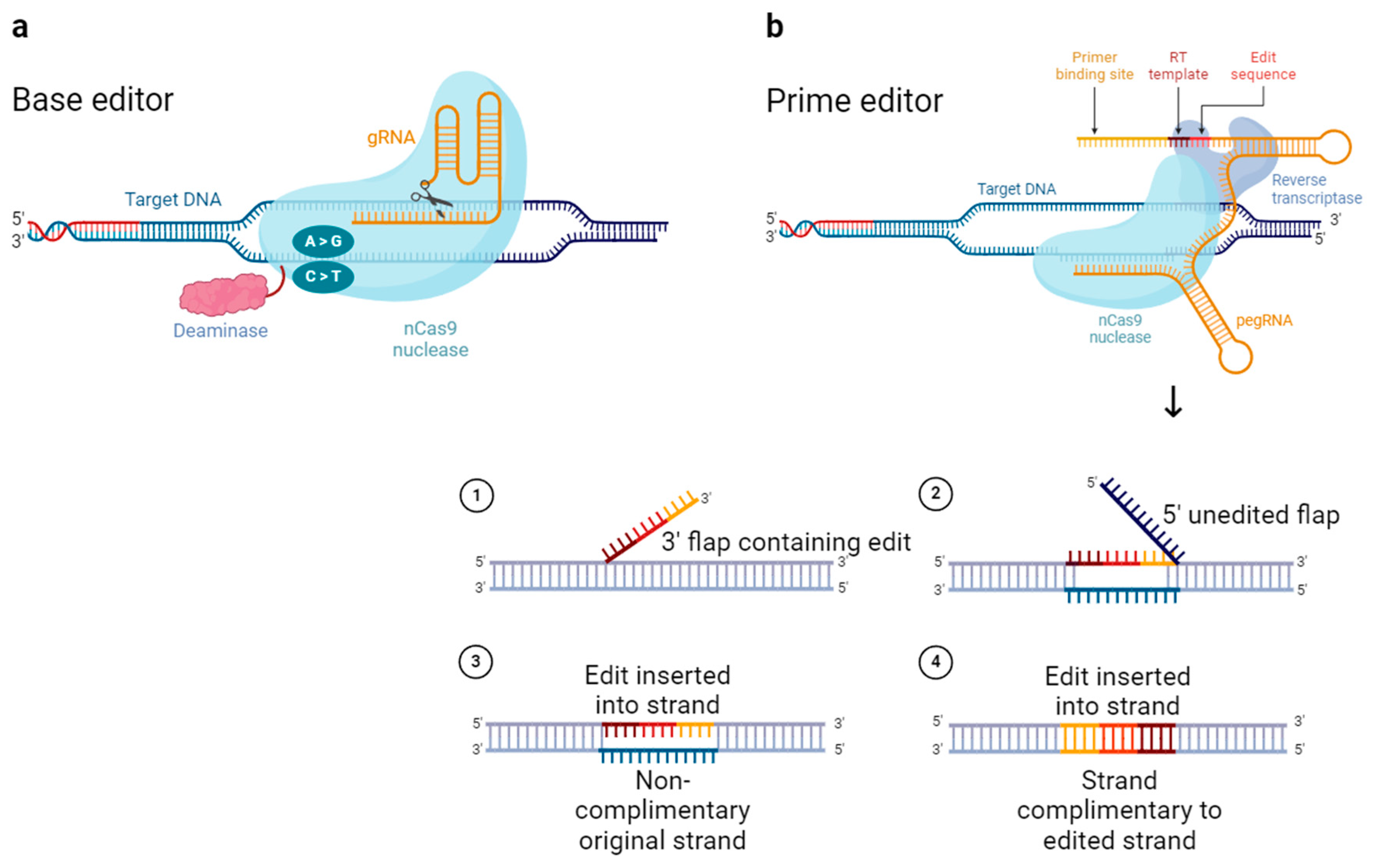
3. Advanced Gene Editing Tools Based on CRISPR/Cas9
4. Miniature CRISPR/Cas9
5. Clinical Trials of CRISPR System
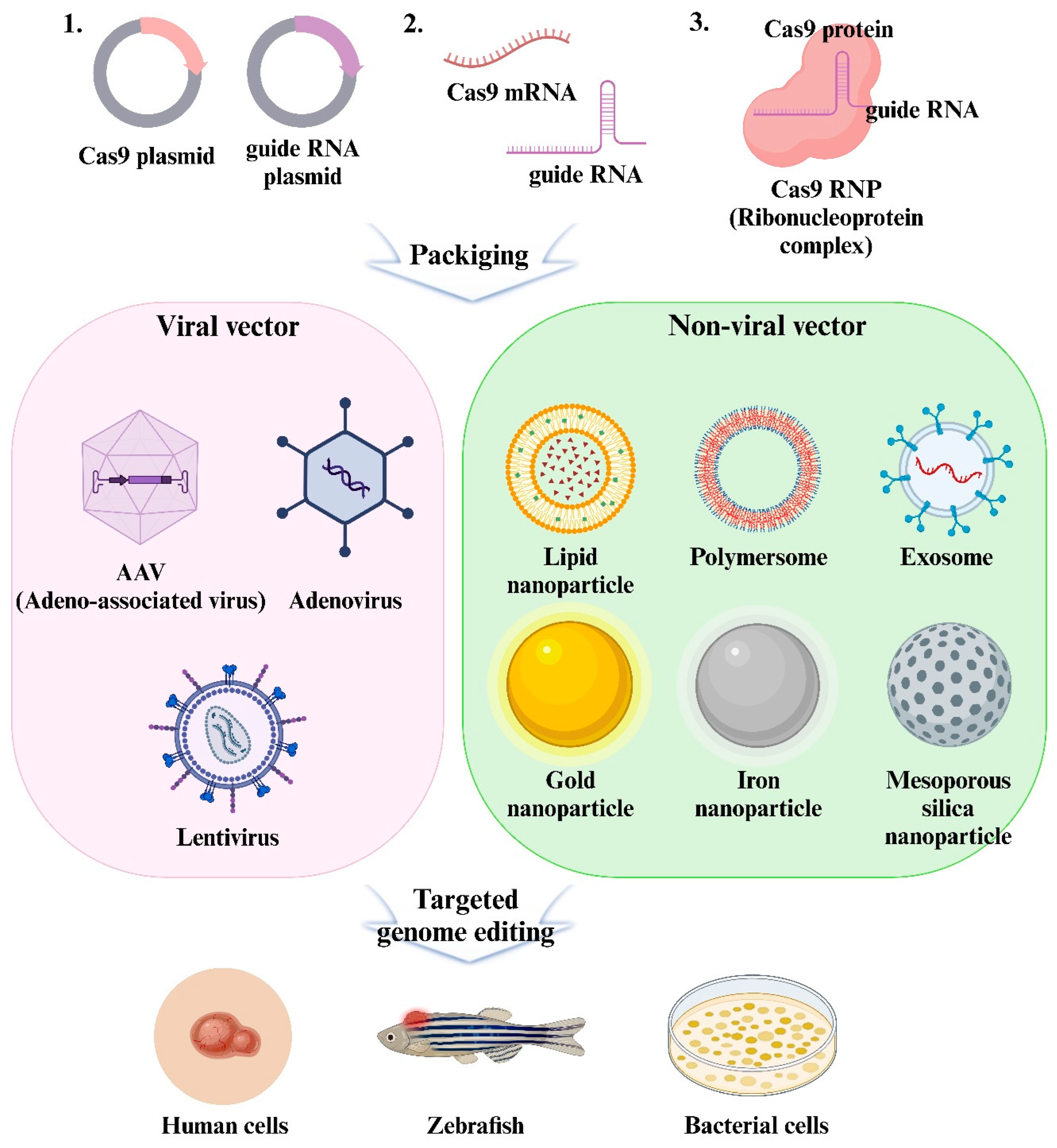
6. Viral Delivery of CRISPR Reagents
6.1. Delivery Format of CRISPR/Cas9
6.2. Viruses Used for Viral Delivery
6.2.1. Adenovirus (AdV)
6.2.2. Adeno-Associated Virus (AAV)
| Type of Delivery System | Delivery Efficiency | Packaging Capacity | Major Advantages | Major Limitations | Reference |
|---|---|---|---|---|---|
| AdV | Medium | 8~36 kb | Large packaging capacity; No-integration | Innate immune response | [52] |
| AAV | Medium | ~4.7 kb | Low Immunogenicity; No-integration | Low packaging capacity | [54] |
| LV | High | ~8 kb | Large packaging capacity; Efficient delivery | Random integration; High off-target effect | [51] |
6.2.3. Lentivirus (LV)
6.3. Non-Viral Vectors to Overcome the Limitations of Viral Vectors
7. Non-Viral Vectors Delivery of CRISPR Reagents
7.1. Non-Viral Delivery System
7.1.1. Lipid-Based Nanoparticle

7.1.2. Polymer-Based Nanoparticles

7.1.3. Inorganic Nanoparticles
Gold Nanoparticles (AuNPs)
Magnetic Nanoparticles

Mesoporous Silica Nanoparticles (MSNs)
7.1.4. Extracellular Vesicles-Based Nanoparticles
8. Conclusions and Future Perspectives
Author Contributions
Funding
Conflicts of Interest
References
- Wang, S.-W.; Gao, C.; Zheng, Y.-M.; Yi, L.; Lu, J.-C.; Huang, X.-Y.; Cai, J.-B.; Zhang, P.-F.; Cui, Y.-H.; Ke, A.-W. Current applications and future perspective of CRISPR/Cas9 gene editing in cancer. Mol. Cancer 2022, 21, 57. [Google Scholar] [CrossRef] [PubMed]
- Khan, S.; Hossain, M.K. Classification and properties of nanoparticles. In Nanoparticle-Based Polymer Composites; Elsevier: Amsterdam, The Netherlands, 2022; pp. 15–54. [Google Scholar]
- Kambhampati, P. Nanoparticles, nanocrystals, and quantum dots: What are the implications of size in colloidal nanoscale materials? J. Phys. Chem. Lett. 2021, 12, 4769–4779. [Google Scholar] [CrossRef] [PubMed]
- Glass, Z.; Lee, M.; Li, Y.; Xu, Q. Engineering the delivery system for CRISPR-based genome editing. Trends Biotechnol. 2018, 36, 173–185. [Google Scholar] [CrossRef]
- Abdellatif, A.A.; Mohammed, H.A.; Khan, R.A.; Singh, V.; Bouazzaoui, A.; Yusuf, M.; Akhtar, N.; Khan, M.; Al-Subaiyel, A.; Mohammed, S.A. Nano-scale delivery: A comprehensive review of nano-structured devices, preparative techniques, site-specificity designs, biomedical applications, commercial products, and references to safety, cellular uptake, and organ toxicity. Nanotechnol. Rev. 2021, 10, 1493–1559. [Google Scholar] [CrossRef]
- Duan, L.; Ouyang, K.; Xu, X.; Xu, L.; Wen, C.; Zhou, X.; Qin, Z.; Xu, Z.; Sun, W.; Liang, Y. Nanoparticle delivery of CRISPR/Cas9 for genome editing. Front. Genet. 2021, 12, 673286. [Google Scholar] [CrossRef]
- Datsenko, K.A.; Pougach, K.; Tikhonov, A.; Wanner, B.L.; Severinov, K.; Semenova, E. Molecular memory of prior infections activates the CRISPR/Cas adaptive bacterial immunity system. Nat. Commun. 2012, 3, 945. [Google Scholar] [CrossRef]
- Nuñez, J.K.; Kranzusch, P.J.; Noeske, J.; Wright, A.V.; Davies, C.W.; Doudna, J.A. Cas1–Cas2 complex formation mediates spacer acquisition during CRISPR–Cas adaptive immunity. Nat. Struct. Mol. Biol. 2014, 21, 528–534. [Google Scholar] [CrossRef]
- Hillary, V.E.; Ceasar, S.A. A review on the mechanism and applications of CRISPR/Cas9/Cas12/Cas13/Cas14 proteins utilized for genome engineering. Mol. Biotechnol. 2023, 65, 311–325. [Google Scholar] [CrossRef]
- Jiang, F.; Doudna, J.A. CRISPR–Cas9 structures and mechanisms. Annu. Rev. Biophys. 2017, 46, 505–529. [Google Scholar] [CrossRef]
- Sharma, G.; Sharma, A.R.; Bhattacharya, M.; Lee, S.-S.; Chakraborty, C. CRISPR-Cas9: A preclinical and clinical perspective for the treatment of human diseases. Mol. Ther. 2021, 29, 571–586. [Google Scholar] [CrossRef]
- Shams, A.; Higgins, S.A.; Fellmann, C.; Laughlin, T.G.; Oakes, B.L.; Lew, R.; Kim, S.; Lukarska, M.; Arnold, M.; Staahl, B.T. Comprehensive deletion landscape of CRISPR-Cas9 identifies minimal RNA-guided DNA-binding modules. Nat. Commun. 2021, 12, 5664. [Google Scholar] [CrossRef] [PubMed]
- Babu, K.; Kathiresan, V.; Kumari, P.; Newsom, S.; Parameshwaran, H.P.; Chen, X.; Liu, J.; Qin, P.Z.; Rajan, R. Coordinated actions of Cas9 HNH and RuvC nuclease domains are regulated by the bridge helix and the target DNA sequence. Biochemistry 2021, 60, 3783–3800. [Google Scholar] [CrossRef]
- Wang, J.; Arantes, P.R.; Ahsan, M.; Sinha, S.; Kyro, G.W.; Maschietto, F.; Allen, B.; Skeens, E.; Lisi, G.P.; Batista, V.S. Twisting and swiveling domain motions in Cas9 to recognize target DNA duplexes, make double-strand breaks, and release cleaved duplexes. Front. Mol. Biosci. 2023, 9, 1072733. [Google Scholar] [CrossRef]
- Li, Y.; Glass, Z.; Huang, M.; Chen, Z.-Y.; Xu, Q. Ex vivo cell-based CRISPR/Cas9 genome editing for therapeutic applications. Biomaterials 2020, 234, 119711. [Google Scholar] [CrossRef] [PubMed]
- Gilbert, L.A.; Larson, M.H.; Morsut, L.; Liu, Z.; Brar, G.A.; Torres, S.E.; Stern-Ginossar, N.; Brandman, O.; Whitehead, E.H.; Doudna, J.A. CRISPR-mediated modular RNA-guided regulation of transcription in eukaryotes. Cell 2013, 154, 442–451. [Google Scholar] [CrossRef]
- Chavez, A.; Tuttle, M.; Pruitt, B.W.; Ewen-Campen, B.; Chari, R.; Ter-Ovanesyan, D.; Haque, S.J.; Cecchi, R.J.; Kowal, E.J.; Buchthal, J. Comparison of Cas9 activators in multiple species. Nat. Methods 2016, 13, 563–567. [Google Scholar] [CrossRef] [PubMed]
- Zhu, H.; Liu, D.; Sui, M.; Zhou, M.; Wang, B.; Qi, Q.; Wang, T.; Zhang, G.; Wan, F.; Zhang, B. CRISPRa-based activation of Fgf21 and Fndc5 ameliorates obesity by promoting adipocytes browning. Clin. Transl. Med. 2023, 13, e1326. [Google Scholar] [CrossRef]
- Han, J.L.; Heinson, Y.W.; Chua, C.J.; Liu, W.; Entcheva, E. CRISPRi gene modulation and all-optical electrophysiology in post-differentiated human iPSC-cardiomyocytes. Commun. Biol. 2023, 6, 1236. [Google Scholar] [CrossRef]
- Fu, Y.-W.; Dai, X.-Y.; Wang, W.-T.; Yang, Z.-X.; Zhao, J.-J.; Zhang, J.-P.; Wen, W.; Zhang, F.; Oberg, K.C.; Zhang, L. Dynamics and competition of CRISPR–Cas9 ribonucleoproteins and AAV donor-mediated NHEJ, MMEJ and HDR editing. Nucleic Acids Res. 2021, 49, 969–985. [Google Scholar] [CrossRef]
- Ran, F.; Hsu, P.D.; Wright, J.; Agarwala, V.; Scott, D.A.; Zhang, F. Genome engineering using the CRISPR-Cas9 system. Nat. Protoc. 2013, 8, 2281–2308. [Google Scholar] [CrossRef]
- Yang, H.; Ren, S.; Yu, S.; Pan, H.; Li, T.; Ge, S.; Zhang, J.; Xia, N. Methods favoring homology-directed repair choice in response to CRISPR/Cas9 induced-double strand breaks. Int. J. Mol. Sci. 2020, 21, 6461. [Google Scholar] [CrossRef] [PubMed]
- Anzalone, A.V.; Koblan, L.W.; Liu, D.R. Genome editing with CRISPR–Cas nucleases, base editors, transposases and prime editors. Nat. Biotechnol. 2020, 38, 824–844. [Google Scholar] [CrossRef] [PubMed]
- Lahr, W.S.; Sipe, C.J.; Skeate, J.G.; Webber, B.R.; Moriarity, B.S. CRISPR-Cas9 base editors and their current role in human therapeutics. Cytotherapy 2023, 25, 270–276. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Z.; Shang, P.; Mohanraju, P.; Geijsen, N. Prime editing: Advances and therapeutic applications. Trends Biotechnol. 2023, 41, 1000–1012. [Google Scholar] [CrossRef]
- Yourik, P.; Fuchs, R.T.; Mabuchi, M.; Curcuru, J.L.; Robb, G.B. Staphylococcus aureus Cas9 is a multiple-turnover enzyme. RNA 2019, 25, 35–44. [Google Scholar] [CrossRef] [PubMed]
- Friedland, A.E.; Baral, R.; Singhal, P.; Loveluck, K.; Shen, S.; Sanchez, M.; Marco, E.; Gotta, G.M.; Maeder, M.L.; Kennedy, E.M. Characterization of Staphylococcus aureus Cas9: A smaller Cas9 for all-in-one adeno-associated virus delivery and paired nickase applications. Genome Biol. 2015, 16, 257. [Google Scholar] [CrossRef]
- Nishimasu, H.; Cong, L.; Yan, W.X.; Ran, F.A.; Zetsche, B.; Li, Y.; Kurabayashi, A.; Ishitani, R.; Zhang, F.; Nureki, O. Crystal structure of Staphylococcus aureus Cas9. Cell 2015, 162, 1113–1126. [Google Scholar] [CrossRef]
- Lee, C.M.; Cradick, T.J.; Bao, G. The Neisseria meningitidis CRISPR-Cas9 system enables specific genome editing in mammalian cells. Mol. Ther. 2016, 24, 645–654. [Google Scholar] [CrossRef]
- Sun, W.; Yang, J.; Cheng, Z.; Amrani, N.; Liu, C.; Wang, K.; Ibraheim, R.; Edraki, A.; Huang, X.; Wang, M. Structures of Neisseria meningitidis Cas9 complexes in catalytically poised and anti-CRISPR-inhibited states. Mol. Cell 2019, 76, 938–952.e935. [Google Scholar] [CrossRef]
- Kim, E.; Koo, T.; Park, S.W.; Kim, D.; Kim, K.; Cho, H.-Y.; Song, D.W.; Lee, K.J.; Jung, M.H.; Kim, S. In vivo genome editing with a small Cas9 orthologue derived from Campylobacter jejuni. Nat. Commun. 2017, 8, 14500. [Google Scholar] [CrossRef]
- Schmidheini, L.; Mathis, N.; Marquart, K.F.; Rothgangl, T.; Kissling, L.; Böck, D.; Chanez, C.; Wang, J.P.; Jinek, M.; Schwank, G. Continuous directed evolution of a compact CjCas9 variant with broad PAM compatibility. Nat. Chem. Biol. 2024, 20, 333–343. [Google Scholar] [CrossRef] [PubMed]
- Ran, F.A.; Cong, L.; Yan, W.X.; Scott, D.A.; Gootenberg, J.S.; Kriz, A.J.; Zetsche, B.; Shalem, O.; Wu, X.; Makarova, K.S. In vivo genome editing using Staphylococcus aureus Cas9. Nature 2015, 520, 186–191. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Wang, Y.; Mao, D.; Wang, Y.; Zhang, H.; Pan, Y.; Wang, Y.; Teng, S.; Huang, P. Current trends of clinical trials involving CRISPR/Cas systems. Front. Med. 2023, 10, 1292452. [Google Scholar] [CrossRef]
- Forget, B.G. Molecular basis of hereditary persistence of fetal hemoglobin. Ann. N. Y. Acad. Sci. 1998, 850, 38–44. [Google Scholar] [CrossRef]
- Traxler, E.A.; Yao, Y.; Wang, Y.-D.; Woodard, K.J.; Kurita, R.; Nakamura, Y.; Hughes, J.R.; Hardison, R.C.; Blobel, G.A.; Li, C. A genome-editing strategy to treat β-hemoglobinopathies that recapitulates a mutation associated with a benign genetic condition. Nat. Med. 2016, 22, 987–990. [Google Scholar] [CrossRef] [PubMed]
- Pavani, G.; Fabiano, A.; Laurent, M.; Amor, F.; Cantelli, E.; Chalumeau, A.; Maule, G.; Tachtsidi, A.; Concordet, J.-P.; Cereseto, A. Correction of β-thalassemia by CRISPR/Cas9 editing of the α-globin locus in human hematopoietic stem cells. Blood Adv. 2021, 5, 1137–1153. [Google Scholar] [CrossRef]
- Zhang, H.; Sun, R.; Fei, J.; Chen, H.; Lu, D. Correction of beta-thalassemia IVS-II-654 mutation in a mouse model using prime editing. Int. J. Mol. Sci. 2022, 23, 5948. [Google Scholar] [CrossRef]
- Liang, P.; Ding, C.; Sun, H.; Xie, X.; Xu, Y.; Zhang, X.; Sun, Y.; Xiong, Y.; Ma, W.; Liu, Y. Correction of β-thalassemia mutant by base editor in human embryos. Protein Cell 2017, 8, 811–822. [Google Scholar] [CrossRef]
- Ashley-Koch, A.; Yang, Q.; Olney, R.S. Sickle hemoglobin (Hb S) allele and sickle cell disease: A HuGE review. Am. J. Epidemiol. 2000, 151, 839–845. [Google Scholar] [CrossRef]
- Akinsheye, I.; Alsultan, A.; Solovieff, N.; Ngo, D.; Baldwin, C.T.; Sebastiani, P.; Chui, D.H.; Steinberg, M.H. Fetal hemoglobin in sickle cell anemia. Blood J. Am. Soc. Hematol. 2011, 118, 19–27. [Google Scholar] [CrossRef]
- Chu, S.H.; Ortega, M.; Feliciano, P.; Winton, V.; Xu, C.; Haupt, D.; McDonald, T.; Martinez, S.; Liquori, A.; Marshall, J. Conversion of HbS to Hb G-makassar by adenine base editing is compatible with normal hemoglobin function. Blood 2021, 138, 951. [Google Scholar] [CrossRef]
- Newby, G.A.; Yen, J.S.; Woodard, K.J.; Mayuranathan, T.; Lazzarotto, C.R.; Li, Y.; Sheppard-Tillman, H.; Porter, S.N.; Yao, Y.; Mayberry, K. Base editing of haematopoietic stem cells rescues sickle cell disease in mice. Nature 2021, 595, 295–302. [Google Scholar] [CrossRef] [PubMed]
- Han, X.; Wang, M.; Duan, S.; Franco, P.J.; Kenty, J.H.-R.; Hedrick, P.; Xia, Y.; Allen, A.; Ferreira, L.M.; Strominger, J.L. Generation of hypoimmunogenic human pluripotent stem cells. Proc. Natl. Acad. Sci. USA 2019, 116, 10441–10446. [Google Scholar] [CrossRef] [PubMed]
- Sintov, E.; Nikolskiy, I.; Barrera, V.; Kenty, J.H.-R.; Atkin, A.S.; Gerace, D.; Sui, S.J.H.; Boulanger, K.; Melton, D.A. Whole-genome CRISPR screening identifies genetic manipulations to reduce immune rejection of stem cell-derived islets. Stem Cell Rep. 2022, 17, 1976–1990. [Google Scholar] [CrossRef]
- Ma, S.; Viola, R.; Sui, L.; Cherubini, V.; Barbetti, F.; Egli, D. β cell replacement after gene editing of a neonatal diabetes-causing mutation at the insulin locus. Stem Cell Rep. 2018, 11, 1407–1415. [Google Scholar] [CrossRef]
- Pacesa, M.; Lin, C.-H.; Cléry, A.; Saha, A.; Arantes, P.R.; Bargsten, K.; Irby, M.J.; Allain, F.H.-T.; Palermo, G.; Cameron, P. Structural basis for Cas9 off-target activity. Cell 2022, 185, 4067–4081.e4021. [Google Scholar] [CrossRef] [PubMed]
- Cohen, D.; Melamed, S.; Millman, A.; Shulman, G.; Oppenheimer-Shaanan, Y.; Kacen, A.; Doron, S.; Amitai, G.; Sorek, R. Cyclic GMP–AMP signalling protects bacteria against viral infection. Nature 2019, 574, 691–695. [Google Scholar] [CrossRef]
- Xu, X.; Wan, T.; Xin, H.; Li, D.; Pan, H.; Wu, J.; Ping, Y. Delivery of CRISPR/Cas9 for therapeutic genome editing. J. Gene Med. 2019, 21, e3107. [Google Scholar] [CrossRef]
- Yip, B.H. Recent advances in CRISPR/Cas9 delivery strategies. Biomolecules 2020, 10, 839. [Google Scholar] [CrossRef]
- Cheng, H.; Zhang, F.; Ding, Y. CRISPR/Cas9 delivery system engineering for genome editing in therapeutic applications. Pharmaceutics 2021, 13, 1649. [Google Scholar] [CrossRef]
- Bulcha, J.T.; Wang, Y.; Ma, H.; Tai, P.W.; Gao, G. Viral vector platforms within the gene therapy landscape. Signal Transduct. Target. Ther. 2021, 6, 53. [Google Scholar] [CrossRef] [PubMed]
- Hayashi, H.; Kubo, Y.; Izumida, M.; Matsuyama, T. Efficient viral delivery of Cas9 into human safe harbor. Sci. Rep. 2020, 10, 21474. [Google Scholar] [CrossRef] [PubMed]
- Daya, S.; Berns, K.I. Gene therapy using adeno-associated virus vectors. Clin. Microbiol. Rev. 2008, 21, 583–593. [Google Scholar] [CrossRef] [PubMed]
- Finn, J.D.; Smith, A.R.; Patel, M.C.; Shaw, L.; Youniss, M.R.; van Heteren, J.; Dirstine, T.; Ciullo, C.; Lescarbeau, R.; Seitzer, J. A single administration of CRISPR/Cas9 lipid nanoparticles achieves robust and persistent in vivo genome editing. Cell Rep. 2018, 22, 2227–2235. [Google Scholar] [CrossRef] [PubMed]
- Sur, S.; Rathore, A.; Dave, V.; Reddy, K.R.; Chouhan, R.S.; Sadhu, V. Recent developments in functionalized polymer nanoparticles for efficient drug delivery system. Nano-Struct. Nano-Objects 2019, 20, 100397. [Google Scholar] [CrossRef]
- Hoang Thi, T.T.; Pilkington, E.H.; Nguyen, D.H.; Lee, J.S.; Park, K.D.; Truong, N.P. The importance of poly (ethylene glycol) alternatives for overcoming PEG immunogenicity in drug delivery and bioconjugation. Polymers 2020, 12, 298. [Google Scholar] [CrossRef]
- Yue, H.; Zhou, X.; Cheng, M.; Xing, D. Graphene oxide-mediated Cas9/sgRNA delivery for efficient genome editing. Nanoscale 2018, 10, 1063–1071. [Google Scholar] [CrossRef]
- Wang, H.-X.; Song, Z.; Lao, Y.-H.; Xu, X.; Gong, J.; Cheng, D.; Chakraborty, S.; Park, J.S.; Li, M.; Huang, D. Nonviral gene editing via CRISPR/Cas9 delivery by membrane-disruptive and endosomolytic helical polypeptide. Proc. Natl. Acad. Sci. USA 2018, 115, 4903–4908. [Google Scholar] [CrossRef]
- Zhang, L.; Wang, P.; Feng, Q.; Wang, N.; Chen, Z.; Huang, Y.; Zheng, W.; Jiang, X. Lipid nanoparticle-mediated efficient delivery of CRISPR/Cas9 for tumor therapy. NPG Asia Mater. 2017, 9, e441. [Google Scholar] [CrossRef]
- Chen, K.; Han, H.; Zhao, S.; Xu, B.; Yin, B.; Trinidad, M.; Burgstone, B.W.; Murthy, N.; Doudna, J.A. Lung and liver editing by lipid nanoparticle delivery of a stable CRISPR-Cas9 RNP. bioRxiv, 2023; Preprint. [Google Scholar] [CrossRef]
- Yin, H.; Song, C.-Q.; Suresh, S.; Wu, Q.; Walsh, S.; Rhym, L.H.; Mintzer, E.; Bolukbasi, M.F.; Zhu, L.J.; Kauffman, K. Structure-guided chemical modification of guide RNA enables potent non-viral in vivo genome editing. Nat. Biotechnol. 2017, 35, 1179–1187. [Google Scholar] [CrossRef]
- Miller, J.B.; Zhang, S.; Kos, P.; Xiong, H.; Zhou, K.; Perelman, S.S.; Zhu, H.; Siegwart, D.J. Non-viral CRISPR/Cas gene editing in vitro and in vivo enabled by synthetic nanoparticle co-delivery of Cas9 mRNA and sgRNA. Angew. Chem. 2017, 129, 1079–1083. [Google Scholar] [CrossRef]
- Liu, Y.; Zhao, G.; Xu, C.-F.; Luo, Y.-L.; Lu, Z.-D.; Wang, J. Systemic delivery of CRISPR/Cas9 with PEG-PLGA nanoparticles for chronic myeloid leukemia targeted therapy. Biomater. Sci. 2018, 6, 1592–1603. [Google Scholar] [CrossRef] [PubMed]
- Sun, W.; Ji, W.; Hall, J.M.; Hu, Q.; Wang, C.; Beisel, C.L.; Gu, Z. Self-assembled DNA nanoclews for the efficient delivery of CRISPR–Cas9 for genome editing. Angew. Chem. 2015, 127, 12197–12201. [Google Scholar] [CrossRef]
- Liu, B.-Y.; He, X.-Y.; Xu, C.; Xu, L.; Ai, S.-L.; Cheng, S.-X.; Zhuo, R.-X. A dual-targeting delivery system for effective genome editing and in situ detecting related protein expression in edited cells. Biomacromolecules 2018, 19, 2957–2968. [Google Scholar] [CrossRef] [PubMed]
- Lee, B.; Lee, K.; Panda, S.; Gonzales-Rojas, R.; Chong, A.; Bugay, V.; Park, H.M.; Brenner, R.; Murthy, N.; Lee, H.Y. Nanoparticle delivery of CRISPR into the brain rescues a mouse model of fragile X syndrome from exaggerated repetitive behaviours. Nat. Biomed. Eng. 2018, 2, 497–507. [Google Scholar] [CrossRef]
- Lee, K.; Conboy, M.; Park, H.M.; Jiang, F.; Kim, H.J.; Dewitt, M.A.; Mackley, V.A.; Chang, K.; Rao, A.; Skinner, C. Nanoparticle delivery of Cas9 ribonucleoprotein and donor DNA in vivo induces homology-directed DNA repair. Nat. Biomed. Eng. 2017, 1, 889–901. [Google Scholar] [CrossRef]
- Zhu, H.; Zhang, L.; Tong, S.; Lee, C.M.; Deshmukh, H.; Bao, G. Spatial control of in vivo CRISPR–Cas9 genome editing via nanomagnets. Nat. Biomed. Eng. 2019, 3, 126–136. [Google Scholar] [CrossRef] [PubMed]
- Noureddine, A.; Maestas-Olguin, A.; Saada, E.A.; LaBauve, A.E.; Agola, J.O.; Baty, K.E.; Howard, T.; Sabo, J.K.; Espinoza, C.R.S.; Doudna, J.A. Engineering of monosized lipid-coated mesoporous silica nanoparticles for CRISPR delivery. Acta Biomater. 2020, 114, 358–368. [Google Scholar] [CrossRef]
- LaBauve, A.E.; Saada, E.A.; Jones, I.K.; Mosesso, R.; Noureddine, A.; Techel, J.; Gomez, A.; Collette, N.; Sherman, M.B.; Serda, R.E. Lipid-coated mesoporous silica nanoparticles for anti-viral applications via delivery of CRISPR-Cas9 ribonucleoproteins. Sci. Rep. 2023, 13, 6873. [Google Scholar] [CrossRef]
- Zhang, X.; Xu, Q.; Zi, Z.; Liu, Z.; Wan, C.; Crisman, L.; Shen, J.; Liu, X. Programmable extracellular vesicles for macromolecule delivery and genome modifications. Dev. Cell 2020, 55, 784–801.e789. [Google Scholar] [CrossRef]
- Usman, W.M.; Pham, T.C.; Kwok, Y.Y.; Vu, L.T.; Ma, V.; Peng, B.; Chan, Y.S.; Wei, L.; Chin, S.M.; Azad, A. Efficient RNA drug delivery using red blood cell extracellular vesicles. Nat. Commun. 2018, 9, 2359. [Google Scholar] [CrossRef] [PubMed]
- McAndrews, K.M.; Xiao, F.; Chronopoulos, A.; LeBleu, V.S.; Kugeratski, F.G.; Kalluri, R. Exosome-mediated delivery of CRISPR/Cas9 for targeting of oncogenic KrasG12D in pancreatic cancer. Life Sci. Alliance 2021, 4, e202000875. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.M.; Yang, Y.; Oh, S.J.; Hong, Y.; Seo, M.; Jang, M. Cancer-derived exosomes as a delivery platform of CRISPR/Cas9 confer cancer cell tropism-dependent targeting. J. Control Release 2017, 266, 8–16. [Google Scholar] [CrossRef] [PubMed]
- Aziz, A.; Rehman, U.; Sheikh, A.; Abourehab, M.A.; Kesharwani, P. Lipid-based nanocarrier mediated CRISPR/Cas9 delivery for cancer therapy. J. Biomater. Sci. Polym. Ed. 2023, 34, 398–418. [Google Scholar] [CrossRef]
- Mout, R.; Ray, M.; Tay, T.; Sasaki, K.; Yesilbag Tonga, G.; Rotello, V.M. General strategy for direct cytosolic protein delivery via protein–nanoparticle co-engineering. ACS Nano 2017, 11, 6416–6421. [Google Scholar] [CrossRef]
- Hall, R. Design and Evaluation of Peptide Lipid-Associated Nucleic Acids (PLANAs) for siRNA and CRISPR/Cas9 Delivery and Protein Silencing. Master’s Thesis, Chapman University, Orange, CA, USA, 2020. [Google Scholar]
- Sinclair, F.; Begum, A.A.; Dai, C.C.; Toth, I.; Moyle, P.M. Recent advances in the delivery and applications of nonviral CRISPR/Cas9 gene editing. Drug Deliv. Transl. Res. 2023, 13, 1500–1519. [Google Scholar] [CrossRef]
- Agarwal, R.; Iezhitsa, I.; Agarwal, P.; Abdul Nasir, N.A.; Razali, N.; Alyautdin, R.; Ismail, N.M. Liposomes in topical ophthalmic drug delivery: An update. Drug Deliv. 2016, 23, 1075–1091. [Google Scholar] [CrossRef]
- Ma, X.; Zhang, Y.; Huang, K.; Zhu, L.; Xu, W. Multifunctional rolling circle transcription-based nanomaterials for advanced drug delivery. Biomaterials 2023, 301, 122241. [Google Scholar] [CrossRef]
- Ha, J.S.; Lee, J.S.; Jeong, J.; Kim, H.; Byun, J.; Kim, S.A.; Lee, H.J.; Chung, H.S.; Lee, J.B.; Ahn, D.-R. Poly-sgRNA/siRNA ribonucleoprotein nanoparticles for targeted gene disruption. J. Control Release 2017, 250, 27–35. [Google Scholar] [CrossRef]
- Ran, F.A.; Hsu, P.D.; Lin, C.-Y.; Gootenberg, J.S.; Konermann, S.; Trevino, A.E.; Scott, D.A.; Inoue, A.; Matoba, S.; Zhang, Y. Double nicking by RNA-guided CRISPR Cas9 for enhanced genome editing specificity. Cell 2013, 154, 1380–1389. [Google Scholar] [CrossRef]
- Stefanovic, S.; McCormick, K.; Fattah, S.; Brannigan, R.; Cryan, S.-A.; Heise, A. Star-shaped poly (l-lysine) with polyester bis-MPA dendritic core as potential degradable nano vectors for gene delivery. Polym. Chem. 2023, 14, 3151–3159. [Google Scholar] [CrossRef]
- Sargazi, S.; Siddiqui, B.; Qindeel, M.; Rahdar, A.; Bilal, M.; Behzadmehr, R.; Mirinejad, S.; Pandey, S. Chitosan nanocarriers for microRNA delivery and detection: A preliminary review with emphasis on cancer. Carbohydr. Polym. 2022, 290, 119489. [Google Scholar] [CrossRef] [PubMed]
- Kheraldine, H.; Rachid, O.; Habib, A.M.; Al Moustafa, A.-E.; Benter, I.F.; Akhtar, S. Emerging innate biological properties of nano-drug delivery systems: A focus on PAMAM dendrimers and their clinical potential. Adv. Drug Deliv. Rev. 2021, 178, 113908. [Google Scholar] [CrossRef] [PubMed]
- Gallops, C.; Ziebarth, J.; Wang, Y. A polymer physics perspective on why PEI is an effective nonviral gene delivery vector. In Polymers in Therapeutic Delivery; ACS Publications: Washington, DC, USA, 2020; pp. 1–12. [Google Scholar]
- Xie, R.; Wang, X.; Wang, Y.; Ye, M.; Zhao, Y.; Yandell, B.S.; Gong, S. pH-responsive polymer nanoparticles for efficient delivery of Cas9 ribonucleoprotein with or without donor DNA. Adv. Mater. 2022, 34, 2110618. [Google Scholar] [CrossRef]
- Javed, M.N.; Akhter, M.H.; Taleuzzaman, M.; Faiyazudin, M.; Alam, M.S. Cationic nanoparticles for treatment of neurological diseases. In Fundamentals of Bionanomaterials; Elsevier: Amsterdam, The Netherlands, 2022; pp. 273–292. [Google Scholar]
- Jung, K.; Corrigan, N.; Wong, E.H.; Boyer, C. Bioactive synthetic polymers. Adv. Mater. 2022, 34, 2105063. [Google Scholar] [CrossRef] [PubMed]
- Chen, F.; Alphonse, M.; Liu, Q. Strategies for nonviral nanoparticle-based delivery of CRISPR/Cas9 therapeutics. Wiley Interdiscip. Rev. Nanomed. Nanobiotechnol. 2020, 12, e1609. [Google Scholar] [CrossRef]
- O’Keeffe Ahern, J.; Lara-Sáez, I.; Zhou, D.; Murillas, R.; Bonafont, J.; Mencía, Á.; García, M.; Manzanares, D.; Lynch, J.; Foley, R. Non-viral delivery of CRISPR–Cas9 complexes for targeted gene editing via a polymer delivery system. Gene Ther. 2022, 29, 157–170. [Google Scholar] [CrossRef]
- Kang, Y.K.; Kwon, K.; Ryu, J.S.; Lee, H.N.; Park, C.; Chung, H.J. Nonviral genome editing based on a polymer-derivatized CRISPR nanocomplex for targeting bacterial pathogens and antibiotic resistance. Bioconjugate Chem. 2017, 28, 957–967. [Google Scholar] [CrossRef]
- Guo, S.; Xiao, P.; Li, B.; Wang, W.; Wang, S.; Lv, T.; Xu, X.; Chen, C.; Huang, L.; Li, Z. Co-immunizing with PD-L1 induces CD8+ DCs-mediated anti-tumor immunity in multiple myeloma. Int. Immunopharmacol. 2020, 84, 106516. [Google Scholar] [CrossRef]
- Tarach, P.; Janaszewska, A. Recent advances in preclinical research using PAMAM dendrimers for cancer gene therapy. Int. J. Mol. Sci. 2021, 22, 2912. [Google Scholar] [CrossRef]
- Tomalia, D.A.; Baker, H.; Dewald, J.; Hall, M.; Kallos, G.; Martin, S.; Roeck, J.; Ryder, J.; Smith, P. A new class of polymers: Starburst-dendritic macromolecules. Polym. J. 1985, 17, 117–132. [Google Scholar] [CrossRef]
- Trigueros, S.; Domènech, E.B.; Toulis, V.; Marfany, G. In vitro gene delivery in retinal pigment epithelium cells by plasmid DNA-wrapped gold nanoparticles. Genes 2019, 10, 289. [Google Scholar] [CrossRef] [PubMed]
- Heuer-Jungemann, A.; Feliu, N.; Bakaimi, I.; Hamaly, M.; Alkilany, A.; Chakraborty, I.; Masood, A.; Casula, M.F.; Kostopoulou, A.; Oh, E. The role of ligands in the chemical synthesis and applications of inorganic nanoparticles. Chem. Rev. 2019, 119, 4819–4880. [Google Scholar] [CrossRef] [PubMed]
- Fratoddi, I.; Venditti, I.; Cametti, C.; Russo, M.V. Gold nanoparticles and gold nanoparticle-conjugates for delivery of therapeutic molecules. Progress and challenges. J. Mater. Chem. B 2014, 2, 4204–4220. [Google Scholar] [CrossRef]
- Elahi, N.; Kamali, M.; Baghersad, M.H. Recent biomedical applications of gold nanoparticles: A review. Talanta 2018, 184, 537–556. [Google Scholar] [CrossRef]
- Qiu, J.; Xie, M.; Wu, T.; Qin, D.; Xia, Y. Gold nanocages for effective photothermal conversion and related applications. Chem. Sci. 2020, 11, 12955–12973. [Google Scholar] [CrossRef]
- Wang, J.; Ni, Q.; Wang, Y.; Zhang, Y.; He, H.; Gao, D.; Ma, X.; Liang, X.-J. Nanoscale drug delivery systems for controllable drug behaviors by multi-stage barrier penetration. J. Control Release 2021, 331, 282–295. [Google Scholar] [CrossRef]
- Vaughan, H.J.; Green, J.J.; Tzeng, S.Y. Cancer-targeting nanoparticles for combinatorial nucleic acid delivery. Adv. Mater. 2020, 32, 1901081. [Google Scholar] [CrossRef]
- Wang, P.; Zhang, L.; Xie, Y.; Wang, N.; Tang, R.; Zheng, W.; Jiang, X. Genome editing for cancer therapy: Delivery of Cas9 protein/sgRNA plasmid via a gold nanocluster/lipid core–shell nanocarrier. Adv. Sci. 2017, 4, 1700175. [Google Scholar] [CrossRef]
- Sani, A.; Cao, C.; Cui, D. Toxicity of gold nanoparticles (AuNPs): A review. Biochem. Biophys. Rep. 2021, 26, 100991. [Google Scholar] [CrossRef]
- Gavilán, H.; Avugadda, S.K.; Fernández-Cabada, T.; Soni, N.; Cassani, M.; Mai, B.T.; Chantrell, R.; Pellegrino, T. Magnetic nanoparticles and clusters for magnetic hyperthermia: Optimizing their heat performance and developing combinatorial therapies to tackle cancer. Chem. Soc. Rev. 2021, 50, 11614–11667. [Google Scholar] [CrossRef] [PubMed]
- Hryhorowicz, M.; Grześkowiak, B.; Mazurkiewicz, N.; Śledziński, P.; Lipiński, D.; Słomski, R. Improved delivery of CRISPR/Cas9 system using magnetic nanoparticles into porcine fibroblast. Mol. Biotechnol. 2019, 61, 173–180. [Google Scholar] [CrossRef]
- Rohiwal, S.; Dvorakova, N.; Klima, J.; Vaskovicova, M.; Senigl, F.; Slouf, M.; Pavlova, E.; Stepanek, P.; Babuka, D.; Benes, H. Polyethylenimine based magnetic nanoparticles mediated non-viral CRISPR/Cas9 system for genome editing. Sci. Rep. 2020, 10, 4619. [Google Scholar] [CrossRef]
- Li, T.; Geng, Y.; Zhang, H.; Wang, J.; Feng, Y.; Chen, Z.; Xie, X.; Qin, X.; Li, S.; Wu, C. A versatile nanoplatform for synergistic chemo-photothermal therapy and multimodal imaging against breast cancer. Expert Opin. Drug Deliv. 2020, 17, 725–733. [Google Scholar] [CrossRef]
- Lu, M.; Huang, Y. Bioinspired exosome-like therapeutics and delivery nanoplatforms. Biomaterials 2020, 242, 119925. [Google Scholar] [CrossRef]
- Liang, Y.; Duan, L.; Lu, J.; Xia, J. Engineering exosomes for targeted drug delivery. Theranostics 2021, 11, 3183. [Google Scholar] [CrossRef] [PubMed]
- Wan, T.; Zhong, J.; Pan, Q.; Zhou, T.; Ping, Y.; Liu, X. Exosome-mediated delivery of Cas9 ribonucleoprotein complexes for tissue-specific gene therapy of liver diseases. Sci. Adv. 2022, 8, eabp9435. [Google Scholar] [CrossRef] [PubMed]
- Chen, R.; Huang, H.; Liu, H.; Xi, J.; Ning, J.; Zeng, W.; Shen, C.; Zhang, T.; Yu, G.; Xu, Q. Friend or foe? Evidence indicates endogenous exosomes can deliver functional gRNA and Cas9 protein. Small 2019, 15, 1902686. [Google Scholar] [CrossRef] [PubMed]
- Melzer, C.; Rehn, V.; Yang, Y.; Bähre, H.; von der Ohe, J.; Hass, R. Taxol-loaded MSC-derived exosomes provide a therapeutic vehicle to target metastatic breast cancer and other carcinoma cells. Cancers 2019, 11, 798. [Google Scholar] [CrossRef]
- Duan, L.; Xu, L.; Xu, X.; Qin, Z.; Zhou, X.; Xiao, Y.; Liang, Y.; Xia, J. Exosome-mediated delivery of gene vectors for gene therapy. Nanoscale 2021, 13, 1387–1397. [Google Scholar] [CrossRef]
- Ye, Y.; Zhang, X.; Xie, F.; Xu, B.; Xie, P.; Yang, T.; Shi, Q.; Zhang, C.-Y.; Zhang, Y.; Chen, J. An engineered exosome for delivering sgRNA: Cas9 ribonucleoprotein complex and genome editing in recipient cells. Biomater. Sci. 2020, 8, 2966–2976. [Google Scholar] [CrossRef] [PubMed]
- Lin, Y.; Wu, J.; Gu, W.; Huang, Y.; Tong, Z.; Huang, L.; Tan, J. Exosome–liposome hybrid nanoparticles deliver CRISPR/Cas9 system in MSCs. Adv. Sci. 2018, 5, 1700611. [Google Scholar] [CrossRef] [PubMed]
- Liang, Y.; Xu, X.; Xu, L.; Iqbal, Z.; Ouyang, K.; Zhang, H.; Wen, C.; Duan, L.; Xia, J. Chondrocyte-specific genomic editing enabled by hybrid exosomes for osteoarthritis treatment. Theranostics 2022, 12, 4866. [Google Scholar] [CrossRef] [PubMed]



| Name | Bacteria Species | PAM Site | Gene Size | Reference |
|---|---|---|---|---|
| SpCas9 | Streptococcus pyogenes | 5′-NGG-3′ | 4.2 kb | [33] |
| SaCas9 | Staphylococcus aureus | 5′-NNGRRT-3′ | 3.1 kb | [33] |
| St1Cas9 | Streptococcus thermophilus LMD-9 | 5′-NNAGAAW-3′ | 3.3 kb | [33] |
| NmCas9 | Neisseria meningitides | 5′-NNNNGATT-3′ | 4.2 kb | [29] |
| SjCas9 | Campylobacter jejuni | 5′-NNNVRYM-3′ | 3.3 kb | [31] |
| Disease | Disease Condition | Target Gene | Intervention | Method | Reference |
|---|---|---|---|---|---|
| β-thalassemia | β-thalassemia (β0/β0, β+/β0, βE/β0) | BCL11A | Gene disruption | CRISPR/Cas9 | [33] |
| SCD (Sickle cell disease) | SCD (βs/βs, βs/βc, βs/β0) | HBB | Gene disruption | CRISPR/Cas9 | [33] |
| β-thalassemia and SCD | Transfusion-dependent β-thalassemia (β0/β0, β+/β0, βE/β0, β+/β+) | BCL11A | Gene disruption | CRISPR/Cas9 | [33] |
| Type of Delivery System | Nanoparticle Formulation | CRISPR/Cas9 Cargo | Efficiency | Application | Reference |
|---|---|---|---|---|---|
| Lipid-based nanoparticle | polyethylene glycol phospholipid-modified cationic lipid nanoparticle (PLNP) | gRNA and Plasmid DNA encoding CRISPR/Cas9 | 67%> | In vitro, In vivo | [60] |
| pegylated lipid and ADP–2k | iGeoCRISPR/Cas9 | 35–56% efficiency in the liver or lungs | In vitro, In vivo | [61] | |
| Cholesterol, C14-PEG 2000, DOPE and ionizable lipid cKK-E12 | Pcsk9 (gRNA and CRISPR/Cas9 mRNA) | >80% editing of Pcsk9 in the liver | In vitro, In vivo | [62] | |
| Zwitterionic amino lipids (ZALs), | CRISPR/Cas9/gRNA mRNA | 95 % decrease in protein expression (HeLa-Luc-CRISPR/Cas9) | In vitro, In vivo | [63] | |
| Biodegradable ionizable lipid (LP01), cholesterol, DSPC and PEG2k-DMG | CRISPR/Cas9/gRNA mRNA | >97% reduction in the liver | In vitro, In vivo | [55] | |
| Polymer-based nanoparticle | PEG5K-b-PLGA11K | Plasmid DNA (pCRISPR/Cas9) | 74.6% (EGFP-positive K562 cells for CLANpCRISPR/Cas9-EGFP) | In vitro | [64] |
| Cationic polymer polyethylenimine (PEI) coated self-assembled DNA nanoclews | CRISPR/Cas9/gRNA RNP | 80% (EGFP in U2OS) | In vitro | [65] | |
| Graphene oxide (GO)-polyethylene glycol (PEG)-polyethylenimine (PEI) nanocarrier | CRISPR/Cas9/gRNA RNP | ∼39% (gene editing in human AGS cells with an efficiency) | In vitro | [58] | |
| Carboxymethyl chitosan (biotinylated carboxymethyl chitosan with biotin ligands and aptamer-incorporated carboxymethyl chitosan with AS1411 ligands) | pDNA | >90% (CDK11 protein) | In vitro | [66] | |
| PEGylated nanoparticles (named P-HNPs) based on the cationic α-helical polypeptide poly(γ–4–((2-(piperidin–1–yl)ethyl)aminomethyl)benzyl-L-glutamate) | CRISPR/Cas9/gRNA pDNA | 60% (CRISPR/Cas9 transfection efficiency), 67.4% (gRNA uptake efficiency), >71% (suppressing the tumor growth) | In vitro, In vivo | [59] | |
| Inorganic nanoparticles | CRISPR-Gold | RNP | 40–50% (Reduced mRNA levels and the protein levels of mGluR5) | In vitro, In vivo | [67] |
| CRISPR-Gold | RNP | 61.5% (encapsulation efficiency), 11.3% (BFP-HEK cells to express GFP via HDR) | In vivo | [68] | |
| Complexing magnetic nanoparticles (MNPs) with recombinant baculoviral vectors (BVs) | Recombinant baculoviral vector (BV) | ~50% indel rate in mouse liver cells | In vitro, In vivo | [69] | |
| monosized lipid-coated mesoporous silica nanoparticle (LC-MSN) | RNP | 70% (release within cancer cells), 10% (gene editing) | In vitro, In vivo | [70] | |
| Lipid-coated mesoporous silica nanoparticles (LCMSNs) | RNP | 20% (Edition against both targets) | In vitro | [71] | |
| Extracellular Vesicles-Based nanoparticles | Gectosomes (Virus G protein; VSV-G, split GFP) | CRISPR/Cas9 protein and RNP | 40% (Reduction in the number of cells positive for Parkin recruitment) | In vitro, In vivo | [72] |
| Red blood cells (RBCs)-derived EV | CRISPR/Cas9 mRNA | ~32% (loss of eGFP in ~32% NOMO1-eGFP cells) | In vitro, In vivo | [73] | |
| Non-autologous exosomes | CRISPR/Cas9 plasmid DNA | ~58% suppression (moderate knockdown) | In vitro, In vivo | [74] | |
| Tumor-derived exosomes (SKOV3-Exo) | CRISPR/Cas9/gRNA plasmid DNA | 27% (Indel at electroporated into SKOV3-Exo), 57% (Inhibited the cellular proliferation at co-treatment with cisplatin and iPARP–1/SKOV3-Exo) | In vitro, In vivo | [75] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kim, M.; Hwang, Y.; Lim, S.; Jang, H.-K.; Kim, H.-O. Advances in Nanoparticles as Non-Viral Vectors for Efficient Delivery of CRISPR/Cas9. Pharmaceutics 2024, 16, 1197. https://doi.org/10.3390/pharmaceutics16091197
Kim M, Hwang Y, Lim S, Jang H-K, Kim H-O. Advances in Nanoparticles as Non-Viral Vectors for Efficient Delivery of CRISPR/Cas9. Pharmaceutics. 2024; 16(9):1197. https://doi.org/10.3390/pharmaceutics16091197
Chicago/Turabian StyleKim, Minse, Youngwoo Hwang, Seongyu Lim, Hyeon-Ki Jang, and Hyun-Ouk Kim. 2024. "Advances in Nanoparticles as Non-Viral Vectors for Efficient Delivery of CRISPR/Cas9" Pharmaceutics 16, no. 9: 1197. https://doi.org/10.3390/pharmaceutics16091197
APA StyleKim, M., Hwang, Y., Lim, S., Jang, H.-K., & Kim, H.-O. (2024). Advances in Nanoparticles as Non-Viral Vectors for Efficient Delivery of CRISPR/Cas9. Pharmaceutics, 16(9), 1197. https://doi.org/10.3390/pharmaceutics16091197








