An Ontology-Driven Personalized Faceted Search for Exploring Knowledge Bases of Capsicum
Abstract
:1. Introduction
1.1. Motivation
- The PO can be used to describe plant characteristics, from anatomy and morphology to the stages of plant development. It is suitable to share knowledge among scientists but not necessarily with non-expert users.
- For non-expert users, when describing a less familiar object (for example, a flower of a plant), they tend to describe it based on generic properties or attributes. For example, to describe the petal of a flower, they would describe it based on familiar properties such as color, size, texture, etc.
1.2. Challenges
- How to start to explore a knowledge base of Capsicum by describing a generic morphological character. Searching should start from a point, for example, by defining at least one plant character. The start point could be any point in the knowledge base, regardless of its generality or specificity.
- How to refine the search results by selecting the most relevant criteria/group. Finding the most relevant criteria/group is the main challenge.
- How to sort multiple results to be presented to the users. When multiple criteria/groups are identified as relevant, they need to be sorted to provide users with the most relevant first. Finding the way to sort the results is the next challenge.
2. Related Work
- Development of an ontology that intends to communicate knowledge to non-experts users. Instead of using existing ontologies, we developed a small yet powerful ontology to describe the characteristics of Capsicum. The ontology was not intended to be complete but to be easily consumed by non-expert users.
- Utilization of faceted search technique to drive search process. This technique has been widely used to overcome information overload or searching from a large amount of data. In contrast, our faceted search was intended to search from an unfamiliar database where the amount of data is not necessarily significant.
3. Method
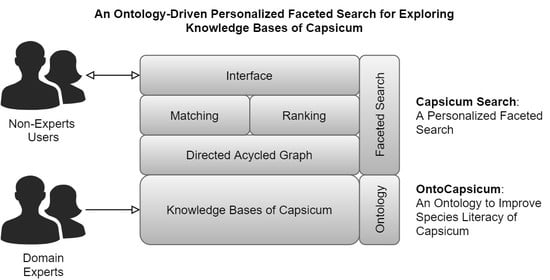
3.1. Capsicum Search: A Personalized Faceted Search
- A domain ontology is represented as a Directed Acyclic Graph (DAG) that consists of vertices and edges starting from the most generic part of a plant. The leaves of the graph are the most specific parts of the plant.
- A list of queries is represented as path traversal procedures, where the encoded entities and properties can be located correctly in the graph.
- Based on the graph and path traversal procedures, relevant entities are identified based on entities’ relationships in the graph, for example, based on relationships of siblings, sub-graphs, etc.
- All relevant entities become the list of facets to be presented to the users that can be used to refine their search results further.
3.2. Research Procedures
3.3. Knowledge Modeling and Query Formulation
- Identification of the purpose and scope of the ontology. As mentioned in Section 1, we share knowledge about the characteristics of Capsicum with non-expert users. Therefore, we expected that the ontology should cover characteristics of Capsicum identifiable by this group of users.
- Building the ontology, which covers the ontology capture, coding, and integration with existing ontologies. We identified entities, properties, and data types, including how entities are related to each other. After that, we represented the identified objects using the Resource Description Framework (RDF) [47] and Ontology Web Language (OWL) [48]. For coding the ontology, we actively used the Protégé ontology editor [49]. For integration with existing ontologies, we adopted the terms from the Plant Ontology [8].
- Evaluation. We evaluated the ontology by using competence questions [46] to carry out reasoning with different characteristics of Capsicum. This evaluation ensures that a list of correct entities can be obtained when a common characteristic is provided. To order the obtained entities as facets, we use a ranking mechanism explained in Section 3.5.
- Documentation. We generated the documentation of our ontology by using the WIDOCO tool [50]. It generated human-readable descriptions of terms and summaries with integration with other external information.
3.4. Matching
- Familiar with only one part of the plant. Users in this type only provide the description of a specific part and ignore other generic or more specific parts.
- Familiar with the generic parts of the plant. Users in this type provide a relatively generic description of the whole plant without focusing on a specific part.
- Focused on small parts of the plant. Users in this type provide more specific descriptions that are related to each other.
- Combination of generic and focused. Users in this type provide random descriptions of the plant.
- Matching #1, users describe a plant using only one entity. In this case, we selected the entities at the same level as well as entities in the same branch, as shown in Figure 2a. We called this matching mechanism a single-entity personalization method.
- Matching #2, users describe a plant using two entities located at the same level in the graph. In this case, we selected entities that are located at the same level, as shown in Figure 2b. We called this matching mechanism a level-based personalization method.
- Matching #3, users describe a plant using two entities not located at the same level but the same branch. In this case, we selected entities that are located at the same branch, as shown in Figure 2c. We called this matching mechanism a branch-based personalization method.
- Matching #4, users describe a plant using two entities not located at the same level or the same branch. In this case, we selected entities at the same level from both entities as well as entities from the branches, as shown in Figure 2d. We called this matching mechanism a level-and-branch-based personalization method.
3.5. Ranking
- Ranking #1: Select the matched entities with similar properties to the provided question. For example, if the query contains the property “Color”, then all entities with “Color” are ordered first.
- Ranking #2: Select the matched entities with a higher number of properties. An entity with a more detailed description (based on the number of available properties) is ordered first.
- Ranking #3: Select the more generic entity first. Since the generality of entities can be obtained through their levels, a lower-level entity is ordered first.
3.6. Evaluation
4. Result
4.1. Ontology
4.2. Search Algorithm
| Algorithm 1: Capsicum Search Algorithm. |
 |
5. Evaluation
- Case 1. The query contains only one node (fit with the matching mechanism #1). Testing case 1 uses permutation of three entities, namely “Fruit”, “Leaf”, and “Stem”. All of them belong to level 1, and they are suitable for case 1.
- Case 2. The query contains two nodes, where both nodes are at the same level (fit with the matching mechanism #2). Testing case 2 is conducted by using a combination of “Petals”, and “Seed”.
- Case 3. The query contains two nodes, where both nodes are at the same branch (fit with the matching mechanism #3). Testing case 3 uses a combination of nodes in the same branch with multiples levels from three entities, such as “Fruit”, “Stamen”, and “Flower”.
- Case 4. The query contains two nodes, where both nodes are neither at the same level nor at the same branch (fit with the matching mechanism #4). Testing case 4 is conducted with a combination of entities as nodes.
6. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Farley, S.S.; Dawson, A.; Goring, S.J.; Williams, J.W. Situating Ecology as a Big-Data Science: Current Advances, Challenges, and Solutions. BioScience 2018, 68, 563–576. [Google Scholar] [CrossRef] [Green Version]
- König, C.; Weigelt, P.; Schrader, J.; Taylor, A.; Kattge, J.; Kreft, H. Biodiversity data integration—The significance of data resolution and domain. PLoS Biol. 2019, 17, e3000183. [Google Scholar] [CrossRef] [PubMed]
- Bell, G.; Hey, T.; Szalay, A. Beyond the Data Deluge. Science 2009, 323, 1297–1298. [Google Scholar] [CrossRef] [PubMed]
- Das Gupta, M.; Tsiantis, M. Gene networks and the evolution of plant morphology. Curr. Opin. Plant Biol. 2018, 45, 82–87. [Google Scholar] [CrossRef]
- Antonio, A.S.; Wiedemann, L.S.M.; Veiga Junior, V.F. The genus Capsicum: A phytochemical review of bioactive secondary metabolites. RSC Adv. 2018, 8, 25767–25784. [Google Scholar] [CrossRef] [Green Version]
- Peter, M.; Diekötter, T.; Kremer, K. Participant Outcomes of Biodiversity Citizen Science Projects: A Systematic Literature Review. Sustainability 2019, 11, 2780. [Google Scholar] [CrossRef] [Green Version]
- Gruber, T.R. Toward principles for the design of ontologies used for knowledge sharing? Int. J. Hum. Comput. Stud. 1995, 43, 907–928. [Google Scholar] [CrossRef]
- Cooper, L.; Walls, R.L.; Elser, J.; Gandolfo, M.A.; Stevenson, D.W.; Smith, B.; Preece, J.; Athreya, B.; Mungall, C.J.; Rensing, S.; et al. The plant ontology as a tool for comparative plant anatomy and genomic analyses. Plant Cell Physiol. 2013, 54, e1. [Google Scholar] [CrossRef]
- Consortium, T.G.O. The Gene Ontology project in 2008. Nucleic Acids Res. 2007, 36, D440–D444. [Google Scholar] [CrossRef] [Green Version]
- Walls, R.L.; Cooper, L.; Elser, J.; Gandolfo, M.A.; Mungall, C.J.; Smith, B.; Stevenson, D.W.; Jaiswal, P. The Plant Ontology Facilitates Comparisons of Plant Development Stages Across Species. Front. Plant Sci. 2019, 10, 631. [Google Scholar] [CrossRef]
- Burgess, H.; DeBey, L.; Froehlich, H.; Schmidt, N.; Theobald, E.; Ettinger, A.; HilleRisLambers, J.; Tewksbury, J.; Parrish, J. The science of citizen science: Exploring barriers to use as a primary research tool. Biol. Conserv. 2017, 208, 113–120. [Google Scholar] [CrossRef] [Green Version]
- Mahdi, M.N.; Ahmad, A.R.; Ismail, R.; Natiq, H.; Mohammed, M.A. Solution for Information Overload Using Faceted Search–A Review. IEEE Access 2020, 8, 119554–119585. [Google Scholar] [CrossRef]
- Lens, F.; Cooper, L.; Gandolfo, M.A.; Groover4, A.; Jaiswal, P.; Lachenbruch, B.; Spicer, R.; Staton, M.E.; Stevenson, D.W.; Walls, R.L.; et al. An extension of the Plant Ontology project supporting wood anatomy and development research. IAWA J. 2012, 33, 113–117. [Google Scholar] [CrossRef] [Green Version]
- Meng, X.; Xu, C.; Liu, X.; Bai, J.; Zheng, W.; Chang, H.; Chen, Z. An Ontology-Underpinned Emergency Response System for Water Pollution Accidents. Sustainability 2018, 10, 546. [Google Scholar] [CrossRef] [Green Version]
- Padilla-Cuevas, J.; Reyes-Ortiz, J.A.; Bravo, M. Ontology-Based Context Event Representation, Reasoning, and Enhancing in Academic Environments. Future Internet 2021, 13, 151. [Google Scholar] [CrossRef]
- Ziemba, P.; Wątróbski, J.; Jankowski, J.; Wolski, W. Construction and Restructuring of the Knowledge Repository of Website Evaluation Methods. In Information Technology for Management; Ziemba, E., Ed.; Springer International Publishing: Cham, Switzerland, 2016; pp. 29–52. [Google Scholar] [CrossRef]
- Avraham, S.; Tung, C.W.; Ilic, K.; Jaiswal, P.; Kellogg, E.A.; McCouch, S.; Pujar, A.; Reiser, L.; Rhee, S.Y.; Sachs, M.M.; et al. The Plant Ontology Database: A community resource for plant structure and developmental stages controlled vocabulary and annotations. Nucleic Acids Res. 2008, 36, D449–D454. [Google Scholar] [CrossRef] [Green Version]
- Arnaud., E.; Cooper., L.; Shrestha., R.; Menda., N.; Nelson., R.T.; Matteis., L.; Skofic., M.; Bastow., R.; Jaiswal., P.; Mueller., L.; et al. Towards a Reference Plant Trait Ontology for Modeling Knowledge of Plant Traits and Phenotypes. In Proceedings of the International Conference on Knowledge Engineering and Ontology Development, Barcelona, Spain, 4–7 October 2012. [Google Scholar]
- Akbar, Z.; Kartika, Y.A.; Ridwan Saleh, D.; Mustika, H.F.; Parningotan Manik, L. On Using Declarative Generation Rules To Deliver Linked Biodiversity Data. In Proceedings of the 2020 International Conference on Radar, Antenna, Microwave, Electronics, and Telecommunications (ICRAMET), Tangerang, Indonesia, 18–20 November 2020; pp. 267–272. [Google Scholar] [CrossRef]
- McLaren, C.G.; Bruskiewich, R.M.; Portugal, A.M.; Cosico, A.B. The International Rice Information System. A Platform for Meta-Analysis of Rice Crop Data. Plant Physiol. 2005, 139, 637–642. [Google Scholar] [CrossRef] [Green Version]
- Schomburg, I.; Jeske, L.; Ulbrich, M.; Placzek, S.; Chang, A.; Schomburg, D. The BRENDA enzyme information system–From a database to an expert system. J. Biotechnol. 2017, 261, 194–206. [Google Scholar] [CrossRef]
- Kim, H.J.; Baek, K.H.; Lee, S.W.; Kim, J.; Lee, B.W.; Cho, H.S.; Kim, W.T.; Choi, D.; Hur, C.G. Pepper EST database: Comprehensive in silico tool for analyzing the chili pepper (Capsicum annuum) transcriptome. BMC Plant Biol. 2008, 8, 101. [Google Scholar] [CrossRef] [Green Version]
- Silalahi, M.; Cahyani, D.E.; Sensuse, D.I.; Budi, I. Developing indonesian medicinal plant ontology using socio-technical approach. In Proceedings of the 2015 International Conference on Computer, Communications, and Control Technology (I4CT), Kuching, Malaysia, 21–23 April 2015; pp. 39–43. [Google Scholar] [CrossRef]
- Kaewboonma, N.; Supnithi, T.; Panawong, J. Developing ontology for Thai Zingiberaceae: From taxonomies to ontologies. In Proceedings of the 2017 14th International Conference on Electrical Engineering/Electronics, Computer, Telecommunications and Information Technology (ECTI-CON), Phuket, Thailand, 27–30 June 2017; pp. 596–599. [Google Scholar] [CrossRef]
- Stoyanova-Doycheva, A.; Ivanova, V.; Doychev, E.; Spassova, K. Development of an Ontology in Plant Genetic Resources. In Proceedings of the 2020 IEEE 10th International Conference on Intelligent Systems (IS), Varna, Bulgaria, 28–30 August 2020; pp. 246–251. [Google Scholar] [CrossRef]
- Ilic, K.; Kellogg, E.A.; Jaiswal, P.; Zapata, F.; Stevens, P.F.; Vincent, L.P.; Avraham, S.; Reiser, L.; Pujar, A.; Sachs, M.M.; et al. The Plant Structure Ontology, a Unified Vocabulary of Anatomy and Morphology of a Flowering Plant. Plant Physiol. 2006, 143, 587–599. [Google Scholar] [CrossRef] [Green Version]
- Hoehndorf, R.; Alshahrani, M.; Gkoutos, G.V.; Gosline, G.; Groom, Q.; Hamann, T.; Kattge, J.; de Oliveira, S.M.; Schmidt, M.; Sierra, S.; et al. The flora phenotype ontology (FLOPO): Tool for integrating morphological traits and phenotypes of vascular plants. J. Biomed. Semant. 2016, 7, 65. [Google Scholar] [CrossRef] [Green Version]
- Simonini, G.; Zhu, S. Big data exploration with faceted browsing. In Proceedings of the 2015 International Conference on High Performance Computing Simulation (HPCS), Amsterdam, The Netherlands, 20–24 July 2015; pp. 541–544. [Google Scholar] [CrossRef]
- Roy, S.B.; Wang, H.; Nambiar, U.; Das, G.; Mohania, M. DynaCet: Building Dynamic Faceted Search Systems over Databases. In Proceedings of the 2009 IEEE 25th International Conference on Data Engineering, Shanghai, China, 29 March–2 April 2009; pp. 1463–1466. [Google Scholar] [CrossRef] [Green Version]
- Chakraborty, T.; Krishna, A.; Singh, M.; Ganguly, N.; Goyal, P.; Mukherjee, A. FeRoSA: A Faceted Recommendation System for Scientific Articles. In Advances in Knowledge Discovery and Data Mining; Bailey, J., Khan, L., Washio, T., Dobbie, G., Huang, J.Z., Wang, R., Eds.; Springer International Publishing: Cham, Switzerland, 2016; pp. 528–541. [Google Scholar] [CrossRef]
- Wongsuphasawat, K.; Moritz, D.; Anand, A.; Mackinlay, J.; Howe, B.; Heer, J. Voyager: Exploratory Analysis via Faceted Browsing of Visualization Recommendations. IEEE Trans. Vis. Comput. Graph. 2016, 22, 649–658. [Google Scholar] [CrossRef]
- Farazi, F.; Chapman, C.; Raju, P.; Melville, L. Ontology-based faceted semantic search with automatic sense disambiguation for bioenergy domain. Int. J. Big Data Intell. 2018, 5, 62–72. [Google Scholar] [CrossRef]
- De Maio, C.; Fenza, G.; Loia, V.; Parente, M. Biomedical data integration and ontology-driven multi-facets visualization. In Proceedings of the 2015 International Joint Conference on Neural Networks (IJCNN), Killarney, Ireland, 12–17 July 2015; pp. 1–8. [Google Scholar] [CrossRef]
- Sánchez-Cervantes, J.L.; Colombo-Mendoza, L.O.; Alor-Hernández, G.; García-Alcaráz, J.L.; Álvarez-Rodríguez, J.M.; Rodríguez-González, A. LINDASearch: A faceted search system for linked open datasets. Wirel. Netw. 2020, 26, 5645–5663. [Google Scholar] [CrossRef]
- Mauro, N.; Ardissono, L.; Lucenteforte, M. Faceted search of heterogeneous geographic information for dynamic map projection. Inf. Process. Manag. 2020, 57, 102257. [Google Scholar] [CrossRef]
- Thadeu Ferreira da Silva, S.; Apolonio, S.d.O.; Vivacqua, A.S.; Oliveira, J.; Xexéo, G.B.; Campos, M.L.M. Ontoogle: Enhancing retrieval with ontologies and facets. In Proceedings of the 2011 15th International Conference on Computer Supported Cooperative Work in Design (CSCWD), Laussane, Switzerland, 8–10 June 2011; pp. 192–199. [Google Scholar] [CrossRef]
- Le, T.; Vo, B.; Duong, T.H. Personalized Facets for Semantic Search Using Linked Open Data with Social Networks. In Proceedings of the 2012 Third International Conference on Innovations in Bio-Inspired Computing and Applications, Kaohsiung, Taiwan, 26–28 September 2012; pp. 312–317. [Google Scholar] [CrossRef]
- Niu, X.; Fan, X.; Zhang, T. Understanding Faceted Search from Data Science and Human Factor Perspectives. ACM Trans. Inf. Syst. 2019, 37, 1–27. [Google Scholar] [CrossRef]
- Bondy, J.; Murty, U. Graph Theory, 1st ed.; Springer Publishing Company, Incorporated: London, UK, 2008. [Google Scholar]
- Suárez-Figueroa, M.C.; Gómez-Pérez, A.; Fernández-López, M. The NeOn Methodology framework: A scenario-based methodology for ontology development. Appl. Ontol. 2015, 10, 107–145. [Google Scholar] [CrossRef]
- Spoladore, D.; Pessot, E. Collaborative Ontology Engineering Methodologies for the Development of Decision Support Systems: Case Studies in the Healthcare Domain. Electronics 2021, 10, 1060. [Google Scholar] [CrossRef]
- Fernández-López, M.; Gómez-Pérez, A.; Juristo, N. METHONTOLOGY: From Ontological Art Towards Ontological Engineering. In Proceedings of the Ontological Engineering AAAI-97 Spring Symposium Series, Palo Alto, CA, USA, 24–26 March 1997. [Google Scholar]
- Corcho, O.; Fernández-López, M.; Gómez-Pérez, A.; López-Cima, A. Building Legal Ontologies with METHONTOLOGY and WebODE. In Law and the Semantic Web: Legal Ontologies, Methodologies, Legal Information Retrieval, and Applications; Benjamins, V.R., Casanovas, P., Breuker, J., Gangemi, A., Eds.; Springer: Berlin/Heidelberg, Germany, 2005; pp. 142–157. [Google Scholar] [CrossRef] [Green Version]
- Villazón-Terrazas, B.; Ramírez, J.; Suárez-Figueroa, M.C.; Gómez-Pérez, A. A network of ontology networks for building e-employment advanced systems. Expert Syst. Appl. 2011, 38, 13612–13624. [Google Scholar] [CrossRef] [Green Version]
- Ziemba, P.; Jankowski, J.; Wątróbski, J.; Becker, J. Knowledge Management in Website Quality Evaluation Domain. In Computational Collective Intelligence; Núñez, M., Nguyen, N.T., Camacho, D., Trawiński, B., Eds.; Springer International Publishing: Cham, Switzerland, 2015; pp. 75–85. [Google Scholar] [CrossRef]
- Uschold, M.; Gruninger, M. Ontologies: Principles, methods and applications. Knowl. Eng. Rev. 1996, 11, 93–136. [Google Scholar] [CrossRef] [Green Version]
- Pan, J.Z. Resource Description Framework. In Handbook on Ontologies; Staab, S., Studer, R., Eds.; Springer: Berlin/Heidelberg, Germany, 2009; pp. 71–90. [Google Scholar] [CrossRef]
- Antoniou, G.; van Harmelen, F. Web Ontology Language: OWL. In Handbook on Ontologies; Staab, S., Studer, R., Eds.; Springer: Berlin/Heidelberg, Germany, 2004; pp. 67–92. [Google Scholar] [CrossRef] [Green Version]
- Musen, M.A. The Protégé Project: A Look Back and a Look Forward. AI Matters 2015, 1, 4–12. [Google Scholar] [CrossRef] [PubMed]
- Garijo, D. WIDOCO: A Wizard for Documenting Ontologies. In The Semantic Web—ISWC 2017; d’Amato, C., Fernandez, M., Tamma, V., Lecue, F., Cudré-Mauroux, P., Sequeda, J., Lange, C., Heflin, J., Eds.; Springer International Publishing: Cham, Switzerland, 2017; pp. 94–102. [Google Scholar] [CrossRef]
- Díaz Rodríguez, N.; Cuéllar, M.P.; Lilius, J.; Delgado Calvo-Flores, M. A fuzzy ontology for semantic modelling and recognition of human behaviour. Knowl. Based Syst. 2014, 66, 46–60. [Google Scholar] [CrossRef]





| Level 0 | Level 1 | Level 2 | Level 3 | Level 4 |
|---|---|---|---|---|
| Plant | Stem | |||
| Leaf | Apex | |||
| Base | ||||
| Petiole | ||||
| Flower | Flower Stalk | |||
| Sepal | ||||
| Petals | Base Petals | |||
| Pistil | Pistil Stalk | |||
| Stamen | Anther | |||
| Filament | ||||
| Fruit | Pedicel | |||
| Calyx | ||||
| Seed | ||||
| Ripe Fruit | ||||
| Raw Fruit |
| No. | Property | # Entities as Domain |
|---|---|---|
| 1 | Color | 13 |
| 2 | Diameter | 1 |
| 3 | Length | 7 |
| 4 | Number of Seed | 1 |
| 5 | Number of Stalk Segment | 1 |
| 6 | Position | 4 |
| 7 | Shape | 9 |
| 8 | Spot | 2 |
| 9 | Surface | 1 |
| 10 | Texture | 2 |
| 11 | Width | 2 |
| Property | Pre-Defined Values |
|---|---|
| Texture | bare; coarse; hairy; hairless; slippery; |
| Color | blue; bluish; dark green; green; greenish-white; greenish-yellow; pale green; purple; red; slightly purplish; white; yellow; yellowish; |
| Shape | bell shape; cuff; elongated; lanceolate; star-like; rounded; rounded eggs; triangular-like; |
| Surface | smooth; |
| Position | hanging; fixed; upright; |
| No. | Case | Search queries | Individuals |
|---|---|---|---|
| 1 | Case 1 | Elongated fruit shape | |
| 2 | Case 1 | Lanceolate leaf shape | |
| 3 | Case 1 | Hairy stems | |
| 4 | Case 2 | Greenish-yellow petals and yellowish seeds | |
| 5 | Case 2 | Elongated fruit shape and yellowish seeds | |
| 6 | Case 3 | Bell fruit shape and yellowish seeds | |
| 7 | Case 3 | Hanging flower position and a few centimeters steam length | |
| 8 | Case 3 | Star-like flower shape and a few millimeters pistil length | |
| 9 | Case 3 | Star-like flower shape and yellow anthers | |
| 10 | Case 3 | Greenish-yellow petals and star-like flower shape | |
| 11 | Case 4 | Elongated fruit shape and greenish-white petals | |
| 12 | Case 4 | Elongated fruit shape and triangular-like leaf shape | |
| 13 | Case 4 | Elongated fruit shape and upright fruit position | |
| 14 | Case 4 | Elongated fruit shape and hanging fruit position | |
| 15 | Case 4 | Star-like flower shape and rounded leaf shape | |
| 16 | Case 4 | Green leafy ripe fruit and lanceolate leaf shape | |
| 17 | Case 4 | Dark green leaves and white petals | |
| 18 | Case 4 | Smooth leaf surface and bluish anthers | |
| 19 | Case 4 | Light green leaves and yellowish seeds |
| No. | Results after Matching Step |
|---|---|
| 1 | Stem, Leaf, Flower, Ripe Fruit, Raw Fruit, Seed, Calyx, Pedicel |
| 2 | Stem, Fruit, Flower, Apex, Base, Petiole |
| 3 | Leaf, Fruit, Flower |
| 4 | Apex, Base, Petiole, Ripe Fruit, Raw Fruit, Seed, Calyx, Pedicel, Sepal, Flower Stalk |
| 5 | Apex, Base, Petiole, Ripe Fruit, Raw Fruit, Flower Stalk, Calyx, Pedicel, Petals |
| 6 | Ripe Fruit, Raw Fruit, Calyx, Pedicel |
| 7 | Sepal, Flower Stalk, Petals, Pistil, Anthers, Filament, Base Petals, Pistil Stalk |
| 8 | Sepal, Flower Stalk, Petals, Stamen, Anthers, Filament, Base Petals, Pistil Stalk |
| 9 | Sepal, Flower Stalk, Petals, Pistil, Stamen, Filament, Base Petals, Pistil Stalk |
| 10 | Sepal, Flower Stalk, Pistil, Stamen, Anthers, Filament, Base Petals, Pistil Stalk |
| 11 | Stem, Leaf, Flower, Ripe Fruit, Raw Fruit, Seed, Calyx, Pedicel, Sepal, Flower Stalk, Base Petals, Pistil Stalk, Pistil, Stamen, Anthers, Filament |
| 12 | Stem, Leaf, Flower, Ripe Fruit, Raw Fruit, Seed, Calyx, Pedicel, Petals, Flower Stalk, Base Petals, Pistil Stalk, Pistil, Stamen, Anthers, Filament |
| 13 | Stem, Leaf, Flower, Ripe Fruit, Raw Fruit, Seed, Calyx, Pedicel, Petals, Flower Stalk, Base Petals, Pistil Stalk, Sepal, Stamen, Anthers, Filament |
| 14 | Stem, Leaf, Fruit, Ripe Fruit, Seed, Calyx, Pedicel, Petals, Flower Stalk, Base Petals, Pistil Stalk, Sepal, Stamen, Anthers, Filament, Pistil |
| 15 | Stem, Leaf, Fruit, Ripe Fruit, Raw Fruit, Calyx, Pedicel, Petals, Flower Stalk, Base Petals, Pistil Stalk, Sepal, Stamen, Anthers, Filament, Pistil |
| 16 | Stem, Leaf, Fruit, Ripe Fruit, Seed, Calyx, Raw Fruit, Petals, Flower Stalk, Base Petals, Pistil Stalk, Sepal, Stamen, Anthers, Filament, Pistil |
| 17 | Stem, Fruit, Flower, Apex, Base, Petiole, Sepal, Flower Stalk, Base Petals, Pistil Stalk, Stamen, Anthers, Filament, Pistil |
| 18 | Stem, Fruit, Flower, Apex, Base, Petiole, Sepal, Flower Stalk, Base Petals, Pistil Stalk, Stamen, Petals, Filament, Pistil |
| 19 | Stem, Fruit, Flower, Apex, Base, Petiole, Ripe Fruit, Raw Fruit, Calyx, Pedicel |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Akbar, Z.; Mustika, H.F.; Rini, D.S.; Manik, L.P.; Indrawati, A.; Fefirenta, A.D.; Djarwaningsih, T. An Ontology-Driven Personalized Faceted Search for Exploring Knowledge Bases of Capsicum. Future Internet 2021, 13, 172. https://doi.org/10.3390/fi13070172
Akbar Z, Mustika HF, Rini DS, Manik LP, Indrawati A, Fefirenta AD, Djarwaningsih T. An Ontology-Driven Personalized Faceted Search for Exploring Knowledge Bases of Capsicum. Future Internet. 2021; 13(7):172. https://doi.org/10.3390/fi13070172
Chicago/Turabian StyleAkbar, Zaenal, Hani Febri Mustika, Dwi Setyo Rini, Lindung Parningotan Manik, Ariani Indrawati, Agusdin Dharma Fefirenta, and Tutie Djarwaningsih. 2021. "An Ontology-Driven Personalized Faceted Search for Exploring Knowledge Bases of Capsicum" Future Internet 13, no. 7: 172. https://doi.org/10.3390/fi13070172
APA StyleAkbar, Z., Mustika, H. F., Rini, D. S., Manik, L. P., Indrawati, A., Fefirenta, A. D., & Djarwaningsih, T. (2021). An Ontology-Driven Personalized Faceted Search for Exploring Knowledge Bases of Capsicum. Future Internet, 13(7), 172. https://doi.org/10.3390/fi13070172